Abstract
FANCI is integral to the Fanconi anemia (FA) pathway of DNA damage repair. Upon the occurrence of DNA damage, FANCI becomes monoubiquitinated on Lys-523 and relocalizes to chromatin, where it functions with monoubiquitinated FANCD2 to facilitate DNA repair. We show that FANCI and its C-terminal fragment possess a DNA binding activity that prefers branched structures. We also demonstrate that FANCI can be ubiquitinated on Lys-523 by the UBE2T-FANCL pair in vitro. These findings should facilitate future efforts directed at elucidating molecular aspects of the FA pathway.
Fanconi anemia (FA)4 is characterized by developmental defects, bone marrow failure, and a strong predisposition to cancer. FA cells exhibit exquisite sensitivity to DNA cross-linking agents and marked genomic instability, indicative of a failure to repair damaged DNA (1–3). Thirteen FA proteins have been identified, of which eight, FANC-A, -B, -C, -E, -F, -G, -L, and -M, form part of a nuclear core complex that is required to monoubiquitinate two other FA proteins, FANCD2 and FANCI. When monoubiquitinated, FANCD2 and FANCI become chromatin-associated in foci that contain various factors, including the RAD51 recombinase BRCA2 (also known as FANCD1) and PALB2 (also called FANCN), which mediate DNA repair via RAD51-catalyzed homologous recombination (4).
Monoubiquitination of FANCD2 appears to be a key event for proper repair of exogenous DNA damage but also occurs during an unperturbed S phase, likely in response to stalled replication forks (4–7). FANCD2 monoubiquitination depends on the E3 ligase activity of FANCL (8) and on the E2 ubiquitin-conjugating enzyme, UBE2T (9). In vitro, FANCL and UBE2T can monoubiquitinate chicken FANCD2 (10).
FANCI was identified recently as a target protein for the ATM/ATR kinase. FANCI is also monoubiquitinated, in a manner that is dependent on the FA core complex (11). In cells, a fraction of FANCD2 and FANCI associates in a complex. Moreover, the amount and monoubiquitination of these two FA proteins are co-dependent in human cells, i.e. the quantity and monoubiquitination of FANCD2 are diminished in FANCI-deficient cells and vice versa (11–14). These observations suggest that FANCI and FANCD2 form a complex integral to cellular DNA repair capacity. Mutating the ubiquitinated target lysine of FANCI (Lys-523) renders cells sensitive to DNA damage and impairs the assembly of DNA damage-induced nuclear foci of FANCD2 and FANCI (11, 14). Herein, we document studies that reveal several biochemical attributes of FANCI, including DNA binding, and its monoubiquitination, that are relevant for understanding the biological role of this key FA protein.
EXPERIMENTAL PROCEDURES
Cloning for Protein Expression
Oligonucleotides and detailed procedures used for cloning are provided in supplemental Table 1 and supplemental Materials and Methods, respectively. The cDNA for human FANCI (isoform 1, KIAA1794) in the pCMV6-XL4 vector was subcloned into pFastBacHT(B) (Invitrogen) to add an N-terminal His6 epitope to the FANCI protein. The FANCI K523R mutation was generated by QuikChange mutagenesis. Human FANCL cDNA was first inserted into the pENTR/D-TEV-Topo vector followed by an LR Clonase reaction (Invitrogen) to transfer the FANCL DNA into pDest20, thereby adding an N-terminal GST epitope to the FANCL coding sequence. The FANCL C307A mutation was generated by QuikChange mutagenesis. Bacmids harboring the FA protein expression constructs were generated in DH10Bac cells (Invitrogen). Sf9 insect cells (Invitrogen) were transfected with bacmids to produce baculoviruses, as well as to amplify the baculoviral titer.
FANCI fragments (codons 985–1328 and 965–1328) and human UBE2T were inserted into pGEX-6P-1 (GE Healthcare) to generate FIΔN1, FIΔN2, and UBE2T with an N-terminal GST epitope. pGEX-6P-1-UBE2T was subject to QuikChange mutagenesis to introduce the UBE2T C86A mutation.
