Abstract
Upon virus infection, the cell mounts an innate type I interferon (IFN) response to limit the spread. This response is orchestrated by the constitutively expressed IFN regulatory factor (IRF)-3 protein, which becomes post-translationally activated. Although the activation events are understood in detail, the negative regulation of this innate response is less well understood. Many viruses, including Kaposi sarcoma-associated herpesvirus (KSHV), have evolved defense strategies against this IFN response. Thus, KSHV encodes a viral IRF (vIRF)-2 protein, sharing homology with cellular IRFs and is a known inhibitor of the innate IFN response. Here, we show that vIRF-2 mediates IRF-3 inactivation by a mechanism involving caspase-3, although vIRF-2 itself is not pro-apoptotic. Importantly, we also show that caspase-3 participates in normal IRF-3 turnover in the absence of vIRF-2, during the antiviral response induced by poly(I:C) transfection. These data provide unprecedented insight into negative regulation of IRF-3 following activation of the type I IFN antiviral response and the mechanism by which KSHV vIRF-2 inhibits this innate response.
The earliest response at the cellular level to virus infection is the establishment of the antiviral state that results from induction of type I interferon (IFN)2 expression. The objective of this antiviral state is containment of the virus infection and elimination of the infected cell. It operates in multiple ways, including inhibiting cell growth by blocking proliferation and modulating apoptosis, and augmenting adaptive immunological surveillance and responses (see Ref. 1). Sensing of the virus infection to initiate the antiviral response occurs in different ways, in part depending upon whether the virus enters the cell by endocytosis or by fusion with the plasma membrane. One of the most important components transducing these virus-sensing signals is IFN regulatory factor (IRF)-3. It participates in transcribing genes that contribute to establishing the antiviral state.
Upon virus infection, IRF-3 is post-translationally modified by C-terminal phosphorylation by a “virus-activated kinase” (2, 3) that promotes translocation of the protein from the cytoplasm to the nucleus. There, it is assimilated into the IFN-β enhancesome, a multiprotein complex that facilitates transcription of IFN and IFN-responsive genes. This enhancesome, whose structure is well characterized (4, 5), represents the paradigm for understanding the molecular basis behind regulation of gene transactivation in response to virus infection. The components of virus-activated kinase that phosphorylate IRF-3 include IκB kinase-ϵ and TANK-binding kinase-1 (6, 7). Depending on the pathway leading to IRF-3 activation, other kinases may also participate, including phosphatidylinositol 3-kinase (8).
Although post-translational activation of IRF-3 is understood in detail, less is known of its deactivation that negatively regulates the type I IFN response. Until now, only phosphorylation-dependent ubiquitination of IRF-3, leading to its proteasomal degradation has been recognized. Hiscott and colleagues showed that C-terminal phosphorylation of IRF-3 is necessary for degradation and is followed by Cullin1 interaction, ubiquitination, and proteasomal degradation (9; see Ref. 10). Poly-ubiquitination and concomitant degradation of IRF-3 are regulated by the peptidylprolyl isomerase Pin-1 (11).
Inhibiting the IFN antiviral response is an important component of the biology of many viruses (1). Studying the molecular interactions of viruses with the immune system, including their strategies of evasion, has provided deeper understanding of its operation. Recent examples set a precedent in the context of the innate immune system. First, the study of the human immunodeficiency virus Vif protein identified a new innate immune response to retroviruses (12) mediated by the cellular protein, CEM15 or APOBEC3G, a DNA deaminase, which destroys or mutates the virus genome (13). Second, the recognition through its binding to paramyxovirus V proteins that mda-5 is a central player in the signal transduction cascade leading to IFN-β expression (14).
Through studying modulation of the IFN response by Kaposi sarcoma-associated herpesvirus (KSHV), we now demonstrate a unique cellular mechanism inhibiting IRF-3 function by a caspase-3-dependent process. KSHV is the etiologic agent of the most common malignancy affecting AIDS patients, Kaposi sarcoma (KS), which is also the most common tumor of men in certain African countries (15). Approximately one quarter of the KSHV genome encodes proteins with either demonstrated or putative immunomodulatory activity (16), and one of these viral genes encodes the vIRF-2 protein (17), which inhibits the type I IFN response to viral infection (18, 19, 20). Here, using a model system of activation of the antiviral response by transfection of synthetic double-stranded RNA, we have identified a novel caspase-3-dependent turnover of cellular IRF-3 that is involved in the normal negative feedback loop to help terminate the antiviral response. This caspase-3-dependent mechanism is targeted by vIRF-2 to accelerate cellular IRF-3 turnover and thereby down-modulate the antiviral response.
