Abstract
Analysis of genome and expressed sequence tag data bases at the turn of the millennium unveiled a new protease family named the type II transmembrane serine proteases (TTSPs) in a Journal of Biological Chemistry minireview (Hooper, J. D., Clements, J. A., Quigley, J. P., and Antalis, T. M. (2001) J. Biol. Chem. 276, 857–860). Since then, the number of known TTSPs has more than doubled, and more importantly, our understanding of the physiological functions of individual TTSPs and their contribution to human disease has greatly increased. Progress has also been made in identifying molecular substrates and endogenous inhibitors. This minireview summarizes the current knowledge of the rapidly advancing TTSP field.
Type II Transmembrane Serine Protease Family
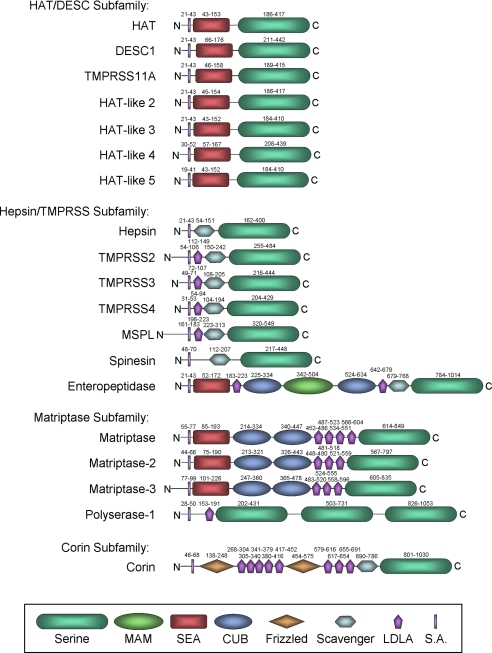
The defining features of TTSPs2 are an N-terminal transmembrane domain, a C-terminal extracellular serine protease domain of the chymotrypsin (S1) fold that contains the catalytic histidine, aspartic acid, and serine residues, and a “stem region” that may contain an assortment of 1–11 protein domains of six different types (Fig. 1) (1, 2). The human TTSP family has 17 members (Fig. 1). Supplemental Table S1 shows the standard name of each TTSP as listed in the Mammalian Degradome Database (3), alternative names, the name and ID of the gene encoding each TTSP, and the chromosomal location of human and mouse TTSP orthologs. TTSPs belong to one of four subfamilies (Fig. 1 and supplemental Table S1) (2). The HAT/DESC (human airway trypsin-like protease/differentially expressed in squamous cell carcinoma) subfamily comprises HAT, DESC1, TMPRSS11A, HAT-like 2, HAT-like 3, HAT-like 4, and HAT-like 5. The stem region of all HAT/DESC proteases is composed of a single SEA (sea urchin sperm protein/enteropeptidase/agrin) domain. The hepsin/TMPRSS (transmembrane protease/serine) subfamily has seven members, comprising hepsin, TMPRSS2, TMPRSS3, TMPRSS4, TMPRSS5/spinesin, MSPL (mosaic serine protease large-form), and enteropeptidase. All have a group A scavenger receptor domain in their stem region, preceded by a single LDLA (low-density lipoprotein receptor class A) domain in TMPRSS2, TMPRSS3, TMPRSS4, and MSPL or by an array of SEA, LDLA, CUB (Cls/Clr, urchin embryonic growth factor, bone morphogenetic protein-1), and MAM (meprin/A5 antigen/receptor protein phosphatase mu) domains in enteropeptidase. The matriptase subfamily has four members: matriptase, matriptase-2, matriptase-3, and the unique polyserase-1. The matriptases have a SEA domain, two CUB domains, and three (matriptase-2 and matriptase-3) to four (matriptase) LDLA domains in their stem region. The polyserase stem region consists of an LDLA domain and two active and one catalytically inactive serine protease domains. Corin displays a complex stem region composed of two frizzled domains, eight LDLA domains, and one group A scavenger receptor domain. TTSP genes are found in all vertebrates (4), and the TTSP family may have originated from two ancestral genes: one giving rise to the HAT/DESC, hepsin/TMPRSS, and matriptase subfamilies and one to the corin subfamily (2). Possibly reflecting this, arthropod genomes encode only two TTSPs: stubble-stubbloid and corin. The former is most closely related to HAT/DESC, hepsin/TMPRSS, and matriptase proteases, and the latter is most closely related to corin (4). Mice and rats have two HAT/DESC subfamily members (Desc4 and Tmprss11c, encoding HAT-like 2 and HAT-like 3) that are not found in humans and chimpanzees. This divergence is caused by gene loss in primates, as orthologs of Desc4 and Tmprss11c exist as pseudogenes in the human