Protein Expression and Purification
Detailed expression and purification protocols are given in the supplemental Materials and Methods. All FA proteins and their mutant variants, except the GST-tagged FANCI fragments FIΔN1 and FIΔN2, were expressed in High Five insect cells (Invitrogen) with the appropriate baculovirus(es) for 36–50 h at 27 °C, whereupon cells were harvested by centrifugation. FIΔN1, FIΔN2, and UBE2T were produced in Escherichia coli (Rosetta DE3 pLysS), inducing with 0.1 mm isopropyl-1-thio-β-d-galactopyranoside (for 20–23 h to produce FIΔN1 and FIΔN2 and 6 h to produce UBE2T) at 16 °C. Insect and bacterial cell pellets were stored at −80 °C. For protein purification, cells were thawed, lysed by sonication, and cleared by high speed centrifugation.
FANCI and the K523R variant were purified from the clarified cell lysate with a combination of Q Sepharose fast flow (Amersham Biosciences), nickel-nitrilotriacetic acid agarose (Qiagen), and Mono Q steps. FANCL was purified with a combination of Q Sepharose fast flow, glutathione-Sepharose, and Mono Q steps.
For the purification of UBE2T, clarified bacterial cell lysate containing GST-tagged UBE2T was subject to affinity chromatography on glutathione-Sepharose followed by fractionation in a Source Q column. The GST tag was cleaved from the tagged UBE2T protein with PreScission protease (GE Healthcare) followed by removal of the free GST using glutathione-Sepharose. The UBE2T C86A mutant protein was purified with the same protocol as the wild type protein.
DNA Substrates
ϕX 174 replicative form I and viral (+) strand DNAs were purchased from Invitrogen. The replicative form I DNA was linearized with StuI to generate the linear form. The oligonucleotides used in the construction of DNA substrates are listed in supplemental Table 2 (15). The 32P-labeled DNA substrates were resolved from other reaction components by polyacrylamide gel electrophoresis, extracted from gel slices, and concentrated, as described previously (15).
DNA Binding
For FANCI binding to radiolabeled DNA, DNA substrates (0.15 or 0.3 pmol, as noted in the legends for Figs. 1–3) were mixed with the indicated amounts of protein in 12–15 μl of DNA binding buffer (15 mm HEPES, pH 7.5, 50 mm KCl, 1 mm MgCl2, 10% glycerol), incubated for 10 min at 37 °C, placed on ice, and mixed with 0.2 volumes of gel loading buffer (20 mm Tris, pH 7.5, 50% glycerol, Orange G). Samples were electrophoresed at 4 °C in 6, 10, or 12% non-denaturing Tris borate-EDTA-acrylamide gels, dried, and visualized by phosphorimaging. ImageQuant software (GE Healthcare) was used for quantification.
FIGURE 1.
FANCI purification and demonstration of its DNA binding activity. A, FANCI purification scheme. Nickel-NTA, nickel-nitrilotriacetic acid. B, Coomassie Blue-stained gel of FANCI and the FANCI K523R mutant. M, molecular mass markers. C, FANCI was tested for binding to pairs of DNA substrates. The percentage of the DNA probes shifted by FANCI is quantified and shown in the right panels. D, H3 was tested in competition with dsDNA H3/H4 (H3 annealed to its complement H4) for FANCI binding. The right panel shows the quantification of the results. In C and D, 0.3 pmol of each DNA substrate was used. Note that treatment of nucleoprotein complexes with SDS and proteinase K (SDS+PK) released the DNA substrates. Error bars indicate S.E.
FIGURE 2.
FANCI prefers branched DNA structures. A, the DNA substrates used. B and C, the indicated combinations of DNA substrates (0.3 pmol each) were incubated with FANCI. Quantifications of the phosphorimaging data are presented in the right panels. Note that treatment of nucleoprotein complexes with SDS and proteinase K (SDS+PK) released the DNA substrates. Error bars indicate S.E.
FIGURE 3.
