Abstract
Genomic islands have been shown to harbor functional traits that differentiate ecologically distinct populations of environmental bacteria. A comparative analysis of the complete genome sequences of the marine Actinobacteria Salinispora tropica and S. arenicola reveals that 75% of the species-specific genes are located in 21 genomic islands. These islands are enriched in genes associated with secondary metabolite biosynthesis providing evidence that secondary metabolism is linked to functional adaptation. Secondary metabolism accounts for 8.8% and 10.9% of the genes in the S. tropica and S. arenicola genomes, respectively, and represents the major functional category of annotated genes that differentiates the two species. Genomic islands harbor all 25 of the species-specific biosynthetic pathways, the majority of which occur in S. arenicola and may contribute to the cosmopolitan distribution of this species. Genome evolution is dominated by gene duplication and acquisition, which in the case of secondary metabolism provide immediate opportunities for the production of new bioactive products. Evidence that secondary metabolic pathways are exchanged horizontally, coupled with prior evidence for fixation among globally distributed populations, supports a functional role and suggests that the acquisition of natural product biosynthetic gene clusters represents a previously unrecognized force driving bacterial diversification. Species-specific differences observed in CRISPR (clustered regularly interspaced short palindromic repeat) sequences suggest that S. arenicola may possess a higher level of phage immunity, while a highly duplicated family of polymorphic membrane proteins provides evidence of a new mechanism of marine adaptation in Gram-positive bacteria.
Keywords: comparative genomics, genomic islands, marine actinobacteria, Salinispora, secondary metabolites
Introduction
Linking functional traits to bacterial phylogeny remains a fundamental but elusive goal of microbial ecology (Hunt et al 2008). Without this information, it becomes difficult to resolve meaningful units of diversity and the mechanisms by which bacteria interact with each other and adapt to environmental change. Most bacterial diversity is delineated among clusters of sequences that share >99% 16S rRNA gene sequence identity (Acinas et al 2004). These sequence clusters are believed to represent fundamental units of diversity, while intra-cluster microdiversity is thought to persist due to weak selective pressures (Acinas et al 2004) suggesting little ecological or taxonomic relevance. Recently, progress has been made in terms of delineating units of diversity that possess the fundamental properties of species by linking genetic diversity with ecology and evolutionary theory (Achtman and Wagner 2008, Fraser et al 2009). Despite these advances, there remains no widely accepted species concept for prokaryotes (Gevers et al 2005), and sequence-based analyses reveal widely varied levels of diversity within assigned species boundaries.
The comparative analysis of bacterial genome sequences has revealed considerable differences among closely related strains (Joyce et al 2002, Thompson et al 2005, Welch et al 2002) and provides a new perspective on genome evolution and prokaryotic species concepts. Genomic differences among closely related strains are concentrated in islands, strain-specific regions of the chromosome that are generally acquired by horizontal gene transfer (HGT) and harbor functionally adaptive traits (Dobrindt et al 2004) that can be linked to niche adaptation. The pelagic cyanobacterium Prochlorococcus is an important model for the study of island genes, which in this case are differentially expressed under low nutrient and high light stress in ecologically distinct populations (Coleman et al 2006). Despite convincing evidence for the adaptive significance of island genes among environmental bacteria, the precise functions of their products have seldom been characterized and their potential role in the evolution of independent bacterial lineages remains poorly understood.
The marine sediment inhabiting genus Salinispora belongs to the Order Actinomycetales, a group of Actinobacteria commonly referred to as actinomycetes. Actinomycetes are a rich source of structurally diverse secondary metabolites and account for the majority of antibiotics discovered as of 2002 (Berdy 2005). Salinispora spp. have likewise proven to be a rich source of secondary metabolites (Fenical and Jensen 2006) including salinosporamide A, which is currently in clinical trials for the treatment of cancer (Fenical et al 2009). At present, the genus is comprised of three species that collectively constitute a microdiverse sequence cluster (sensu (Acinas et al 2004), i.e., they share ≥99% 16S rRNA gene sequence identity (Jensen and Mafnas 2006). Although the microdiversity within this cluster has been formally delineated into species-level taxa (Maldonado et al 2005), it remains to be determined if these taxa represent ecologically or functionally distinct lineages.
Here we report the comparative analysis of the complete genome sequences of S. tropica (strain CNB-440, the type strain for the species and thus a contribution to the Genomic Encyclopedia of Bacteria and Archaea project), hereafter referred to as ST, and S. arenicola (strain CNS-205), hereafter referred to as SA, the first obligately marine Actinobacteria to be obtained in culture (Mincer et al 2002). The aims of this study were to describe, compare, and contrast the gene content and organization of the two genomes in the context of prevailing species concepts, identify the functional attributes that differentiate the two species, assess the processes that have driven genome evolution, and search for evidence of marine adaptation in this unusual group of Gram-positive marine bacteria.
Methods and Materials
Sequencing and ortholog identification
The sequencing and annotation of the SA genome was as previously reported for ST (Udwary et al 2007). Both genomes were sequenced as part of the Department of Energy, Joint Genome Institute, Community Sequencing Program. Orthologs within the two genomes were predicted using the Reciprocal Smallest Distance (RSD) method (Wall et al 2003), which includes a maximum likelihood estimate of amino acid substitutions. A linear alignment of positional orthologs was created and the positions of rearranged orthologs and species-specific genes identified. Genomic islands were defined as regions >20 kb that are flanked by regions of conservation and within which <40% of the island genes possess a positional ortholog in the reciprocal genome. Paralogs within each genome were identified using the blastclust algorithm (Dondoshansky and Wolf 2000) with a cut-off of 30% identity over 40% of the sequence length. The automated phylogenetic inference system (APIS) was used to identify recent gene duplications (Badger et al 2005).