EXPERIMENTAL PROCEDURES
Reagents
Poly(I:C), MG132, and etoposide were obtained from Sigma-Aldrich, and Z-VAD-FMK was from Calbiochem. SYTO® 16 green-fluorescent nucleic acid stain was a generous gift of Gemma Kelly.
Cell Culture
HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with non-essential amino acids, penicillin, and streptomycin and 10% heat-inactivated fetal bovine serum. MCF-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with penicillin and streptomycin and 10% fetal bovine serum.
Plasmid Constructs and Site-directed Mutagenesis
Full-length KSHV vIRF-2 cDNA was subcloned into the pcDNA4/HisMax expression vector as previously described (20). The resulting vIRF-2 protein contains contiguous N-terminal polyhistidine and XpressTM epitope tags and is referred to as Xpress-tagged vIRF-2. The caspase-3 (pcDNA3/caspase-3) and FLAG epitope-tagged IRF-3 wild-type (IRF-3WT) (21) expression plasmids were kindly provided by Reiner U. Jänicke and John Hiscott, respectively. The p125-luc luciferase plasmid contains the full-length IFN-β promoter upstream of the firefly luciferase gene and was generously provided by Takashi Fujita (22). Firefly luciferase levels were normalized against Renilla luciferase constitutively expressed from the co-transfected plasmid pRLSV40 (Stratagene).
To mutate the serine and threonine residues in the C-terminal region of IRF-3 to alanine, site-directed mutagenesis was performed with the QuikChangeTM system (Stratagene), according to the manufacturer's instructions. Thus, to generate the 2A(S385/386A), 5A(ST396/398/402/404/405AA), and 2A5A constructs, PCR was performed with the following primers: 2A(S385/386A), 5′-GGGTGCCGCCGCCCTGGAG-3′ and 5′-CTCCAGGGCGGCGGCACCC-3′; 5A(ST396/398/402/404/405AA), 5′-CATTGCCAACGCCCACCCACTCGCCCTCGCCGCCGACC-3′ and 5′-GGTCGGCGGCGAGGGCGAGTGGGTGGGCGTTGGCAATG-3′. The 2A5A mutant was derived with the 2A PCR primers on the 5A mutant. All constructs were verified by sequencing.
Transient Transfections
Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions on 75% confluent HEK293 or MCF-7 cells grown in 6-well plates. Plasmid quantities were: FLAG-tagged IRF-3WT or mutant expression plasmids (300 ng/well), pcDNA3/caspase-3 expression plasmid (300 ng/well), pcDNA4/vIRF-2 expression plasmid (500 ng/well), p125-luc (100 ng/well), and pRLSV40 (5 ng/well). The corresponding empty vectors were added to equalize DNA concentration (see Ref. 20).
Cell Treatments
MG132 (10 μm) or Z-VAD-FMK (10 μm) treatment (30 min, 37 °C) was performed 24 h following plasmid transfection. Negative controls were treated with the equivalent volume of DMSO. Next, without changing the medium, the cells were transfected with poly(I:C) (10 μg/ml). Caspase-3 knockdown was performed with caspase-3 ShortCut® siRNA (25 nm, New England Biolabs); the corresponding negative control was the nonspecific Lit28i Polylinker ShortCut® siRNA mix (25 nm, New England Biolabs). Plasmid transfection was performed at the same time, as required. Twenty-four hours later, transfection with poly(I:C) (10 μg/ml) was performed as necessary.