and chimpanzee genomes (3). Detailed information on the cellular location of hepsin, TMPRSS2, spinesin, enteropeptidase, matriptase, matriptase-2, and corin is available. The relative mRNA abundance in tissues is known for TMPRSS3, TMPRSS4, MSPL, matriptase-3, and polyserase, whereas the sites of expression of TMPRSS11A, HAT-like 3, HAT-like 4, and HAT-like 5 are unknown (supplemental Table S2). Mutations in five TTSP genes are now established as the underlying cause of autosomal recessive genetic disorders in humans, and chromosomal rearrangements and altered expression of TTSP genes appear to critically contribute to human carcinogenesis (supplemental Table S3).
FIGURE 1.
The TTSP family is composed of four subfamilies: HAT/DESC, hepsin/TMPRSS, matriptase, and corin. The structure of each TTSP is shown schematically. TTSPs consist of an N-terminal signal anchor domain (S.A.) and a C-terminal trypsin-like serine protease domain (Serine) that flank a stem region composed of 1–11 protein domains of six different types (see box at bottom). The location of each domain (amino acid numbering) is indicated above the domain (4). HAT-like 2 and HAT-like 3 are found in rodents but not in primates. Polyserase has three C-terminal serine protease domains, of which the third is catalytically inactive. Scavenger indicates the group A scavenger receptor domain.
Biochemical Properties of TTSPs
TTSPs are synthesized as inactive single chain proenzymes (zymogens), which require activation by cleavage following a basic amino acid residue (Arg or Lys) in a conserved activation motif preceding the catalytic domain. They remain membrane-associated after activation because of a disulfide bond that links the prodomain and catalytic domain (1). An unusual feature of several TTSPs, including matriptase (5), matriptase-2 (6), hepsin (7), TMPRSS2 (8), TMPRSS3 (9), TMPRSS4 (10), and TMPRSS11C (11), is that they undergo autocatalytic activation in vitro. This suggests that many of the TTSPs could function as initiators of proteolytic cascades. The mechanism of autoactivation is not known but likely involves oligomerization. Matriptase and enteropeptidase also undergo proteolytic cleavage within the SEA domain, believed to occur by spontaneous conformation-driven hydrolysis of a specific peptide bond, suggesting that this processing may occur for all of the SEA domain-containing TTSPs (4). The cytoplasmic tail domain may contribute to the targeting of the TTSPs to plasma membrane microdomains, and some tails contain consensus phosphorylation sites. The extracellular domains of TTSPs are critical for cellular localization, activation, inhibition, and/or substrate specificity (4). For example, the ability of corin to activate pro-ANP is dependent on frizzled domain 1 and LDLA domain repeats 1–4 (12). Ectodomain shedding is frequent, providing a mechanism by which TTSPs could be active in the pericellular space (4). TTSPs are regulated by endogenous protease inhibitors, specifically Kunitz domain-containing inhibitors and serpins. The transmembrane Kunitz-type inhibitor HAI-1 is implicated in the inhibition of matriptase (13), hepsin (14), and prostasin (15). The related HAI-2 also displays potent inhibitory activity toward hepsin and matriptase (14, 16). Matriptase is found in human milk in complex with the secreted serpins antithrombin III, α1-antitrypsin, and α2-antiplasmin (17). Inhibitory complexes can be formed in vitro of the serpins PAI-1 and protein C inhibitor with mouse DESC1 (18); PAI-1, protein C inhibitor, α1-proteinase inhibitor, α2-antiplasmin, and antithrombin III with matriptase-3 (19); and PAI-1 and α2-antiplasmin with serase-1B (20).