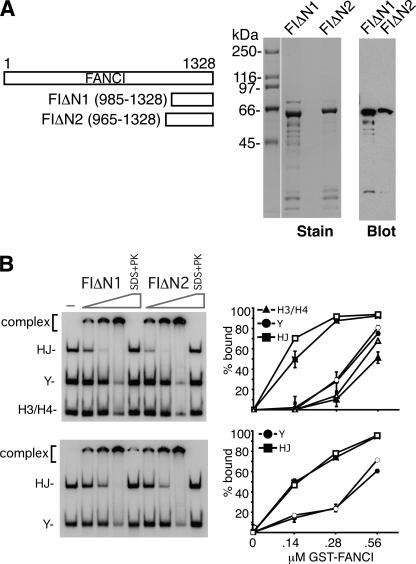
The FANCI C terminus binds branched DNA. A, purified GST-tagged fragments of the FANCI C terminus (FIΔN1 and FIΔN2, as indicated in the schematic) were analyzed by SDS-PAGE with Coomassie Blue staining (Stain) or subject to immunoblotting with anti-GST antibodies (Blot). B, FIΔN1 and FIΔN2 were incubated with the combination of the HJ, Y, and H3/H4 or of the HJ and Y (0.15 pmol each) and then analyzed as in Fig. 1. The right panels show the quantification of results. Filled and open symbols represent substrate binding by FIΔN1 and FIΔN2, respectively. Note that treatment of nucleoprotein complexes with SDS and proteinase K (SDS+PK) released the DNA substrates. Error bars indicate S.E.
Ubiquitination Assays
HA epitope-tagged human ubiquitin and human UBE1 were purchased from Boston Biochem (Cambridge, MA). Ubiquitination reactions were performed in 25 μl and contained 10 nm UBE1, 100–600 nm UBE2T, 0.5–1 μm GST-FANCL, 34 μm HA-Ub, 2 mm ATP, and 0.3–0.6 μm FANCI in buffer containing 50 mm Tris-HCl, pH 7.5, 50–150 mm KCl, 2 mm MgCl2, and 0.5 mm dithiothreitol. Reactions were incubated for 90 min at 25 °C, and following the addition of gel loading buffer, they were subjected to electrophoresis in a 7.5 or 10% denaturing polyacrylamide gel. Proteins were transferred to nitrocellulose, which was probed with anti-HA-horseradish peroxidase (clone 3F10, Roche Applied Science) to detect ubiquitinated proteins.
RESULTS
Purification of FANCI and FANCI K532R
Human FANCI tagged with an N-terminal His6 epitope was expressed in insect cells and purified to near homogeneity with a multistep procedure that we have devised (Fig. 1, A and B). The same expression and purification protocols were suited to obtaining the His6-tagged FANCI K523R mutant (Fig. 1B). The FANCI K523R mutant protein behaves like the wild type protein during purification and in DNA binding assays (see below; data not shown).
FANCI Binds Single-stranded DNA and Double-stranded DNA
FANCI becomes chromatin-bound upon DNA damage (11, 13), and FANCD2 possesses a DNA binding activity (16), so we surmised that FANCI may bind DNA. Consistent with this premise, FANCI clearly bound plasmid-length ssDNA and dsDNA in a DNA mobility shift experiment (supplemental Fig. 1). We further characterized the DNA binding attributes of FANCI with radiolabeled, oligonucleotide-based DNA substrates. Initially, we incubated FANCI with pairs of 32P-labeled DNA substrates, including 83-mer oligo(dT), with either the 80-mer H3 or the 80-mer P1. The results of these experiments revealed that FANCI binds all three species of DNA but possesses a higher affinity for H3 or P1 than oligo(dT) (Fig. 1C). As shown in Fig. 1D, FANCI binds the duplex obtained by hybridizing H3 to its exact complement H4 (H3/H4) with a higher affinity than H3. Thus, FANCI binds both ssDNA and dsDNA but with an apparent preference for the latter.
Preference of FANCI for Branched DNA Structures
Oligonucleotides H3 and P1, which invariably harbor a certain degree of secondary structure, are preferred over oligo(dT) by FANCI (Fig. 1C), suggesting to us that FANCI may have specificity for DNA structures. Additional DNA binding experiments were performed using radiolabeled substrates with a defined DNA structure, namely, a Holliday junction (HJ), a Y structure (Y), and also duplexes with either a 3′ single-stranded overhang (3′-OH) or a 5′ single-stranded overhang (5′-OH) (Fig. 2A). We co-incubated the combination of HJ, Y, and duplex or HJ, Y, and 3′-OH with FANCI. The results, shown in Fig. 2B, revealed a distinct preference of FANCI for the HJ followed by the Y substrate, and lastly, for the duplex or 3′-OH DNA. That FANCI prefers to bind HJ over Y was further confirmed by co-incubating just these two substrates with FANCI (Fig. 2C). We found FANCI to have a slightly higher affinity for the duplex over either 3′-OH or 5′-OH (see supplemental Fig. 2).