Western Blot Analysis
Cell pellets were suspended in lysis buffer E (100 mm Tris-HCl, pH 8, 100 mm NaCl, 2 mm EDTA, 2 mm EGTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.5 mm phenylmethylsulfonyl fluoride, mini protease inhibitor mixture (Roche Diagnostic)). After incubation (10 min, 4 °C), insoluble material was removed by centrifugation (16,000 × g, 5 min, 4 °C). The protein concentration was determined by the Bradford dye-binding procedure (Bio-Rad Laboratories), and 35 μg of total protein was separated by SDS-PAGE on 12% acrylamide gels. Proteins were transferred to Immobilon-P transfer membrane (Millipore) that was then blocked in Tris-buffered saline containing 0.02% Tween-20 (TBS-T) and nonfat milk (5%) and probed with primary antibodies diluted in TBS-T containing 5% nonfat milk as described below. Bands were visualized by probing with appropriate secondary antibodies conjugated to horseradish peroxidase (DAKO) and performing enhanced chemiluminescence. Native PAGE was performed essentially according to the protocol of a previous study (23).
Antibodies for Western Blot and Immunoprecipitation
Primary antibodies were used at the following dilutions and incubation conditions: anti-Pro-caspase-3 polyclonal antibody (Cell Signaling Technologies) diluted 1:5,000 and incubated for 18 h at 4 °C; anti-poly(ADP-ribose) polymerase monoclonal antibody (Sigma-Aldrich) diluted 1:5,000 and incubated for 18 h at 4 °C; anti-phospho-IRF-3 (Ser396) polyclonal antibody (Upstate) diluted 1:5,000 and incubated for 18 h at 4 °C; anti-FLAG® M2 monoclonal antibody (Sigma-Aldrich) diluted 1:50,000 and incubated for 1 h at 4 °C; anti-XpressTM monoclonal antibody (Invitrogen) diluted 1:5,000 and incubated for 1 h at 4 °C; anti-β-actin monoclonal antibody (Sigma-Aldrich) diluted 1:50,000 and incubated for 1 h at 4 °C; anti-IRF-3 monoclonal antibody (Abcam) diluted 1:100 and incubated for 2 h at 4 °C; and for denaturing gels IRF-3 was detected with anti-IRF-3(FL-425) polyclonal antibody (Santa Cruz Biotechnology) diluted 1:1000 and incubated 1 h at 4 °C.
Co-immunoprecipitation
Cell lysates were prepared in buffer E, as described above. They were pre-cleared by incubation with protein G-Sepharose 4B fast flow (Sigma-Aldrich) or protein A-Sepharose 4B fast flow (Sigma-Aldrich), and supernatants were collected by centrifugation (16,000 × g, 1 min, 4 °C). They were then incubated with either 1 μg of anti-Xpress (90 min) or 1 μl of anti-procaspase-3 antibodies (16 h) at 4 °C. Immunoprecipitation was performed with either protein G- or protein A-Sepharose beads, and proteins were analyzed by Western blot. Negative control immunoprecipitations were performed on cells transfected with the plasmid pCEP4-SM (1 μg) expressing the FLAG-tagged Epstein-Barr virus protein BSLF2/BMLF, a generous gift of Martin Rowe.
Transient Transfections and Luciferase Assays in HEK293 and MCF-7 Cells
Dual luciferase assays were performed according to a previous study (20).
Quantification of Apoptosis
Apoptosis was quantified by flow cytometry on cells stained with Syto-16 and propidium iodide according to the method of a previous study (24). Data for ∼25,000 cells were collected for each treatment condition.
Caspase Activity Assay
Caspase activity was measured with the Caspase-Glo®-3/7 assay kit (Promega) that makes use of the luminogenic caspase-3/-7 substrate Z-DEVD-aminoluciferin. Measurements were performed according to the manufacturer's instructions.