Physiology and Pathobiology of TTSPs
Enteropeptidase in Digestion
The role of enteropeptidase in digestion was first recognized by Ivan Pavlov in canine models in the early 1900s and subsequently studied by Moses Kunitz with purified proteins in the 1930s (21). Enteropeptidase is expressed on the brush-border membrane of the duodenum, where it converts pancreatic trypsinogen to trypsin. This function is essential for initiating proteolytic reactions of digestive enzymes in the small intestine. Patients with congenital enteropeptidase deficiency, which is caused by mutations in the PRSS7 gene encoding enteropeptidase (21), are unable to digest food efficiently and suffer from malnutrition and growth delay.
Corin in Cardiac Function and Blood Pressure Regulation
Corin is expressed primarily in cardiomyocytes. Corin converts pro-ANP to active ANP, a cardiac hormone that regulates blood pressure and cardiac function by promoting natriuresis, dieresis, and vasodilation (22). In mice, lack of corin prevents conversion of pro-ANP to ANP (23). Corin null mice develop hypertension that is exacerbated by a high-salt diet and exhibit cardiac hypertrophy. Single nucleotide polymorphisms in the corin gene are found in African Americans with a history of high blood pressure and cardiac hypertrophy (24, 25). These single nucleotide polymorphisms alter amino acids in corin frizzled domain 2 and impair its zymogen activation and pro-ANP processing activity (26), suggesting that genetic defects in corin contribute to hypertensive disease in humans. Corin is also expressed in other tissues (supplemental Table S2) and has a role in pigmentation of mouse fur (27).
TTSPs in Hearing
Several TTSPs play a role in normal hearing (supplemental Table S3). The TMPRSS3 gene was first identified in a locus on chromosome 21 that was associated with familiar congenital deafness. An array of mutations in the TMPRSS3 gene have been found in patients with nonsyndromic recessive hearing loss (28). Mutations in the TMPRSS5 gene may also contribute to hearing loss (29). Both TMPRSS3 and TMPRSS5 are expressed in inner ear tissues. Many naturally occurring mutations prevent zymogen activation of these enzymes. Recently, hepsin was also found to be important for normal hearing (30). Hepsin null mice have abnormal cochlear structures and reduced myelin protein expression in the auditory nerve. Hepsin null mice also have low levels of thyroxine. As thyroid hormone is required for the development of the inner ear, this suggests that the impaired hearing may be secondary to a defect in thyroid hormone metabolism.
Matriptase-2 in Iron Metabolism
Matriptase-2 plays an important role in iron homeostasis. In chemically induced or gene knock-out mouse models, disruption of the Tmprss6 gene caused severe iron deficiency anemia (31, 32). Similarly, mutations in the TMPRSS6 gene have been identified in patients with microcytic anemia who respond poorly to iron therapy (33). Matriptase-2 is expressed primarily in the liver, where it appears to suppress hepcidin expression. Hepcidin binds and internalizes the iron export protein, ferroportin, on the surface of enterocytes and macrophages, thereby reducing iron supply. Recently, matriptase-2 was shown to degrade hemojuvelin, a membrane-anchored protein that acts as a cofactor for bone morphogenetic protein-mediated activation of hepcidin gene expression (34). Matriptase-2 deficiency leads to high levels of hemojuvelin and hence increased hepcidin expression, which in turn suppresses iron absorption and causes iron deficiency anemia.