The C-terminal Region of FANCI Harbors a DNA Binding Activity
We wished to identify the DNA binding domain in FANCI (1,328 amino acid residues in size) by biochemical mapping. Although we were able to express protein fragments that encompass different portions of FANCI in either E. coli or insect cells, the vast majority of them were, unfortunately, insoluble (data not shown), with the exception of the FANCI fragments harboring residues 985–1328 and 965–1328, referred to as FIΔN1 and FIΔN2, respectively (Fig. 3A), that proved to be soluble when expressed in E. coli. These FANCI fragments were purified to near homogeneity from E. coli extracts and examined for DNA binding with the HJ, Y structure, H3, and H3/H4 duplex as substrates. Importantly, both FANCI fragments possess DNA binding activity with a clear preference for the HJ substrate (Fig. 3B). Thus, the FANCI C terminus encompassing amino acids 985–1328 harbors a DNA binding activity similar in substrate preference to that of full-length FANCI.
Monoubiquitination of FANCI by the UBE2T-FANCL Pair
FANCL, indispensable for FANCD2 monoubiquitination in vivo (8, 17), interacts with two E2 enzymes, UBE2T and UBE2W (9, 10), the former of which is required for FANCD2 monoubiquitination in vivo (9). To test whether FANCI can be ubiquitinated by the FANCL-UBE2T pair, we purified from insect cells infected with a recombinant baculovirus human FANCL or the FANCL C307A mutant that is incapable of UBE2T interaction (9). Human UBE2T and a catalytically defective C86A variant (9) were expressed in E. coli and purified to near homogeneity (Fig. 4A). Purified FANCL and UBE2T are active for ubiquitin conjugation, as deduced from the UBE2T-dependent autoubiquitination of FANCL in the presence of the E1-conjugating enzyme, ubiquitin, and ATP (Fig. 4B, lane 2). As expected, no autoubiquitination of FANCL occurred with the FANCL C307A mutant (supplemental Fig. 3, lane 4) or the UBE2T C86A mutant (Fig. 4B, lane 3).
FIGURE 4.
Monoubiquitination of FANCI by FANCL and UBE2T. A, purified GST-FANCL, GST-FANCL C307A, UBE2T, and UBE2T C86A were analyzed by SDS-PAGE with Coomassie Blue staining. M, molecular mass markers. B, Western blot probed with anti-HA showing that autoubiquitination of GST-FANCL (Ub-FANCL) occurred upon incubation with UBE2T, ATP, and HA-ubiquitin (HA-Ub). Incubation of FANCI with FANCL, UBE2T, ATP, and HA-Ub resulted in FANCL autoubiquitination and an additional doublet of monoubiquitinated FANCI species (Ub-FANCI). The FANCI K523R substrate (KR) was ubiquitinated much less efficiently than FANCI and gave only the faster migrating monoubiquitinated species. No autoubiquitination of FANCL or monoubiquitination of FANCI was seen upon substituting UBE2T with the catalytically inactive C86A variant. The ubiquitin-activating enzyme UBE1 was present in all the reactions. WT, wild type.
Importantly, FANCI became monoubiquitinated when UBE2T and FANCL were present, producing a doublet of modified species migrating ∼10 kDa above the position of unmodified FANCI (Fig. 4B, lane 5). As expected, FANCI ubiquitination in this system is dependent on ATP and FANCL, and neither UBE2T C86A (Fig. 4B, lanes 3, 6, and 11) nor FANCL C307A (supplemental Fig. 3, lane 8) was active. In vivo, lysine 523 of FANCI is the ubiquitin acceptor site (11, 13, 14). In vitro, the level of monoubiquitination becomes 3–4-fold lower with the FANCI K532R mutant (Fig. 4B, lane 10), and importantly, instead of a doublet of monoubiquitinated products (Fig. 4B, lane 5), a single monoubiquitinated species is seen (Fig. 4B, compare lanes 5 and 10). Thus, the UBE2T-FANCL-mediated monoubiquitination of FANCI in vitro appears to occur largely with the same site specificity as FANCI ubiquitination in vivo.