Confocal Microscopy
MCF-7 cells were plated on coverslips in 24-well plates. The next day, they were transfected with expression vectors for FLAG-tagged IRF-3 wild-type (IRF-3WT), or Xpress-tagged vIRF-2 (vIRF-2) or caspase-3, as required. The vIRF-2 protein is dually tagged with both Xpress and His epitopes. Twenty-four hours later, cells were transfected with poly(I:C) (10 μg/ml) for 16 h, as required. The cells were fixed with a final concentration of 2% of formaldehyde solution (methanol-free, Pierce) added directly to the medium for 20 min at room temperature. After fixation, the cells were washed three times (PBS, 15 min, room temperature) and were then permeabilized (2% bovine serum albumin, 0.5% Triton X-100 in PBS, 10 min, at room temperature) and blocked (blocking buffer: 2% bovine serum albumin, 0.1% Triton X-100 in PBS, 60 min at room temperature). Primary antibodies were mouse anti-His (Sigma-Aldrich, 1/100) to detect vIRF-2, goat anti-IRF-3 (RandD, 10 μg/ml), and rabbit anti-caspase-3 (AbCam) diluted 1/50. All primary antibody incubations were performed in blocking buffer overnight at 4 °C. After primary antibody incubation, cells were washed (PBS, four times for 15 min, at room temperature) and incubated with appropriate secondary antibodies: anti-mouse AlexaFluor®-555 (Invitrogen, 1/500 dilution, kindly provided by Dr. Claire Shannon-Lowe), anti-goat AlexaFluor®-633 (Invitrogen, diluted 1/100), anti-rabbit-fluorescein isothiocyanate (Sigma-Aldrich, at 1/100 dilution). All secondary antibody incubations were performed in blocking buffer for 1 h at room temperature. Cells were then washed (PBS, four times for 15 min, at room temperature) and incubated with 4′,6-diamidino-2-phenylindole (1/1000 in PBS, 5 min, at room temperature). The coverslips were mounted with ProLong® Gold Antifade reagent (Molecular Probes, Invitrogen).
Establishment of the Clone #3–9 Cell Line Stably Transfected with an Inducible vIRF-2 Expression Cassette
The vIRF-2 gene was subcloned in-frame into the doxycycline-inducible expression vector pTRE2-pur-Myc (Clontech), creating a 5′-Myc-labeled vIRF-2 derivative. This plasmid was transfected into HEK293-Tet-On cells (Clontech) that stably express the tetracycline-regulated transactivator rtTA. From these cells, a clonal puromycin-resistant cell line was derived with minimal basal vIRF-2 expression; it is referred to as “clone #3–9.” A negative control cell line (“Empty Vector” (EV)), isogenic to clone 3–9 cells except that it lacked vIRF-2, was established in the same way.
Measuring Endogenous IRF-3 Activation
Active endogenous IRF-3 was measured by TransAMTM IRF-3 transcription factor ELISA (Active Motif) that was performed according to the manufacturer's directions. Nuclear extracts for this assay were prepared with the Nuclear Extract Kit (Active Motif).
RESULTS
KSHV vIRF-2 Represses IFN-β Promoter Transactivation and Reduces Wild-type IRF-3 Protein Levels
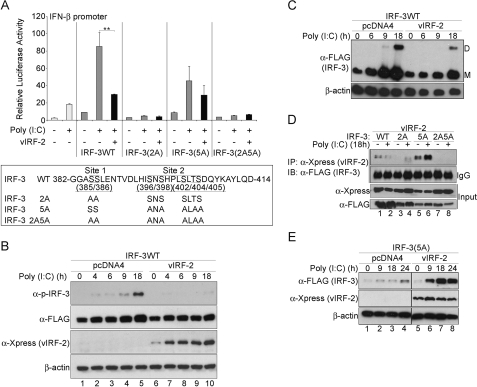
Ectopic expression studies of vIRF-2 were performed to investigate the function of the vIRF-2 protein without confounding its activity by the presence of other KSHV immunomodulatory proteins. Thus, the cDNA encoding the spliced vIRF-2 gene was cloned into the pcDNA4/HisMax vector (Invitrogen) to generate pcDNA/vIRF-2 expressing the full-length vIRF-2 protein containing contiguous N-terminal polyhistidine and Xpress epitope tags (20). When measuring the activity of the full-length IFN-β promoter of the transiently co-transfected reporter plasmid p125-luc, vIRF-2 inhibited IFN-β promoter transactivation by wild-type FLAG-tagged IRF-3 (IRF-3WT), by up to 70% (Fig. 1A, upper panel). In this model system, the antiviral response and IRF-3 phosphorylation are activated by transfection of synthetic double-stranded RNA (poly(I:C)), as opposed to using the constitutively active phosphomimetic IRF-3 mutant, IRF-3(5D) (21). These data are consistent with our previous findings on vIRF-2 inhibition of IRF-3(5D) function (20).
FIGURE 1.