Horizontal Gene Transfer
All genes were assessed for evidence of HGT based on abnormal DNA composition, phylogenetic, taxonomic, and sequence-based relationships, and comparisons to known Mobile Genetic Elements (MGEs). Genes identified by ≥2 different methodologies were counted as positive for HGT. To reflect confidence in the assignments, genes displaying positive evidence of HGT were color coded from yellow to red corresponding to total scores from 2 to 6. The results were mapped onto the genome to reveal HGT clustering patterns and adjacent clusters were merged (Figure 1a). Four DNA compositional analyses included G+C content (obtained from the JGI annotation), codon adaptive index, calculated with the CAI calculator (Wu et al 2005) using a suite of housekeeping genes as reference, dinucleotide frequency differences (δ*), calculated using IslandPath (Hsiao et al 2003), and DNA composition, calculated using Alien_Hunter (Vernikos and Parkhill 2006). G+C content or codon usage values >1.5 standard deviations from the genomic mean and dinucleotide frequency differences >1 standard deviation from the mean were scored positive for HGT. Taxonomic relationships in the form of lineage probability index (LPI) values for all protein coding genes were assigned using the Darkhorse algorithm (Podell and Gaasterland 2007). Genes with an LPI of <0.5, indicating the orthologs are not in closely related genomes, were scored positive for HGT. A reciprocal Darkhorse analysis (Podell et al 2008) was then performed on the orthologs of all positives, and if these genes had an LPI score >0.5, indicating the match sequence is phylogenetically typical within its own lineage, they were assigned an additional positive score.
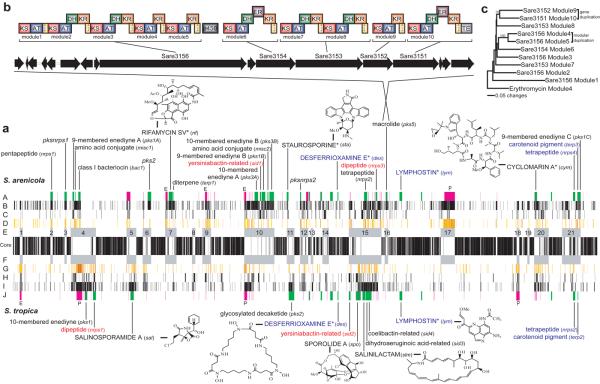
Figure 1.
Linear alignment of the S. tropica and S. arenicola genomes starting with the origins of replication. (a) Positional orthologs (core) flanked by islands (E, F), heat-mapped HGT genes (D, G), rearranged orthologs (C, H), species-specific genes (B, I), secondary metabolite genes (green), MGEs (pink) with prophage (P) and AICES (E) indicated (A, J). For genomic islands, predicted (lower case) and isolated (uppercase with structures) secondary metabolites are given (not shown are six non-island secondary metabolic gene clusters of unknown function). Shared positional (blue) and rearranged (red) secondary metabolite clusters are indicated. *Previously isolated from other bacteria. (b) Expanded view of SA pks5 revealing gene and modular architecture. (c) Neighbor-joining phylogenetic tree of KS domains from SA pks5 revealing gene and modular duplication events (erythromycin root, % bootstrap values from 1000 re-samplings).
A phylogenetic approach using the APIS program (Badger et al 2005) was also employed to assess HGT. Using this program, bootstrapped neighbor-joining trees of all predicted protein coding genes within each genome were created. All genes cladding with non-Actinobacterial homologs were binned into their respective taxonomic groups and given a positive HGT score. Evidence of HGT was also inferred from RSD analyses of each genome against a compiled set of 27 finished Actinobacterial genomes that included at least two representatives of each genus for which sequences were available. Genes present in SA and/or ST and not observed among the 27 Actinobacterial genomes were assigned a positive HGT score. Bacteriophage were identified using Prophage (Bose 2006) and Phage Finder (Fouts 2006). Other insertion elements were identified as prophage or transposon in origin through blastX homology searches. Gene annotation based on searches for identity across PFAM, SPTR, KEGG and COG databases was also used to help identify mobile genetic elements (MGEs). Each gene associated with an MGE was assigned a positive HGT score. Test scores were amalgamated and those genes showing evidence of HGT in two or more tests (maximum score 6) were classified as horizontally acquired. The results were mapped onto the genome and genes identified by only one test but associated with clusters of genes that scored in two or more tests were added to the total HGT pool. Adjacent clusters were merged.
CRISPRs were identified using CRISPR finder (http://crispr.u-psud.fr/Server/CRISPRfinder.php) while repeats larger than 35 bases were identified using Reputer (Kurtz et al 2001). Secondary metabolite gene clusters were manually annotated as in (Udwary et al 2007). Cluster boundaries were predicted using previously reported gene clusters when available as in the case of rifamycin. For unknown clusters, loss of gene conservation across the Actinobacteria was used to aid boundary predictions. In the future, programs such as “ClustScan” may prove useful for pathway annotation and product prediction (Starcevic et al 2008). However, many biosynthetic genes are large (5-10 kb) and highly repetitive creating challenges associated with gene calling and assembly, eg., (Udwary et al 2007) and the interpretation of operon structure. The ratio of non-synonymous to synonymous mutations (dN/dS) for all orthologs was calculated using the perl progam SNAP (http://www.hiv.lanl.gov) with the alignments for all values >1 checked manually.