TTSPs in Epithelial Homeostasis
HAT, HAT-like 2, TMPRSS2, and matriptase are widely expressed in epithelial tissues (supplemental Table S2). Other TTSPs may have a similar expression pattern, as judged by mRNA abundance in tissues (supplemental Table S2). HAT is expressed in airway epithelium, suprabasal layers of the epidermis, and multiple other organs (supplemental Table S2). HAT is proposed to execute a diverse array of functions, including fibrinogenolysis, PAR-2 activation, amphiregulin expression, and urokinase plasminogen activator receptor cleavage (4, 35). However, these candidate proteolytic targets of HAT and the roles of HAT in epithelial physiology and pathology were designated only from the study of in vitro cleavage reactions and cell-based assays. HAT-like 2, encoded by the Desc4 gene, is found in epithelial cells of the circumvallate papillae of adult rats, embryonic salivary gland ducts, nasal epithelial cells, and tear gland ducts, and the mRNA has been detected in a large number of other tissues but has not been localized to specific cell types (supplemental Table S2). Desc4 is a pseudogene in both humans and chimpanzees (3), indicating that the functions of HAT-like 2 are rodent-specific or are redundant with other proteases in higher mammalian species. TMPRSS2 was first found in basal cells of the prostate epithelium (36) and later in the epithelial component of a number of other tissues, including kidney tubules, upper airway epithelium and alveoli, colonic epithelium, bile duct, and ovaries. TMPRSS2 transcripts have also been detected in many other tissues without cellular localization (supplemental Table S2). Expression of the TMPRSS2 gene in the prostate is androgen- and androgen receptor-dependent (8, 36). The physiological function of TMPRSS2 is unclear, as Tmprss2 null mice did not present with an obvious phenotype, and no compensatory up-regulation of other TTSPs with overlapping patterns of expression was apparent (37). TMPRSS2 has been proposed to regulate epithelial sodium currents in the lung through proteolytic cleavage of the epithelial sodium channel (38) and inflammatory responses in the prostate via the proteolytic activation of PAR-2 (39). Matriptase is widely, although not uniformly, expressed in epithelial tissues. Human monocytes, B-cells, and mast cells also express matriptase (supplemental Table S2). The recent identification of individuals with homozygosity for null and hypomorphic mutations in the ST14 gene (encoding matriptase) and the study of St14 null and hypomorphic mice revealed a critical role for matriptase in terminal epidermal differentiation, epidermal barrier formation, and hair follicle development (supplemental Table S2). Matriptase-deficient individuals and mice display ichthyosis (dry, thickened, and scaly skin) and hypotrichosis (sparse and abnormal hair) (40–43). Corneal opacity, photophobia, and abnormal primary and deciduous teeth are other manifestations of matriptase deficiency. Matriptase may be part of an epidermal proteolytic cascade that involves the glycosylphosphatidylinositol-anchored serine protease prostasin as based on the identical phenotype of mice with epidermal ablation of either the St14 or Prss8 gene (encoding prostasin), the efficient activation of pro-prostasin by matriptase in vitro, and the greatly reduced levels of active prostasin in the epidermis of matriptase-deficient mice and humans (40, 44).
TTSPs and Epithelial Carcinogenesis
Hepsin, TMPRSS2, and matriptase have attracted considerable attention in the context of epithelial carcinogenesis. General interest in hepsin in cancer was kindled when analysis of high-density cDNA and tissue microarrays revealed that the HPN gene (encoding hepsin) was among the most consistently and quantitatively overexpressed genes in human prostate cancer and was the most reliable single discriminator of prostatic intraepithelial neoplasia from benign prostate hyperplasia (45, 46). The association between hepsin and prostate cancer progression was further strengthened when overexpression of Hpn in prostate epithelium was shown to induce metastasis in a non-metastatic murine model of prostate carcinoma (47). More recently, expression of hepsin, but not a catalytically inactive version of hepsin, was reported to promote the progression of two ovarian carcinoma cell lines after engraftment in mice (48). Overexpression of hepsin in basal prostate epithelial cells of mice leads to the disorganization of the basement membrane (47). Substrates linked to carcinogenesis that can be cleaved by hepsin ex vivo are coagulation factor VII, pro-hepatocyte growth factor, laminin 332, and pro-urokinase plasminogen activator (46, 49).