KSHV vIRF-2 expression represses IFN-β promoter transactivation and reduces wild type IRF-3 protein levels. In A: upper panel: transcriptional activity of the full-length IFN-β promoter (p125-luc) reporter vector. HEK293 cells were transiently co-transfected with p125-luc and an expression plasmid of either FLAG epitope-tagged IRF-3 wild type (IRF-3WT), or the mutants IRF-3(2A), IRF-3(5A), and IRF-3(2A5A). Each transfection also included the Xpress epitope-tagged vIRF-2-expressing plasmid, pcDNA4/vIRF-2 (vIRF-2), or the empty parental plasmid backbone, pcDNA4. The pRLSV40 plasmid constitutively expressing Renilla luciferase was added as an internal control to which firefly luciferase levels were normalized. Twenty-four hours after plasmid transfection, the cells were transfected with poly(I:C) (10 μg/ml), and luciferase activity was measured 18 h later. All reporter assays were repeated at least three times and mean values (± S.D.) are presented from one representative experiment (**, p < 0.01, Student's t test). White bars, cells lacking ectopic IRF-3 or vIRF-2; gray bars, cells expressing ectopic IRF-3; black bars, cells co-transfected to express ectopic vIRF-2. Lower panel: the IRF-3 C-terminal phosphorylation sites 1 and 2 (amino acids 382 to 414) are presented with the identities of the serine/threonine substitutes. B, vIRF-2 expression reduces total phosphorylated IRF-3 levels. Lysates were prepared from HEK293 cells that had been transfected with the IRF-3WT expression vector and either the vIRF-2 expression vector or the empty vector control (pcDNA4). Twenty-four hours after plasmid transfection, the cells were transfected with poly(I:C). They were then harvested at the times indicated and separated by SDS-PAGE and immunoblotted with the following antibodies: anti-phospho-IRF-3 (α-p-IRF-3; Ser396), anti-Xpress (vIRF-2), anti-FLAG (total ectopic IRF-3) and anti-β-actin. C, vIRF-2 expression reduces both monomeric and dimeric IRF-3 levels. Lysates prepared as described in B were separated by native PAGE to detect the IRF-3 dimeric (D) and monomeric (M) forms with anti-FLAG antibody. D, interaction of vIRF-2 with IRF-3. Co-immunoprecipitation studies were performed with lysates of HEK293 cells that had been transfected with the expression plasmids and poly(I:C) as described in A. vIRF-2 was immunoprecipitated with anti-Xpress monoclonal antibody. Immunoprecipitates and input extracts were separated by SDS-PAGE before immunoblotting with anti-FLAG polyclonal antibody to detect IRF-3 or anti-Xpress to detect vIRF-2. WT, IRF-3 wild type; 2A, IRF-3(2A) mutant; 5A, IRF-3(5A) mutant; 2A5A, IRF-3(2A5A) mutant; IP, antibody used for immunoprecipitation; IB, antibody used for immunoblot. IgG, provides a measure ensuring equal loading of immunoprecipitate between lanes; heavy chain is shown. Input, these lanes demonstrate the expression levels for vIRF-2 and IRF-3 in the lysates before immunoprecipitation. E, IRF-3(5A) accumulates in the presence of vIRF-2. HEK293 cells were transfected with FLAG-tagged IRF-3(5A)-expression vector in the presence and absence of the Xpress-tagged vIRF-2 expression vector and transfected with poly(I:C) 24 h later. After incubating for the times indicated, protein extracts were prepared and analyzed by SDS-PAGE and immunoblot.
Because these studies (Fig. 1A, upper panel) were performed by activating IRF-3WT with transfection of poly(I:C), the influence of IRF-3 phosphorylation on the control of transactivation by vIRF-2 was investigated. IRF-3 phosphorylation occurs predominantly within two C-terminal domains, sites 1 and 2, containing serine/threonine-rich tracts (Fig. 1A, lower panel) and is essential for the activity of the protein. Although some debate continues as to the precise function of phosphorylation of specific residues within these sites (see Refs. 10, 25), a detailed understanding of the significance of phosphorylation of sites 1 and 2 en masse exists. Thus, IRF-3 autoinhibition is repressed by phosphorylation of the seven serine and threonine residues in these sites that induces a structural change releasing the N terminus and allowing the protein to homodimerize, interact with the transcriptional coactivator CBP/p300, and participate in assembly of the enhancesome (21, 26). Specifically, phosphorylation within IRF-3 site 2 represses autoinhibition, permitting interaction with CBP/p300, and facilitating phosphorylation within site 1 that then enables homodimerization (25).