Results and Discussion
The ST and SA genomes share 3606 orthologs, representing 79.4% and 73.2% of the respective genomes (Table 1). The average nucleotide identity among these orthologs is 87.2%, well below the 94% cut-off that has been suggested to delineate bacterial species (Konstantinidis and Tiedje 2005). Despite differing by only seven nucleotides (99.7% identity) in the 16S rRNA gene, the genome of SA is 603 kb (11.6%) larger and possesses 1505 species-specific genes compared to 987 in ST. Seventy-five percent of these species-specific genes are located in 21 genomic islands (Tables 1, S1), none of which are comprised of genes originating entirely from one genome (Figure 1). The presence of genomic islands in the same location on the chromosomes of closely related bacteria is well recognized (Coleman et al 2006) and facilitated by the presence of tRNAs (Tuanyok et al 2008). Twelve islands in the Salinispora alignment share at least one tRNA between both genomes and of those, four share two or more tRNAs within a single island indicating multiple insertion sites. In addition to tRNAs, direct repeats detected in the same location in both genomes could also act as insertion sites to help create islands. These islands are enriched with large clusters of genes devoted to the biosynthesis of secondary metabolites (Figure 1). They house all 25 of the species-specific secondary metabolic pathways, while eight of the 12 shared pathways occur in the genus-specific core (Tables 2, 3). We have isolated and identified the products of eight of these pathways, which include the highly selective proteasome inhibitor salinosporamide A (Feling et al 2003) as well as sporolide A (Buchanan et al 2005), which is derived from an enediyne polyketide precursor (Udwary et al 2007), one of the most potent classes of biologically active agents discovered to date. A previous analysis of 46 Salinispora strains revealed that secondary metabolite production is the major phenotypic difference among the three species (Jensen et al 2007), an observation supported by the analysis of the S. tropica genome (Udwary et al 2007).
Table 1.
General genome features
| Feature | S. tropica (ST) | ST% | S. arenicola (SA) | SA % |
|---|---|---|---|---|
| No. base pairs | 5183331 | NA | 5786361 | NA |
| % G+C | 69.4 | NA | 69.5 | NA |
| Total genes | 4536 | NA | 4919 | NA |
| Pseudogenes | 57 | 1.26% | 192 | 3.90% |
| Hypotheticals (% genome) | 1140 | 25.10% | 1418 | 28.80% |
| No. rRNA operons (% identity) | 3 | 100% | 3 | 100% |
| Orthologs (% genome) | 3606 | 79.40% | 3606 | 73.20% |
| Positional orthologs (% genome) | 3178 | 70.10% | 3178 | 64.60% |
| Rearranged orthologs (% genome) | 428 | 9.40% | 428 | 8.70% |
| Species-specific genes (% genome) | 987 | 21.80% | 1505 | 30.60% |
| Island genes (% genome) | 1350 | 29.80% | 1690 | 34.30% |
| Total genes with evidence of HGT (% genome) | 652 | 14.30% | 750 | 14.70% |
| Species-specific genes with evidence of HGT (% species-specific) | 405 | 41.00% | 573 | 38.10% |
| Total island genes with evidence of HGT (% HGT) | 473 | 72.50% | 555 | 74.00% |
| Paralogsa (% genome) | 1819 | 39.60% | 2179 | 42.60% |
| Species-specific paralogs (% species-specific genes) | 391 | 39.70% | 647 | 43.00% |
| Secondary metabolism (% genome) | 405 | 8.80% | 556 | 10.90% |
Totals include parental gene.
NA: not applicable.
Table 2.