A breakthrough in the understanding of the molecular pathogenesis of prostate cancer was made with the discovery of somatic fusions of the TMPRSS2 gene to members of the ETS gene family of transcription factors in the majority of prostate cancers (50). Four different ETS fusion partners have been identified, ERG, ETV1, ETV4, and ETV5, of which the TMPRSS2-ERG fusion is the most common. A complex array of chromosomal rearrangements causes the fusion of TMPRSS2 to ERG and ETV genes, leading to the generation of a wide variety of C-terminally truncated TMPRSS2 proteins fused to N-terminally truncated ERG/ETV proteins or to the expression of N-terminally truncated ERG/ETV proteins. In all cases, these gene fusions result in androgen-regulated and overall increased expression of this family of cell growth-regulating proto-oncogenes (supplemental Table S3). TMPRSS2 rearrangements have been detected in high-grade prostatic intraepithelial neoplasia, indicating that they are early events in prostate carcinogenesis (51). Additionally, the presence of TMPRSS2-ERG fusion transcripts is associated with a high rate of prostate cancer recurrence, distant metastasis, and death after prostatectomy (52–54).
Elevated matriptase, high matriptase/HAI-1 ratio, or increased activation of matriptase is associated with poor prognosis in many human carcinomas (55). This is noteworthy because matriptase in many epithelia is found exclusively in terminally differentiated cells but not in cells of the basal compartment from which carcinomas originate. Indeed, matriptase becomes expressed de novo in the epidermal basal compartment during pre-malignant progression (55). Low-level forced expression of the protease in basal keratinocytes of mice suffices to induce squamous cell carcinoma and dramatically potentiates ras-mediated squamous cell carcinogenesis (56). Tumor grafting experiments suggest that matriptase may also promote late stages of tumor dissemination (57, 58). Pro-hepatocyte growth factor, pro-urokinase plasminogen activator, and PAR-2 represent possible downstream targets for matriptase in carcinogenesis (5, 59–61).
Perspectives
The TTSP family is the most recently identified protease family, and much is still to be learned. However, some tentative generalizations can be made at this time: TTSPs have diverse roles in vertebrate physiology, and their structural likeness does not reflect a common biochemical function. TTSPs mostly serve to maintain basic homeostasis rather than re-establish homeostasis after external challenges, such as tissue injury and infection. In this respect, TTSPs may differ from other more extensively studied families of pericellular proteases. Finally, TTSPs appear to be frequently involved in either hormone or growth factor activation or in the initiation of proteolytic cascades. The next decade is likely to bring a wealth of additional information about this fascinating protease family.
Acknowledgments
We thank our many colleagues in the TTSP field for helpful suggestions while preparing this minireview.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Program (to T. H. B.) and National Institutes of Health Grants CA098369 and HL084387 (to T. M. A.) and HL089298 (to Q. W.). This work was also supported by the Mary Kay Ash Charitable Foundation (075-07). This is the eighth article in the Thematic Minireview Series on Proteolytic Enzymes. The first article was published in the November 7, 2008 issue; the second and third articles were published in the May 22, 2009 issue; the fourth and fifth articles were published in the July 31, 2009 issue; and the sixth and seventh articles were published in the August 14, 2009 issue. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and additional references.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and additional references.
- TTSP
- type II transmembrane serine protease
- ANP
- atrial natriuretic peptide
- HAI
- hepatocyte growth factor activator inhibitor
- PAI
- plasminogen activator inhibitor
- PAR
- protease-activated receptor.