Sites 1 and 2 were mutated separately and together, creating two previously described IRF-3 mutants (21, 27) 2A and 5A and the double mutant 2A5A (Fig. 1A, lower panel). As expected, the inherent transactivation capacity of the 5A mutant, IRF-3(5A), was reduced by ∼50% compared with that of IRF-3WT (Fig. 1A, upper panel), consistent with its reduced dimerization capability (27). vIRF-2 reproducibly reduced IRF-3(5A) transactivation of the IFN-β promoter slightly, but not by a statistically significant amount (Fig. 1A, upper panel; p > 0.05, Student's t test). These data suggest vIRF-2 inhibition of IRF-3 transactivation depends on cognate phosphorylation of residues at IRF-3 C-terminal site 2. Neither the 2A mutant, IRF-3(2A), nor the double mutant, IRF-3(2A5A), possess significant transactivation activity (Fig. 1A, upper panel), consistent with their inability to dimerize (27), and therefore the impact of vIRF-2 on their function is indeterminable.
Quantifying the temporal amounts of phosphorylated IRF-3 revealed they were reduced in the presence of vIRF-2. Thus, 18 h after activating the antiviral response by poly(I:C) transfection, phosphorylated IRF-3 levels were reduced in the presence of vIRF-2 (Fig. 1B, upper panel, compare the level of phospho-IRF-3 in the presence of vIRF-2, lanes 9 and 10, with that in the absence of vIRF-2, lanes 4 and 5). Total IRF-3 levels, detected with anti-FLAG antibody, were likewise also reduced to some extent, and this finding was confirmed by non-denaturing gel analyses that revealed vIRF-2 reduced both dimeric (phosphorylated) and monomeric IRF-3 levels (Fig. 1C, upper panel). Repeating these studies up to 48 h post-activation of the antiviral response with poly(I:C) transfection provided further evidence that vIRF-2 reduced IRF-3 levels (data not shown). Thus, these data correlate vIRF-2-mediated inhibition of IRF-3 transactivation of the IFN-β promoter (Fig. 1A) with reduced amounts of phosphorylated IRF-3, following poly(I:C) activation of the antiviral response.
Given these findings, the physical interaction of vIRF-2 and IRF-3 was investigated. vIRF-2 was immunoprecipitated, and the products were analyzed for the presence of IRF-3 by Western blot. Only IRF-3WT and IRF-3(5A) co-precipitated with vIRF-2 to any demonstrable extent (Fig. 1D). Indeed, the amount of IRF-3WT co-precipitating was always less than that of IRF-3(5A). Furthermore, activation of the antiviral response by poly(I:C) transfection consistently reduced the amount of IRF-3WT co-precipitating with vIRF-2 (Fig. 1D, top panel, compare lanes 1 and 2), whereas the level of co-precipitating IRF-3(5A) was unaffected or even increased following poly(I:C) treatment (Fig. 1D, top panel, compare lanes 5 and 6). The faint band appearing in the presence of IRF-3(2A) (Fig. 1D, top panel, lane 4) is a nonspecific protein of lower molecular size than IRF-3.
Like IRF-3WT (Fig. 1B, upper panel, lanes 4 and 5), IRF-3(5A) gradually accumulated after poly(I:C) transfection in the absence of vIRF-2 (Fig. 1E, upper panel, lanes 1–4), most likely as a consequence of phosphorylation within C-terminal site 1. The absolute levels are lower than for IRF-3WT, because the mutant protein is expressed less efficiently than the wild type (compare “input levels” detected by anti-FLAG antibody in Fig. 1D, bottom panel, lanes 5 and 6 for IRF-3(5A) and lanes 1 and 2 for IRF3WT). However, this mutant, whose transactivation of the IFN-β promoter is not significantly inhibited by vIRF-2, but which binds this viral protein, is not depleted from the cell in the presence of the viral protein, unlike IRF-3WT. Rather, IRF-3(5A) accumulates in the presence of vIRF-2 (Fig. 1E, upper panel, compare the IRF-3(5A) level in the presence of vIRF-2, lanes 5–8, with that in its absence, lanes 1–4).
How might vIRF-2 cause the selective turnover of IRF-3WT? Multiple processes, enzymes, and subcellular components drive cellular protein turnover. They include calpain, autophagy, and the proteasome. The stability of IRF-3 in the presence of vIRF-2 was not affected by treatment with a specific inhibitor of calpain (calpain inhibitor I, data not shown), nor by an activator of autophagy (rapamycin, data not shown).