Secondary metabolite gene clusters in S. tropica (ST)
| No. | Cluster name |
Equivalent cluster |
Biosynthetic class | Product | Biological activity/target | Island | Gene start |
Gene stop |
No. genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ST pks1 | none | polyketide | 10-membered enediyne | cytotoxin/DNA | 4 | 586 | 610 | 25 |
| 2 | ST nrps1 | SA nrps3a | non-ribosomal peptide | dipeptide | N/D | 4/15 | 667 | 694 | 28 |
| 3 | ST sal | none | polyketide/non-ribosomal peptide | salinosporamide | cytotoxin/proteasome | 5 | 1012 | 1043 | 32 |
| 4 | ST pks2 | none | polyketide | glycosylated decaketide | N/D | 11 | 2174 | 2227 | 54 |
| 5 | ST amc | SA amc | carbohydrate | aminocyclitol | N/D | NI/NI | 2340 | 2346 | 7 |
| 6 | ST bac1 | SA bac2 | ribosomal peptide | class I bacteriocin (non-lantibiotic) | antimicrobial | NI/NI | 2428 | 2440 | 13 |
| 7 | ST pks3 | SA pks4 | polyketide | aromatic polyketide | N/D | NI/NI | 2486 | 2510 | 25 |
| 8 | ST desb | SA des | hydroxamate | desferrioxamine c | siderophore/iron chelation | NI/NI | 2541 | 2555 | 15 |
| 9 | ST sid2 | SA sid1a | non-ribosomal peptide | yersiniabactin-related | siderophore/iron chelation | 15/10 | 2645 | 2659 | 15 |
| 10 | ST spo | none | polyketide | sporolide | N/D | 15 | 2691 | 2737 | 47 |
| 11 | ST slm | none | polyketide | salinilactam | N/D | 15 | 2757 | 2781 | 25 |
| 12 | ST sid3 | none | non-ribosomal peptide | dihydroaeruginoic acid-related siderophore | siderophore/iron chelation | 15 | 2786 | 2813 | 28 |
| 13 | ST sid4 | none | non-ribosomal peptide | coelibactin-related siderophore | siderophore/iron chelation | 15 | 2814 | 2842 | 29 |
| 14 | ST bac2 | SA bac3 | ribosomal peptide | class I bacteriocin (non-lantibiotic) | antimicrobial | NI/NI | 3042 | 3054 | 13 |
| 15 | ST lym | SA lym | polyketide/non-ribosomal peptide | lymphostin c | immunosuppressant | NI/NI | 3055 | 3066 | 12 |
| 16 | ST terp1 | SA terp2 | terpenoid | carotenoid pigment | antioxidant | NI/NI | 3244 | 3253 | 10 |
| 17 | ST pks4 | SA pks6 | polyketide | phenolic lipids | cell wall lipid | NI/NI | 4264 | 4267 | 4 |
| 18 | ST nrps2 | SA nrps4 | non-ribosomal peptide | tetrapeptide | N/D | 21/21 | 4410 | 4429 | 20 |
| 19 | ST terp2 | SA terp3 | terpenoid | carotenoid pigment | antioxidant | 21/21 | 4437 | 4441 | 5 |
|
| |||||||||
| Total | 407 | ||||||||
NI: non-island. Italics: predicted product or activity. Bold: observed product or activity. N/D: not determined.
Partial cluster.
Previously designated ST Sid1 (32).
Product observed in other bacteria.
Table 3.
Secondary metabolite gene clusters in S. arenicola (SA)
| No. | Cluster name | Equivalent cluster |
Biosynthetic class | Product | Biological activity/target | Island | Gene start |
Gene stop |
No. genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | SA nrps1 | none | non-ribosomal peptide | pentapeptide | N/D | 2 | 345 | 367 | 23 |
| 2 | SA pksnrps1 | none | polyketide/non-ribosomal peptide | N/D | N/D | 3 | 478 | 499 | 22 |
| 3 | SA pks1A | none | polyketide | 9-membered enediyne unit/kedarcidin-related, fragment A | cytotoxin/DNA | 4 | 545 | 560 | 16 |
| 4 | SA misc1 | none | aminoacyl tRNA synthetase-derived | amino acid conjugate | N/D | 4 | 570 | 573 | 4 |
| 5 | SA bac1 | none | ribosomal peptide | class I bacteriocin (lantibiotic) | antimicrobial | 4 | 602 | 623 | 22 |
| 6 | SA pks2 | none | polyketide | N/D | N/D | 6 | 1041 | 1073 | 33 |
| 7 | SA rif | none | polyketide | rifamycin b | antibiotic/RNA polymerase | 7 | 1240 | 1278 | 39 |
| 8 | SA terp1 | none | terpenoid | diterpene | N/D | 7 | 1286 | 1288 | 3 |
| 9 | SA pks3A | none | polyketide | 10-membered enediyne unit/calicheamicin-related, fragment A | cytotoxin/DNA | 10 | 2017 | 2049 | 33 |
| 10 | SA sid1b | ST sid2 | non-ribosomal peptide | yersiniabactin-related | siderophore/iron chelation | 10/15 | 2070 | 2081 | 12 |
| 11 | SA pks1B | none | polyketide-associated | modified tyrosine and deoxysugar units/kedarcidin-related, fragment B | cytotoxin/DNA | 10 | 2088 | 2121 | 34 |
| 12 | SA misc2 | none | aminoacyl tRNA synthetase-derived | amino acid conjugate | N/D | 10 | 2144 | 2151 | 8 |
| 13 | SA pks3B | none | polyketide-related | aryltetrasaccharide unit/calicheamicin-related, fragment B | cytotoxin/DNA | 10 | 2163 | 2206 | 44 |
| 14 | SA sta | none | indolocarbazole | staurosporine b | cytotoxin/protein kinase | 11 | 2326 | 2342 | 17 |
| 15 | SA pksnrps2 | none | polyketide/non-ribosomal peptide | N/D | N/D | 12 | 2400 | 2409 | 10 |
| 16 | SA amc | ST amc | carbohydrate | aminocyclitol | N/D | NI/NI | 2483 | 2491 | 9 |
| 17 | SA bac2 | ST bac1 | ribosomal peptide | class I bacteriocin (non-lantibiotic) | antimicrobial | NI/NI | 2583 | 2595 | 13 |
| 18 | SA pks4 | ST pks3 | polyketide | aromatic polyketide | N/D | NI/NI | 2669 | 2694 | 26 |
| 19 | SA des | ST des | hydroxamate | desferrioxamine b | siderophore/iron chelation | NI/NI | 2728 | 2744 | 17 |
| 20 | SA nrps2 | none | non-ribosomal peptide | tetrapeptide | N/D | 15 | 2939 | 2968 | 30 |
| 21 | SA nrps3a | ST nrps1 | non-ribosomal peptide | dipeptide | N/D | 15/4 | 3051 | 3063 | 13 |
| 22 | SA pks5 | none | polyketide | macrolide | N/D | 16 | 3148 | 3163 | 16 |
| 23 | SA bac3 | ST bac2 | ribosomal peptide | class I bacteriocin (non-lantibiotic) | antimicrobial | NI/NI | 3268 | 3280 | 13 |
| 24 | SA lym | ST lym | polyketide | lymphostin b | immunosuppressant | NI/NI | 3281 | 3293 | 13 |