DISCUSSION
We have found a DNA binding activity in FANCI that has preference for branched structures and have provided evidence that the C-terminal region of FANCI (amino acids 985–1328) is sufficient for DNA binding. Notably, several disease-causing mutations of FANCI map to the C-terminal region within the DNA binding domain that we have identified (11, 13, 18). FANCI forms a dimeric complex (the ID complex) with FANCD2 (11), which also binds DNA (16). Upon DNA damage, FANCD2 and FANCI are dependent upon one another for relocalization to chromatin at DNA repair foci (11, 14). As such, FANCI and FANCD2 are likely to perform their DNA repair functions together and in a manner that requires the DNA binding activity of the ID complex.
Current evidence suggests that the ID complex plays an early role in DNA repair, prior to homologous recombination or translesion synthesis (19, 20). The branched DNA structures that FANCI preferentially binds in vitro resemble intermediates that arise when a replication fork encounters a blocking lesion (20, 21). The ID complex could be involved in the recognition of blocked forks and/or blocked forks that have been processed into four-branched structures resembling HJ recombination intermediates. Once engaged at these structures, the ID complex may help to mediate the DNA repair process by recruiting factors, such as homologous recombination proteins (e.g. BRCA2, PALB2, and RAD51) and/or components of translesion DNA synthesis machinery (e.g. REV1), that perform repair or replicative bypass. Interestingly, both FANCI and FANCD2 have been shown to localize near BLM helicase foci at mitotic anaphase bridges. Anaphase bridges of interwound DNA between sister chromatids are thought to indicate unreplicated DNA or aberrant replication intermediates that persist into anaphase (22, 23). The FA core complex is required for BLM recruitment to these foci (23). BLM functions with topoisomerase IIIα and other partner proteins to process branched DNA structures, including the double Holliday junction, into non-crossover products (24). Our results are consistent with the idea that ID complex binding to branched DNA structures, such as blocked replication forks, aids in BLM recruitment to process these structures.
In human cells, retention of FANCD2 and FANCI in DNA repair foci requires their monoubiquitination (11, 25), implicating a chromatin-bound ubiquitin receptor that serves to anchor the ID complex in chromatin. Monoubiquitination of the ID complex may also enhance its ability to mediate the recruitment of DNA repair proteins to the damage site (26). Consistent with genetic data implicating the FANCL-containing FA core complex in FANCI monoubiquitination (11) and the recent demonstration of in vitro UBE2T-FANCL-catalyzed FANCD2 monoubiquitination (10), we have found that FANCI serves as a substrate for the UBE2T-FANCL pair. Furthermore, using the FANCI K523R mutant, ablated for the known cellular ubiquitin acceptor site, we have furnished evidence that monoubiquitination in vitro occurs mostly on Lys-523 in FANCI. Establishing this minimal in vitro ubiquitination system sets the stage for asking how the other FA core complex components and protein phosphorylation (see below) affect the efficiency of the ubiquitination reaction.
We note that in human cells, the FANCI K523R mutant is defective in DNA damage focus formation, and in addition, has been reported to assert a dominant negative effect on FANCD2 monoubiquitination and DNA damage-induced FANCD2 focus assembly (11). However, in chicken DT40 cells genetically ablated for endogenous FANCI, expression of the K525R mutant (equivalent to the human FANCI K523R mutant) lacking the ubiquitin acceptor site can significantly restore cellular resistance to the DNA cross-linking agent mitomycin C and some FANCD2 chromatin recruitment (14), suggesting that FANCI monoubiquitination plays a subsidiary role in FA pathway activation in these chicken cells. Additional studies are clearly needed to determine the relative importance of FANCI monoubiquitination in the activation of the FA DNA repair pathway in mammalian versus non-mammalian cells.