REFERENCES
- 1.Hooper J. D., Clements J. A., Quigley J. P., Antalis T. M. (2001) J. Biol. Chem. 276,857–860 [DOI] [PubMed] [Google Scholar]
- 2.Szabo R., Wu Q., Dickson R. B., Netzel-Arnett S., Antalis T. M., Bugge T. H. (2003) Thromb. Haemost. 90,185–193 [DOI] [PubMed] [Google Scholar]
- 3.Quesada V., Ordóñez G. R., Sánchez L. M., Puente X. S., López-Otín C. (2009) Nucleic Acids Res. 37,D239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szabo R., Bugge T. H. (2008) Int. J. Biochem. Cell Biol. 40,1297–1316 [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi T., Harris J. L., Huang W., Yan K. W., Coughlin S. R., Craik C. S. (2000) J. Biol. Chem. 275,26333–26342 [DOI] [PubMed] [Google Scholar]
- 6.Velasco G., Cal S., Quesada V., Sánchez L. M., López-Otín C. (2002) J. Biol. Chem. 277,37637–37646 [DOI] [PubMed] [Google Scholar]
- 7.Qiu D., Owen K., Gray K., Bass R., Ellis V. (2007) Biochem. Soc. Trans. 35,583–587 [DOI] [PubMed] [Google Scholar]
- 8.Afar D. E., Vivanco I., Hubert R. S., Kuo J., Chen E., Saffran D. C., Raitano A. B., Jakobovits A. (2001) Cancer Res. 61,1686–1692 [PubMed] [Google Scholar]
- 9.Guipponi M., Vuagniaux G., Wattenhofer M., Shibuya K., Vazquez M., Dougherty L., Scamuffa N., Guida E., Okui M., Rossier C., Hancock M., Buchet K., Reymond A., Hummler E., Marzella P. L., Kudoh J., Shimizu N., Scott H. S., Antonarakis S. E., Rossier B. C. (2002) Hum. Mol. Genet. 11,2829–2836 [DOI] [PubMed] [Google Scholar]
- 10.Andreasen D., Vuagniaux G., Fowler-Jaeger N., Hummler E., Rossier B. C. (2006) J. Am. Soc. Nephrol. 17,968–976 [DOI] [PubMed] [Google Scholar]
- 11.Stallmach R., Gloor S. M. (2008) Biochem. J. 412,81–91 [DOI] [PubMed] [Google Scholar]
- 12.Knappe S., Wu F., Masikat M. R., Morser J., Wu Q. (2003) J. Biol. Chem. 278,52363–52370 [DOI] [PubMed] [Google Scholar]
- 13.Lin C. Y., Anders J., Johnson M., Dickson R. B. (1999) J. Biol. Chem. 274,18237–18242 [DOI] [PubMed] [Google Scholar]
- 14.Kirchhofer D., Peek M., Lipari M. T., Billeci K., Fan B., Moran P. (2005) FEBS Lett. 579,1945–1950 [DOI] [PubMed] [Google Scholar]
- 15.Fan B., Wu T. D., Li W., Kirchhofer D. (2005) J. Biol. Chem. 280,34513–34520 [DOI] [PubMed] [Google Scholar]
- 16.Szabo R., Hobson J. P., List K., Molinolo A., Lin C. Y., Bugge T. H. (2008) J. Biol. Chem. 283,29495–29504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng I. C., Chou F. P., Su S. F., Oberst M., Madayiputhiya N., Lee M. S., Wang J. K., Sloane D. E., Johnson M., Lin C. Y. (2008) Am. J. Physiol. Cell Physiol. 295,C423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobson J. P., Netzel-Arnett S., Szabo R., Réhault S. M., Church F. C., Strickland D. K., Lawrence D. A., Antalis T. M., Bugge T. H. (2004) J. Biol. Chem. 279,46981–46994 [DOI] [PubMed] [Google Scholar]
- 19.Szabo R., Netzel-Arnett S., Hobson J. P., Antalis T. M., Bugge T. H. (2005) Biochem. J. 390,231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okumura Y., Hayama M., Takahashi E., Fujiuchi M., Shimabukuro A., Yano M., Kido H. (2006) Biochem. J. 400,551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q. (2003) Curr. Top. Dev. Biol. 54,167–206 [DOI] [PubMed] [Google Scholar]
- 22.Wu Q. (2007) Front. Biosci. 12,4179–4190 [DOI] [PubMed] [Google Scholar]