| 25 | SA terp2 | ST terp1 | terpenoid | carotenoid pigment | antioxidant | NI/NI | 3471 | 3480 | 10 |
| 26 | SA cym | none | non-ribosomal peptide | cyclomarin b | anti-inflammatory, antiviral | 20 | 4547 | 4569 | 23 |
| 27 | SA pks6 | ST pks4 | polyketide | phenolic lipids | cell wall lipid | NI/NI | 4694 | 4697 | 4 |
| 28 | SA nrps4 | ST nrps2 | non-ribosomal peptide | tetrapeptide | N/D | 21/21 | 4885 | 4904 | 20 |
| 29 | SA terp3 | ST terp2 | terpenoid | carotenoid pigment | antioxidant | 21/21 | 4927 | 4931 | 5 |
| 30 | SA pks1C | none | polyketide | naphthoic acid unit/kedarcidin-related, fragment C | cytotoxin/DNA | 21 | 4932 | 4956 | 25 |
|
| |||||||||
| Total | 540 | ||||||||
NI: non-island. Italics: predicted product or activity. Bold: observed product or activity. N/D: not determined.
Partial cluster.
Product observed in other bacteria.
Of the eight secondary metabolites that have been isolated from the two strains, all but salinosporamide A, sporolide A, and salinilactam have been reported from unrelated taxa (Figure 1), providing strong evidence of HGT. Further evidence for HGT comes from a phylogenetic analysis of the polyketide synthase (PKS) genes associated with the rifamycin biosynthetic gene cluster (rif) in SA and Amycolatopsis mediterranei, the original source of this antibiotic (Yu 1999). This analysis confirms prior observations of HGT in this pathway (Kim et al 2006) and reveals that all 10 of the ketosynthase domains are perfectly interleaved, as would be predicted if the entire PKS gene cluster had been exchanged between the two strains (Figure S1). Evidence of HGT coupled with prior evidence for the fixation of specific pathways such as rif among globally distributed SA populations (Jensen et al 2007) supports vertical inheritance following pathway acquisition (Ochman 2005). This evolutionary history is what might be expected if pathway acquisition fostered ecotype diversification or a selective sweep (Cohan 2002) resulting from strong selection for the acquired pathway, either of which provide compelling evidence that secondary metabolites represent functional traits with important ecological roles. The concept that gene acquisition provides a mechanism for ecological diversification that may ultimately drive the formation of independent bacterial lineages has been previously proposed (Ochman et al 2000). The inclusion of secondary metabolism among the functional categories of acquired genes that may have this effect sheds new light on the functional importance and evolutionary significance of this class of genes. Although the ecological functions of secondary metabolites remain largely unknown, and thus it is not clear how these molecules might facilitate ecological diversification, there is mounting evidence that they play important roles in chemical defense (Haeder et al 2009) or as signaling molecules involved in population or community communication (Yim et al 2007).
Differences between the two species also occur in CRISPR sequences, which are non-continuous direct repeats separated by variable (spacer) sequences that have been shown to confer immunity to phage (Barrangou et al 2007). The ST genome carries three intact prophage and three CRISPRs (35 spacers), while only one prophage has been identified in the genome of SA, which possesses eight different CRISPRs (140 spacers). The SA prophage is unprecedented among bacterial genomes in that it occurs in two adjacent copies that share 100% sequence identity. These copies are flanked by tRNA att sites and separated by an identical 45 bp att site, suggesting double integration as opposed to duplication (te Poele et al 2008). Remarkably, four of the SA CRISPRs possess a spacer that shares 100% identity with portions of three different genes found in ST prophage 1 (Figure 2). These spacer sequences have no similar matches to genes in the SA prophage or in any prophage sequences deposited in the NCBI, CAMERA, or the SDSU Center for Universal Microbial Sequencing databases. The detection of these spacer sequences provides evidence that SA has been exposed to a phage related to one that currently infects ST and that SA now maintains acquired immunity to this phage genotype as has been previously reported in other bacteria (Barrangou et al 2007). This is a rare example in which evidence has been obtained for CRISPR-mediated acquired immunity to a prophage that resides in the genome of a closely related environmental bacterium. Given that SA strain CNS-205 was isolated from Palau while ST strain CNB-440 was recovered 15 years earlier from the Bahamas, it appears that actinophage have broad temporal-spatial distributions or that resistance is maintained on temporal scales sufficient for the global distribution of a bacterial species.
Figure 2.
S. tropica prophage and S. arenicola CRISPRs. Four of 8 SA CRISPRs (1, 5, 7, 8) have spacers (color coded) that share 100% sequence identity with genes (Stro numbers and annotation given) in ST prophage 1 (Table S2, inverted for visual purposes). Other CRISPRs are colored purple. SA CRISPRs 2-3 and 5-6 share the same direct repeats and may have at one time been a single allele. CRISPR associated (CAS) genes (red) and genes interrupting CRISPRs (black) are indicated. None of the spacer sequences possessed 100% identity to prophage in the NCBI non-redundant sequence database, the SDSU Center for Universal Microbial Sequencing database, or the CAMERA metagenomic database.