Both FANCD2 and FANCI are subject to phosphorylation by the DNA damage checkpoint kinase ATR. Mutation of the ATR phosphorylation sites Thr-691 and Ser-717 in FANCD2 compromises its monoubiquitination and sensitizes cells to DNA cross-linking agents (27). Interestingly, the phosphorylation status of FANCI is decisive for FANCD2 monoubiquitination and also DNA damage resistance in the chicken DT40 cell line (14). In DT40 cells, phosphomimic mutations of a conserved cluster of ATR phosphorylation consensus (S/T)Q motifs surrounding the Lys-525 monoubiquitination acceptor site of FANCI act as a “switch” for constitutive FANCD2 monoubiquitination and chromatin localization of FANCI and FANCD2. Nevertheless, the FANCI phosphorylation switch is not sufficient to promote colocalization of FANCD2/FANCI with other DNA repair factors, e.g. RAD51 (14).
The discovery and characterization of a deubiquitinating enzyme, USP1, specific for FANCD2, has revealed the additional importance of FANCD2 deubiquitination for DNA damage repair (28). USP1-deficient cells exhibit an elevated sensitivity to DNA cross-linking agents (29, 30), suggesting that deubiquitination of FANCD2 is necessary to complete DNA repair and/or to relocalize FANCD2 to other DNA damage sites to initiate new repair events. That USP1 is similarly involved in the deubiquitination and regulation of FANCI seems likely. The establishment of in vitro FANCD2 and FANCI ubiquitination systems (Ref. 10 and this study) should facilitate future efforts directed at defining the mechanism and consequence of the USP1-mediated modification of monoubiquitinated FANCD2 and FANCI and for addressing which post-translational modifications of FA proteins are relevant for regulation of different aspects of FA pathway functions.
Supplementary Material
Acknowledgments
We are grateful to Tony Huang (New York University) and Ruhikanta Meetei (Cincinnati Children's Hospital) for materials.
This work was supported, in whole or in part, by National Institutes of Health Grants PO1 CA92584, PO1 CA129186, and RO1 ES015252.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” supplemental Tables 1 and 2, and supplemental Figs. 1–3.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” supplemental Tables 1 and 2, and supplemental Figs. 1–3.
- FA
- Fanconi anemia
- ATM
- ataxia telangiectasia-mutated
- ATR
- ATM and Rad3-related
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- ID
- FANCI-FANCD2
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin-protein isopeptide ligase
- Ub
- ubiquitin
- HJ
- Holliday junction
- Y
- Y structure.
REFERENCES
- 1.Poll E. H., Arwert F., Joenje H., Wanamarta A. H. (1985) Hum. Genet. 71,206–210 [DOI] [PubMed] [Google Scholar]
- 2.German J., Schonberg S., Caskie S., Warburton D., Falk C., Ray J. H. (1987) Blood 69,1637–1641 [PubMed] [Google Scholar]
- 3.Auerbach A. D. (1988) Blood 72,366–367 [PubMed] [Google Scholar]
- 4.Wang W. (2007) Nat. Rev. Genet. 8,735–748 [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi T., Garcia-Higuera I., Andreassen P. R., Gregory R. C., Grompe M., D'Andrea A. D. (2002) Blood 100,2414–2420 [DOI] [PubMed] [Google Scholar]
- 6.Rothfuss A., Grompe M. (2004) Mol. Cell. Biol. 24,123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobeck A., Stone S., Costanzo V., de Graaf B., Reuter T., de Winter J., Wallisch M., Akkari Y., Olson S., Wang W., Joenje H., Christian J. L., Lupardus P. J., Cimprich K. A., Gautier J., Hoatlin M. E. (2006) Mol. Cell. Biol. 26,425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meetei A. R., de Winter J. P., Medhurst A. L., Wallisch M., Waisfisz Q., van de Vrugt H. J., Oostra A. B., Yan Z., Ling C., Bishop C. E., Hoatlin M. E., Joenje H., Wang W. (2003) Nat. Genet. 35,165–170 [DOI] [PubMed] [Google Scholar]