The Accelerated Decay of Activated IRF-3WT by vIRF-2 Is Independent of the Proteasome
The proteasome degrades poly-ubiquitinated proteins. Indeed, as a strategy to evade induction of the innate antiviral response, viruses as diverse as the flavivirus bovine viral diarrhea virus (28), having an RNA genome, and bovine herpesvirus 1 (29) with a DNA genome, can accelerate the decay of IRF-3 by augmenting its poly-ubiquitination and proteasomal degradation (see Ref. 1).
Proteasome degradation is inhibited by treatment with MG132. As expected, treating cells with MG132 stabilized IRF-3 following poly(I:C) transfection (Fig. 2A, top panel, compare the amount of IRF-3 in lanes 4–6 in the presence of MG132 with that in its absence in lanes 1–3; see also supplemental Figs. S1 and supplemental Fig. S2A(i)). These data confirm that IRF-3 can be the target of a proteasome-dependent degradative process, as previously reported (9).
FIGURE 2.
The accelerated decay of activated wild-type IRF-3 by vIRF-2 is independent of the proteasome, but inhibited by a general caspase inhibitor. A, vIRF-2-accelerated decay of wild-type IRF-3 is independent of the proteasome. HEK293 cells were transiently co-transfected with expression vectors for either FLAG epitope-tagged wild type IRF-3 (IRF-3WT) (left panel) or IRF-3(5A) (right panel) and either Xpress epitope-tagged vIRF-2, or the empty parental plasmid backbone, pcDNA4. Twenty-four hours later, cells were treated for 30 min with either DMSO (as the vehicle negative control) or MG132 (10 μm) and then transfected with poly(I:C) (10 μg/ml). Lysates were prepared at various times thereafter, and protein samples were separated by SDS-PAGE and immunoblotted with anti-FLAG (to detect IRF-3), anti-Xpress (to detect vIRF-2), and anti-β-actin antibodies. This experiment is representative of more than three performed independently. A longer exposure of the anti-FLAG (IRF-3) (top) panel is presented in supplemental Fig. S1, to confirm IRF-3 can be visualized in lanes 12-18. B, vIRF-2-accelerated decay of wild-type IRF-3 is repressed by Z-VAD-FMK (Z-VAD) treatment, which inhibits caspase activity. This experiment was repeated essentially as described for A, with the exception that the cells were treated with Z-VAD-FMK (10 μm) in place of MG132 (left panel) and the IRF-3(5A) expression plasmid was not used. Alternatively, cells were treated with DMSO, MG132 (10 μm), or both Z-VAD-FMK (10 μm) and MG132 (10 μm) (right panel). Immunoblotting was performed with anti-FLAG (IRF-3), anti-Xpress (vIRF-2), anti-PARP, and anti-β-actin antibodies. The anti-PARP antibody recognizes uncleaved PARP (Un-Cl, 116 kDa) and one cleaved fragment (Cleaved, 83 kDa). This experiment is representative of more than three performed independently.
Consistent with the previous data (Fig. 1, B–E), the level of IRF-3 was reduced in the presence of vIRF-2 (Fig. 2A, top panel, compare the amount of IRF-3 in lanes 1–3 in the presence of DMSO and absence of vIRF-2 with that in the presence of both DMSO and vIRF-2 at the same times in lanes 8–10; see also supplemental Fig. S2A(ii)). However, importantly and remarkably, IRF-3 turnover was dramatically accelerated in the presence of vIRF-2 and MG132 compared with the absence of vIRF-2 (Fig. 2A, upper panels, compare the amount of IRF-3 in lanes 12–15 in the presence of MG132 and vIRF-2 with that in the absence of vIRF-2 in lanes 4–7). Under the same conditions, the IRF-3(5A) mutant, which accumulates in the presence of vIRF-2 (Fig. 1E), also accumulated in the presence of MG132, and even more so in the presence of both the proteasome inhibitor and the viral protein (Fig. 2A, upper panels, compare the amount of IRF-3(5A) in lanes 19–21 in the presence of MG132 and absence of vIRF-2, with that in the presence of both MG132 and vIRF-2 at the same times in lanes 27–29; see also supplemental Fig. S2A(iii)).