Enhanced phage immunity, as evidenced by 140 relative to 35 CRISPR spacer sequences, coupled with a larger genome size and a greater number of species-specific secondary metabolic pathways may account for the cosmopolitan distribution of SA relative to ST, which to date has only been recovered from the Caribbean (Jensen and Mafnas 2006). Also included among the SA-specific gene pool is a complete phospho-transferase system (PTS, Sare4844-4850). PTSs are centrally involved in carbon source uptake and regulation (Parche et al 2000) and may provide growth advantages that also factor into the relatively broad distribution of SA. However, additional strains will need to be studied before any of these differences can be firmly linked to species distributions.
The 21 genomic islands are not contiguous regions of species-specific DNA but were instead created by a complex process of gene acquisition, loss, duplication, and inactivation (Figure 3). The overall composition, evolutionary history, and function of the island genes are similar in both strains, with duplication and HGT accounting for the majority of genes and secondary metabolism representing the largest functionally annotated category. Remarkably, 42% of the rearranged island orthologs fall within other islands indicating that inter-island movement or “island hopping” is common, thus providing support for the hypothesis that islands undergo continual rearrangement (Coleman et al 2006). There is dramatic, operon-scale evidence of this process in the shared yersiniabactin pathways (ST sid2 and SA sid1), which occur in islands 15 and 10, respectively, and in the unknown dipeptide pathways (ST nrps1 and SA nrps3), which occur in islands 4 and 15, respectively. In both cases, these pathways remain intact yet are located in different islands in the two strains (Figure 1, Table 2, 3). There is also evidence of cluster fragmentation in the 10-membered enediyne gene set SA pks3, which contains the core set of genes associated with calicheamicin biosynthesis (Figure S2) (Ahlert et al 2002), yet is split by the introduction of 145 kb of DNA from three different biosynthetic loci (island 10, Figure 1). The conserved fragments appear to encode the biosynthesis of a calicheamicin anolog, while flanking genes display a high level of gene duplication and rearrangement indicative of active pathway evolution. Cluster fragmentation is also observed in the 9-membered enediyne PKS cluster SA pks1(A-C), which is scattered across the genome in islands 4, 10, and 21 (Figure 1, Table 3).
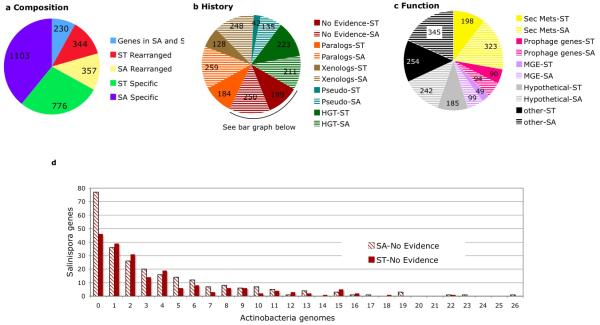
Figure 3.
Composition, evolutionary history, and function of island genes in S. tropica (ST) and S. arenicola (SA). (a) 3040 genes comprising 21 genomic islands were analyzed for positional orthology (ie., the gene is part of the shared “core” genome), re-arranged orthology (ie., the gene is present in the other genome but not in the same position or island), and species-specificity (gene totals presented in wedges). (b) The ST and SA species-specific island genes were analyzed for evidence of parology, xenology, and HGT. Pseudogenes and the number of genes with no evidence for any of these processes were also identified. (c) Functional annotation of the species-specific island genes. (d) Distribution of species-specific island genes that have no evidence for HGT or parology among 27 Actinobacterial genomes.
The genomic islands are also enriched in mobile genetic elements including prophage, integrases, and actinobacterial integrative and conjugative elements (AICEs) (Burrus et al 2002) (Tables S2, S3), the later of which are known to play a role in gene acquisition and rearrangement. The Salinispora AICEs possess traB homologs, which promote conjugal plasmid transfer in mycelial streptomycetes (Reuther 2006), suggesting that hyphal tip fusion is a prominent mechanism driving gene exchange in these bacteria. AICEs have been linked to the acquisition of secondary metabolite gene clusters (te Poele et al 2007) and their occurrence in island 7 (SA AICE1), which includes the entire 90 kb rif cluster, and island 10 (SA AICE3), which contains biosynthetic gene clusters for enediyne, siderophore, and amino acid-derived secondary metabolites, provides a mechanism for the acquisition of these pathways (Figure 1). Six additional secondary metabolite gene clusters (ST nrps1, ST spo, SA nrps3, SA pks5, SA cym, and SA pks2) are flanked by direct repeats, providing further support for HGT. In the case of cym (Schultz 2008), which is clearly inserted into a tRNA, the pseudogenes preceding and following it are all related to transposases or integrases providing a mechanism for chromosomal integration.