- 9.Machida Y. J., Machida Y., Chen Y., Gurtan A. M., Kupfer G. M., D'Andrea A. D., Dutta A. (2006) Mol. Cell 23,589–596 [DOI] [PubMed] [Google Scholar]
- 10.Alpi A. F., Pace P. E., Babu M. M., Patel K. J. (2008) Mol. Cell 32,767–777 [DOI] [PubMed] [Google Scholar]
- 11.Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E. R., 3rd, Hurov K. E., Luo J., Ballif B. A., Gygi S. P., Hofmann K., D'Andrea A. D., Elledge S. J. (2007) Cell 129,289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levitus M., Rooimans M. A., Steltenpool J., Cool N. F., Oostra A. B., Mathew C. G., Hoatlin M. E., Waisfisz Q., Arwert F., de Winter J. P., Joenje H. (2004) Blood 103,2498–2503 [DOI] [PubMed] [Google Scholar]
- 13.Sims A. E., Spiteri E., Sims R. J., 3rd, Arita A. G., Lach F. P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., Auerbach A. D., Huang T. T. (2007) Nat. Struct. Mol. Biol. 14,564–567 [DOI] [PubMed] [Google Scholar]
- 14.Ishiai M., Kitao H., Smogorzewska A., Tomida J., Kinomura A., Uchida E., Saberi A., Kinoshita E., Kinoshita-Kikuta E., Koike T., Tashiro S., Elledge S. J., Takata M. (2008) Nat. Struct. Mol. Biol. 15,1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Komen S., Reddy M. S., Krejci L., Klein H., Sung P. (2003) J. Biol. Chem. 278,44331–44337 [DOI] [PubMed] [Google Scholar]
- 16.Park W. H., Margossian S., Horwitz A. A., Simons A. M., D'Andrea A. D., Parvin J. D. (2005) J. Biol. Chem. 280,23593–23598 [DOI] [PubMed] [Google Scholar]
- 17.Meetei A. R., Yan Z., Wang W. (2004) Cell Cycle 3,179–181 [PubMed] [Google Scholar]
- 18.Dorsman J. C., Levitus M., Rockx D., Rooimans M. A., Oostra A. B., Haitjema A., Bakker S. T., Steltenpool J., Schuler D., Mohan S., Schindler D., Arwert F., Pals G., Mathew C. G., Waisfisz Q., de Winter J. P., Joenje H. (2007) Cell Oncol 29,211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurtan A. M., D'Andrea A. D. (2006) DNA Repair 5,1119–1125 [DOI] [PubMed] [Google Scholar]
- 20.Niedernhofer L. J., Lalai A. S., Hoeijmakers J. H. (2005) Cell 123,1191–1198 [DOI] [PubMed] [Google Scholar]
- 21.Räschle M., Knipscheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Schärer O. D., Walter J. C. (2008) Cell 134,969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan K. L., Palmai-Pallag T., Ying S., Hickson I. D. (2009) Nat. Cell Biol. 11,753–760 [DOI] [PubMed] [Google Scholar]
- 23.Naim V., Rosselli F. (2009) Nat. Cell Biol. 11,761–768 [DOI] [PubMed] [Google Scholar]
- 24.Raynard S., Bussen W., Sung P. (2006) J. Biol. Chem. 281,13861–13864 [DOI] [PubMed] [Google Scholar]
- 25.Wang W. (2008) Nat. Struct. Mol. Biol. 15,1128–1130 [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Andreassen P. R., D'Andrea A. D. (2004) Mol. Cell. Biol. 24,5850–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho G. P., Margossian S., Taniguchi T., D'Andrea A. D. (2006) Mol. Cell. Biol. 26,7005–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nijman S. M., Huang T. T., Dirac A. M., Brummelkamp T. R., Kerkhoven R. M., D'Andrea A. D., Bernards R. (2005) Mol. Cell 17,331–339 [DOI] [PubMed] [Google Scholar]
- 29.Oestergaard V. H., Langevin F., Kuiken H. J., Pace P., Niedzwiedz W., Simpson L. J., Ohzeki M., Takata M., Sale J. E., Patel K. J. (2007) Mol. Cell 28,798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J. M., Parmar K., Huang M., Weinstock D. M., Ruit C. A., Kutok J. L., D'Andrea A. D. (2009) Dev. Cell 16,314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.