- 23.Chan J. C., Knudson O., Wu F., Morser J., Dole W. P., Wu Q. (2005) Proc. Natl. Acad. Sci. U.S.A. 102,785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rame J. E., Drazner M. H., Post W., Peshock R., Lima J., Cooper R. S., Dries D. L. (2007) Hypertension 49,857–864 [DOI] [PubMed] [Google Scholar]
- 25.Dries D. L., Victor R. G., Rame J. E., Cooper R. S., Wu X., Zhu X., Leonard D., Ho S. I., Wu Q., Post W., Drazner M. H. (2005) Circulation 112,2403–2410 [DOI] [PubMed] [Google Scholar]
- 26.Wang W., Liao X., Fukuda K., Knappe S., Wu F., Dries D. L., Qin J., Wu Q. (2008) Circ. Res. 103,502–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enshell-Seijffers D., Lindon C., Morgan B. A. (2008) Development 135,217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guipponi M., Antonarakis S. E., Scott H. S. (2008) Front. Biosci. 13,1557–1567 [DOI] [PubMed] [Google Scholar]
- 29.Guipponi M., Toh M. Y., Tan J., Park D., Hanson K., Ballana E., Kwong D., Cannon P. Z., Wu Q., Gout A., Delorenzi M., Speed T. P., Smith R. J., Dahl H. H., Petersen M., Teasdale R. D., Estivill X., Park W. J., Scott H. S. (2008) Hum. Mutat. 29,130–141 [DOI] [PubMed] [Google Scholar]
- 30.Guipponi M., Tan J., Cannon P. Z., Donley L., Crewther P., Clarke M., Wu Q., Shepherd R. K., Scott H. S. (2007) Am. J. Pathol. 171,608–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folgueras A. R., de Lara F. M., Pendás A. M., Garabaya C., Rodríguez F., Astudillo A., Bernal T., Cabanillas R., López-Otín C., Velasco G. (2008) Blood 112,2539–2545 [DOI] [PubMed] [Google Scholar]
- 32.Du X., She E., Gelbart T., Truksa J., Lee P., Xia Y., Khovananth K., Mudd S., Mann N., Moresco E. M., Beutler E., Beutler B. (2008) Science 320,1088–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finberg K. E., Heeney M. M., Campagna D. R., Aydinok Y., Pearson H. A., Hartman K. R., Mayo M. M., Samuel S. M., Strouse J. J., Markianos K., Andrews N. C., Fleming M. D. (2008) Nat. Genet. 40,569–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silvestri L., Pagani A., Nai A., De Domenico I., Kaplan J., Camaschella C. (2008) Cell Metab. 8,502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chokki M., Eguchi H., Hamamura I., Mitsuhashi H., Kamimura T. (2005) FEBS J. 272,6387–6399 [DOI] [PubMed] [Google Scholar]
- 36.Lin B., Ferguson C., White J. T., Wang S., Vessella R., True L. D., Hood L., Nelson P. S. (1999) Cancer Res. 59,4180–4184 [PubMed] [Google Scholar]
- 37.Kim T. S., Heinlein C., Hackman R. C., Nelson P. S. (2006) Mol. Cell. Biol. 26,965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donaldson S. H., Hirsh A., Li D. C., Holloway G., Chao J., Boucher R. C., Gabriel S. E. (2002) J. Biol. Chem. 277,8338–8345 [DOI] [PubMed] [Google Scholar]
- 39.Wilson S., Greer B., Hooper J., Zijlstra A., Walker B., Quigley J., Hawthorne S. (2005) Biochem. J. 388,967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alef T., Torres S., Hausser I., Metze D., Türsen U., Lestringant G. G., Hennies H. C. (2009) J. Invest. Dermatol. 129,862–869 [DOI] [PubMed] [Google Scholar]
- 41.List K., Currie B., Scharschmidt T. C., Szabo R., Shireman J., Molinolo A., Cravatt B. F., Segre J., Bugge T. H. (2007) J. Biol. Chem. 282,36714–36723 [DOI] [PubMed] [Google Scholar]