Despite exhaustive analyses of HGT, only 22% of the 127 genes in the five biosynthetic pathways (rif, sta, des, lym, cym) whose products have also been observed in other bacteria (Figure 1, Table 3) scored positive for HGT. This observation suggests that the pathways either originated in Salinispora or that the exchange of these biosynthetic genes has occurred largely among closely related bacteria and therefore gone undetected with the HGT methods applied in this study. The latter scenario is supported by the observation that all five of the shared biosynthetic pathways were previously reported in other actinomycetes. The acquisition of genes from closely related bacteria likely accounts for many of the species-specific island genes for which no evidence of evolutionary history could be determined (Figure 3b). These genes were poorly conserved among 27 Actinobacterial genomes (Figure 3d) providing additional support that they were acquired, most likely from environmental Actinobacteria that are not well represented among sequenced genomes. Although gene loss was not quantified, this process is also a likely contributor to island formation. In support of an adaptive role for island genes, 7.6% (44/573) of the orthologs show evidence of positive selection (dN/dS >1) compared to 1.6% (49/3027) of the non-island pairs. Given that the majority of island genes display evidence of HGT, the increased dN/dS ratio is in agreement with the observation that acquired genes experience relaxed functional constraints (Hao and Golding, 2006).
Functional differences between related organisms can be obscured when orthologs are taken out of the context of the gene clusters in which they reside. For example, the PKS genes Sare1250 and Stro2768 are orthologous and likely perform similar functions, yet they reside in the rif and slm pathways, respectively, and thus contribute to the biosynthesis of dramatically different secondary metabolites. Likewise, intra-cluster PKS gene duplication (Sare3151 and Sare3152, Figure 1) has an immediate effect on the product of the pathway by the introduction of an additional acyl group into the carbon skeleton of the macrolide, as opposed to the more traditional concept of parology facilitating mutation-driven functional divergence (Prince 2002). Sub-genic, modular duplications are also observed (Sare3156 modules 4 and 5, Figure 1), which likewise have an immediate effect on the structure of the secondary metabolite produced by the pathway. While HGT is considered a rapid method for ecological adaptation in bacteria (Ochman et al 2000), PKS gene duplication provides a complementary evolutionary strategy (Fischbach et al 2008) that could lead to the rapid production of new secondary metabolites that subsequently drive the creation of new adaptive radiations.
Salinispora species are the first marine Actinobacteria reported to require seawater for growth (Maldonado et al 2005). Unlike Gram-negative marine bacteria, in which seawater requirements are linked to a specific sodium ion requirement (Kogure 1998), Salinispora strains are capable of growth in osmotically adjusted, sodium-free media (Tsueng and Lam 2008). An analysis of the Salinispora core for evidence of genes associated with this unusual osmotic requirement reveals a highly duplicated family of 29 polymorphic membrane proteins (PMPs) that include homologs associated with polymorphic outer membrane proteins (POMPs). POMPs remain functionally uncharacterized however there is strong evidence that they are type V secretory systems (Henderson 2001), making this the first report of type V autotransporters outside of the Proteobacteria (Henderson 2004). Phylogenetic analyses provide evidence that the Salinispora PMPs were acquired from aquatic, Gram-negative bacteria and that they have continued to undergo considerable duplication subsequent to divergence of the two species (Figure S3). The occurrence of this large family of PMP autotransporters in marine Actinobacteria may represent a low nutrient adaptation that renders cells susceptible to lysis in low osmotic environments.
Conclusions
In conclusion, the comparative analysis of two closely related marine Actinobacterial genomes provides new insight into the functional traits associated with genomic islands. It has been possible to assign precise, physiological functions to island genes and link differences in secondary metabolism to fine-scale phylogenetic architecture in two distinct bacterial lineages, which by all available metrics maintain the fundamental characteristics of species-level units of diversity. It is clear that gene clusters devoted to secondary metabolite biosynthesis are dynamic entities that are readily acquired, rearranged, and fragmented in the context of genomic islands, and that the results of these processes create natural product diversity that can have an immediate effect on fitness or niche utilization. The high level of species specificity associated with secondary metabolism suggests that this functional trait may represent a previously unrecognized force driving ecological diversification among closely related, sediment inhabiting bacteria.
Supplementary Material
Acknowledgements
This manuscript is dedicated to Professor William Fenical for his pioneering work on the secondary metabolites of marine actinomycetes. PRJ and BSM were funded by the California Sea Grant Program (R/NMP-98), NOAA grant NAO80AR4170669, and the JGI Community Sequencing Program. Additional funding was from NIH grant CA127622 to BSM and a post-doctoral fellowship from the DAAD to MN. EEA thanks the Gordon and Betty Moore Foundation for funding through CAMERA. We acknowledge Dr. Jonathan Badger for assistance with APIS and Prof. Terry Gaasterland for computational assistance.
