Abstract
New software to browse chemical space, with structures organized by rings, will enable chemical insight.
Chemical space is so enormous that it is hard to look at. This is a problem for medicinal chemists and chemical biologists who seek new molecules for their biological targets. Current tools, often little more than spreadsheets, display only a few molecules at a time, and typically fail to capture the relationships between molecules. The challenge for a chemical space browser is to organize and depict large sets of molecules such as high-throughput screening results intuitively, enabling insight. In this issue, Wetzel et al.1 report new software for just this purpose, and a companion paper2 elaborates how this new tool can be used. The software will be useful for investigators who want to organize, classify and understand sets of thousands of molecules.
In principle, one would like to be able to organize and browse large chemical datasets such as screening libraries as easily as one can today browse maps on the internet. A chemical space map would position similar molecules near each other and less related ones increasingly further apart. Molecules would be linked to external data sources and the scientific literature. A chemical space browser would be a new scientific tool in its own right.
Coping with the size of chemical space is a challenge. One estimate of its size is 1060 molecules3, more than the atoms in a billion Earths. Even tiny subsets, such as the 1,350 small-molecule drugs approved by the US Food and Drug Administration, for example, can be unwieldy to look at all at once. A key insight and enabling simplification of this work is its focus on rings, an intuitive and ubiquitous organizing concept. By focusing on rings, the enormity of the problem is instantly reduced. For instance, in a recent report, Pitt et al.4 calculate that fewer than 25,000 aromatic ring systems are relevant to medicinal chemistry. Though this is still a big number, it is a far more feasible starting point.
A second problem faced by any depiction of chemical space is how close any two molecules should be. Many measures of similarity are in use, based on shape5, physical properties6, topology7 or some combination of these. Some measures will typically be more intuitive than others, depending on which molecules are being compared and what question is being asked. Yet with so many different measures of similarity, reasonable people will likely disagree about what a sensible default method might be, or even whether one exists at all.
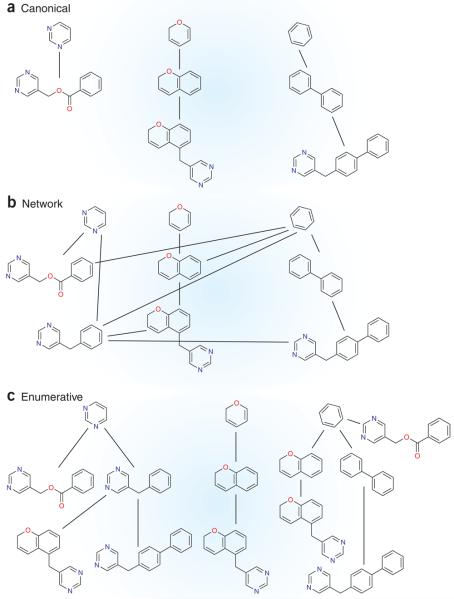
Wetzel et al.1 tackle both of these problems using the simplifying concepts of rings and scaffolds to make depiction of chemical space tractable. Each molecule is simplified by removing acyclic substituents to leave a bare scaffold, which is pruned one ring at a time to arrive at a root ring. The pruning steps are represented as a tree (Fig. 1a), which can be viewed interactively in their program. The authors demonstrate the possibility of prospective exploration of chemical space with PubChem BioAssay screening data, using the program to identify 65 virtual scaffolds, 4 of which were then used to discover 9 new ligands with better than 10 μM affinity.
Figure 1.
Depictions of scaffolds and their relationships using selected dihydrofolate reductase ligands drawn from the MDL Drug Data Report. (a) A canonically rooted tree produced by progressively pruning scaffolds to arrive at a canonical root ring as described in this paper. Note that whereas the largest scaffold in each branch contains pyrimidine, the key group required for binding, only one has pyrimidine as the canonical root ring. (b) A network of related scaffolds, where lines indicate addition of a single ring. Such networks rapidly become unwieldy as the number of scaffolds grows; the number of connections rises much more slowly in trees (a,c) than for networks. (c) Enumeratively rooted trees. Each ring in each molecule is used once as a root so that each molecule appears as many times as it has rings. This facilitates seeing patterns in the data. Here, a common pyrimidine root unites all molecules consistent with their presumed binding mode. These were rooted separately in the canonical scheme (a), obscuring the common key binding group.
Approaches that depend on rings and scaffolds have obvious weaknesses. For instance, the key functional group may not be part of a ring, such as the hydroxamic acid of histone deacetylase ligands or the sulfonamides of carbonic anhydrase ligands. Some molecules, such as lipids, have no rings at all. But perhaps the biggest problem with using rings to organize and classify molecules is that they ignore molecular similarity.
To investigators accustomed to scaffold hopping—the process of finding new ligands by analogy to known ones, often using topological or shape-based methods5—rings might seem misguided, and trees might seem arbitrary. Why not use molecular similarity instead of rings, and a network instead of trees (Fig. 1b)? Trees are practical because of their simplicity, and trees are possible because of rings. As it now stands, the program can handle large and complex structural datasets. If molecular similarity and network data structures were used, the data could rapidly become unwieldy (compare Fig. 1a,b).
Since both the graphical interface and the tree-making program are freely available under the GNU Public License, they can be further cultivated and improved by the community. A few simple changes could help immediately. URLs to link to public databases could connect objects in Scaffold Hunter to the many other data sources that already exist. Tree structures that enumerate all possible tree roots rather than canonical hierarchy trees would also provide more intuitive results in some cases (Fig. 1c). The organization of tree roots could be more intuitive.
Notwithstanding the scope for improvement, Scaffold Hunter offers a new interactive way of looking at and organizing large sets of chemical structures and leverages our natural pattern-perceiving ability. It heralds a new era of graphical representation of complex molecular structure relationships. The software is available now, and should be immediately useful to the many investigators who are willing to develop the skill to use it.
References
- 1.Wetzel S, et al. Nat. Chem. Biol. 2009;5:581–583. doi: 10.1038/nchembio.187. [DOI] [PubMed] [Google Scholar]
- 2.Renner S, et al. Nat. Chem. Biol. 2009;5:585–592. doi: 10.1038/nchembio.188. [DOI] [PubMed] [Google Scholar]
- 3.Bohacek RS, McMartin C, Guida WC. Med. Res. Rev. 1996;16:3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 4.Pitt WR, Parry DM, Perry BG, Groom CR. J. Med. Chem. 2009;52:2952–2963. doi: 10.1021/jm801513z. [DOI] [PubMed] [Google Scholar]
- 5.Rush TS, III, Grant JA, Mosyak L, Nicholls A. J. Med. Chem. 2005;48:1489–1495. doi: 10.1021/jm040163o. [DOI] [PubMed] [Google Scholar]
- 6.Rosén J, et al. J. Comput. Aided Mol. Des. 2009;23:253–259. doi: 10.1007/s10822-008-9255-y. [DOI] [PubMed] [Google Scholar]
- 7.Willett P. Drug Discov. Today. 2006;11:1046–1053. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]