- 42.Basel-Vanagaite L., Attia R., Ishida-Yamamoto A., Rainshtein L., Ben Amitai D., Lurie R., Pasmanik-Chor M., Indelman M., Zvulunov A., Saban S., Magal N., Sprecher E., Shohat M. (2007) Am. J. Hum. Genet. 80,467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.List K., Szabo R., Wertz P. W., Segre J., Haudenschild C. C., Kim S. Y., Bugge T. H. (2003) J. Cell Biol. 163,901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Netzel-Arnett S., Currie B. M., Szabo R., Lin C. Y., Chen L. M., Chai K. X., Antalis T. M., Bugge T. H., List K. (2006) J. Biol. Chem. 281,32941–32945 [DOI] [PubMed] [Google Scholar]
- 45.Dhanasekaran S. M., Barrette T. R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2001) Nature 412,822–826 [DOI] [PubMed] [Google Scholar]
- 46.Wu Q., Parry G. (2007) Front. Biosci. 12,5052–5059 [DOI] [PubMed] [Google Scholar]
- 47.Klezovitch O., Chevillet J., Mirosevich J., Roberts R. L., Matusik R. J., Vasioukhin V. (2004) Cancer Cell 6,185–195 [DOI] [PubMed] [Google Scholar]
- 48.Miao J., Mu D., Ergel B., Singavarapu R., Duan Z., Powers S., Oliva E., Orsulic S. (2008) Int. J. Cancer 123,2041–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tripathi M., Nandana S., Yamashita H., Ganesan R., Kirchhofer D., Quaranta V. (2008) J. Biol. Chem. 283,30576–30584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomlins S. A., Rhodes D. R., Perner S., Dhanasekaran S. M., Mehra R., Sun X. W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J. E., Shah R. B., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2005) Science 310,644–648 [DOI] [PubMed] [Google Scholar]
- 51.Cerveira N., Ribeiro F. R., Peixoto A., Costa V., Henrique R., Jerónimo C., Teixeira M. R. (2006) Neoplasia 8,826–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nam R. K., Sugar L., Wang Z., Yang W., Kitching R., Klotz L. H., Venkateswaran V., Narod S. A., Seth A. (2007) Cancer Biol. Ther. 6,40–45 [DOI] [PubMed] [Google Scholar]
- 53.Nam R. K., Sugar L., Yang W., Srivastava S., Klotz L. H., Yang L. Y., Stanimirovic A., Encioiu E., Neill M., Loblaw D. A., Trachtenberg J., Narod S. A., Seth A. (2007) Br. J. Cancer 97,1690–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demichelis F., Fall K., Perner S., Andrén O., Schmidt F., Setlur S. R., Hoshida Y., Mosquera J. M., Pawitan Y., Lee C., Adami H. O., Mucci L. A., Kantoff P. W., Andersson S. O., Chinnaiyan A. M., Johansson J. E., Rubin M. A. (2007) Oncogene 26,4596–4599 [DOI] [PubMed] [Google Scholar]
- 55.List K., Bugge T. H., Szabo R. (2006) Mol. Med. 12,1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.List K., Szabo R., Molinolo A., Sriuranpong V., Redeye V., Murdock T., Burke B., Nielsen B. S., Gutkind J. S., Bugge T. H. (2005) Genes Dev. 19,1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uhland K. (2006) Cell. Mol. Life Sci. 63,2968–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bugge T. H., List K., Szabo R. (2007) Front. Biosci. 12,5060–5070 [DOI] [PubMed] [Google Scholar]
- 59.Suzuki M., Kobayashi H., Kanayama N., Saga Y., Suzuki M., Lin C. Y., Dickson R. B., Terao T. (2004) J. Biol. Chem. 279,14899–14908 [DOI] [PubMed] [Google Scholar]
- 60.Kilpatrick L. M., Harris R. L., Owen K. A., Bass R., Ghorayeb C., Bar-Or A., Ellis V. (2006) Blood 108,2616–2623 [DOI] [PubMed] [Google Scholar]
- 61.Lee S. L., Dickson R. B., Lin C. Y. (2000) J. Biol. Chem. 275,36720–36725 [DOI] [PubMed] [Google Scholar]