Footnotes
References
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nat. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, et al. The calicheamicin gene cluster and its iterative type I enediyne PKS. Sci. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- Badger JH, Eisen JA, Ward NL. Genomic analysis of Hyphomonas neptunium contradicts 16S rRNA gene-based phylogenetic analysis: implications for the taxonomy of the orders ‘Rhodobacterales’ and Caulobacterales. Int J Syst Evol Microbiol. 2005;55:1021–1026. doi: 10.1099/ijs.0.63510-0. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Sci. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Berdy J. Bioactive microbial metabolites. A personal view. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Bose M, Barber RD. Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences. In Silico Biology. 2006;6:223–227. [PubMed] [Google Scholar]
- Buchanan GO, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. Sporolides A and B: Structurally unprecedented halogenated macrolides from the marine actinomycete Salinispora tropica. Organ Lett. 2005;7:2731–2734. doi: 10.1021/ol050901i. [DOI] [PubMed] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B, GuÈdon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- Cohan FM. What are bacterial species? Ann Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Sci. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- Dondoshansky I, Wolf Y. BLASTCLUST. 2.2 edn. National Institutes of Health; Bethesda, MD: 2000. [Google Scholar]
- Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angewandte Chemie. 2003;115:369–371. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd GK, Potts BC. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorg Med Chem. 2009;17:2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Nat Acad Sci. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucl Acids Res. 2006;34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: Making sense of genetic and ecological diversity. Sci. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, et al. Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Haeder S, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc Natl Acad Sci. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Lam AC. Polymorphic proteins of Chlamydia spp. - autotransporters beyond the Proteobacteria. Trends Microbiol. 2001;9:573–578. doi: 10.1016/s0966-842x(01)02234-x. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathways: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao W, Wan I, Jones SJ, Brinkman FSL. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics. 2003;19:418–420. doi: 10.1093/bioinformatics/btg004. [DOI] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Sci. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Mafnas C. Biogeography of the marine actinomycete Salinispora. Environ Microbiol. 2006;8:1881–1888. doi: 10.1111/j.1462-2920.2006.01093.x. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Williams PG, Oh D-C, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EA, Chan K, Salama NR, Falkow S. Redefining bacterial populations: A post-genomic reformation. Nat Rev Gen. 2002;3:462–473. doi: 10.1038/nrg820. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hewavitharana AK, Shaw PN, Fuerst JA. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl Environ Microbiol. 2006;72:2118–2125. doi: 10.1128/AEM.72.3.2118-2125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure K. Bioenergenics of marine bacteria. Curr Opin Biotechnol. 1998;9:278–282. doi: 10.1016/s0958-1669(98)80059-1. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucl Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, et al. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol. 2005;55:1759–1766. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- Mincer TJ, Jensen PR, Kauffman CA, Fenical W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol. 2002;68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nat. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lerat E, Daublin V. Examining bacterial species under the specter of gene transfer and exchange. Proc Natl Acad Sci. 2005;102:6595–6599. doi: 10.1073/pnas.0502035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parche S, Nothaft H, Kamionka A, Titgemeyer F. Sugar uptake and utilisation in Streptomyces coelicolor: A PTS view to the genome. Antonie van Leeuwenhoek. 2000;78:243–251. doi: 10.1023/a:1010274317363. [DOI] [PubMed] [Google Scholar]
- Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Gaasterland T, Allen EE. A database of phylogenetically atypical genes in archaeal and bacterial genomes, identified using the DarkHorse algorithm. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Gen. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Reuther J, Gekeler C, Tiffert Y, Wohlleben W, Muth G. Unique conjugative mechanism in mycelial streptomycetes: A DNA-binding ATPase translocates unprocessed plasmid DNA at the hyphal tip. Mol Microbiol. 2006;61:436–446. doi: 10.1111/j.1365-2958.2006.05258.x. [DOI] [PubMed] [Google Scholar]
- Schultz AW, Oh D-C, Carney JR, Williamson RT, Udwary DW, Jensen PR, Gould SJ, Fenical W, Moore BS. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J Amer Chem Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- Starcevic A, Zucko J, Simunkovic J, Long PF, Cullum J, Hranueli D. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucl Acids Res. 2008;36:6882–6892. doi: 10.1093/nar/gkn685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Poele E, Bolhuis H, Dijkhuizen L. Actinomycete integrative and conjugative elements. Antonie van Leeuwenhoek. 2008;94:127–143. doi: 10.1007/s10482-008-9255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Poele EM, Habets MN, Tan GYA, Ward AC, Goodfellow M, Bolhuis H, et al. Prevalence and distribution of nucleotide sequences typical for pMEA-like accessory genetic elements in the genus Amycolatopsis. FEMS Microbiol Ecol. 2007;61:285–294. doi: 10.1111/j.1574-6941.2007.00334.x. [DOI] [PubMed] [Google Scholar]
- Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, et al. Genotypic diversity within a natural coastal bacterioplankton population. Sci. 2005;307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- Tsueng G, Lam K. A low-sodium-salt formulation for the fermentation of salinosporamides by Salinispora tropica strain NPS21184. Appl Microbiol Biotechnol. 2008;78:821–826. doi: 10.1007/s00253-008-1357-x. [DOI] [PubMed] [Google Scholar]
- Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, et al. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics. 2008;9:14. doi: 10.1186/1471-2164-9-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, et al. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos GS, Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- Welch RA, Burland V, Plunkett G, Redford P, Roesch P, Rasko D, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Culley DE, Zhang W. Predicted highly expressed genes in the genomes of Streptomyces coelicolor and Streptomyces avermitilis and the implications for their metabolism. Microbiol. 2005;151:2175–2187. doi: 10.1099/mic.0.27833-0. [DOI] [PubMed] [Google Scholar]
- Yim G, Wang HMH, Davies J. Antibiotics as signalling molecules. Philosophical Transactions of the Royal Society B-Biological Sciences. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T-W, Shen Y, Doi-Katayama Y, Tang L, Park C, Moore BS, Hutchinson CR, Floss HG. Direct evidence that rifamycin polyketide synthase assembles polyketide chains processively. Proc Natl Acad Sci. 1999;96:9051–9056. doi: 10.1073/pnas.96.16.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.