These data suggested vIRF-2 might recruit, to fully activated IRF-3WT, either a protein or a protein complex responsible for IRF-3 decay and perhaps normally targeted for degradation by the proteasome, but stabilized in the presence of MG132. In contrast, IRF-3(5A), lacking phosphorylation at C-terminal site 2, resists this degradative process.
To determine if vIRF-2 enhanced IRF-3 ubiquitination and thereby degradation by the proteasome, the extent of poly-ubiquitination of IRF-3 in the presence of vIRF-2 was measured. No effect of the viral protein could be determined (data not shown).
The Accelerated Decay of Activated IRF-3WT Is Inhibited by a General Caspase Inhibitor
Another class of enzymes involved in proteolysis is the caspases, cysteine-dependent proteases that cleave substrates after an aspartate residue. They function during apoptosis, a homeostatic mechanism of programmed cell death. However, caspases are increasingly recognized as functioning in the absence of apoptosis (see Refs. 30–32). For example, caspase-3 was recently implicated in promoting stem cell development (33, 34). Caspase-8 and -10 participate in signaling by the RNA helicases RIG-I and mda-5 following stimulation with double-stranded RNA, and they drive inflammatory cytokine production by activating NF-κB (35). Caspase-8 and -10 are initiator caspases that generally activate the effector caspases, such as caspase-3, -6, and -7 (see Ref. 32).
To determine if caspases might be responsible for vIRF-2-mediated turnover of IRF-3, their activity was suppressed by treatment with a general inhibitor (Z-VAD-FMK). Even in the presence of Z-VAD-FMK and absence of vIRF-2, IRF-3 accumulated (Fig. 2B, top panel, compare the level of IRF-3 in the absence of Z-VAD-FMK in lanes 1–3 with the levels in its presence in lanes 4–6; see also supplemental Fig. S2B(i)). These data suggest that one or more caspases is responsible in part for IRF-3 turnover in the normal cell in which the antiviral response has been invoked by poly(I:C) transfection. IRF-3 also accumulated in the presence of Z-VAD-FMK and vIRF-2 (Fig. 2B, top panel, compare the level of IRF-3 in the absence of Z-VAD-FMK and presence of vIRF-2 in lanes 7–9 with the levels in the presence of Z-VAD-FMK and vIRF-2 in lanes 10–12; see also supplemental Fig. S2B(ii)).
The pronounced accelerated decay of IRF-3 identified in the presence of MG132 and vIRF-2 after poly(I:C) transfection (Fig. 2A, top panel, lanes 12-15) was also inhibited with Z-VAD-FMK (Fig. 2B, top panel, compare the level of IRF-3 in the presence of vIRF-2 and MG132 in lanes 17-20 with the level in the presence of vIRF-2, MG132 and Z-VAD-FMK in lanes 21–24; see also supplemental Fig. S2B(iii)). These data suggest that, when vIRF-2 binds to IRF-3 (Fig. 1D), it might recruit a caspase that then contributes to the decay of IRF-3.
IRF-3 levels in the presence of Z-VAD-FMK and vIRF-2 did not reach those seen in the presence of Z-VAD-FMK alone (Fig. 2B, compare the level of IRF-3 in lanes 10–12 with that in lanes 4–6; see also supplemental Fig. S2B(v)), presumably because caspase inhibition by Z-VAD-FMK was not absolute. vIRF-2 also accumulated in the presence of Z-VAD-FMK (Fig. 2B, lanes 10–12 and 21–24) compared with the absence of the caspase inhibitor (Fig. 2B, lanes 7–9 and 13–16; see also supplemental Fig. S2B(iv)), suggesting the viral protein is a target of one or more caspases.
The stabilization of IRF-3 levels by Z-VAD-FMK treatment, despite the presence of vIRF-2, raised the possibility that this viral protein may be pro-apoptotic and that it was reducing IRF-3 levels as a consequence of activating apoptosis. To investigate this possibility, lysates of cells with and without vIRF-2 were assayed for a hallmark indicator of apoptosis: cleavage of poly(adenosine diphosphate-ribose) polymerase (PARP). PARP is a target of mature caspase-3 cleavage (36). As expected, low levels of cleaved PARP could be detected in cells transfected with poly(I:C) (Fig. 2B, lanes 2 and 3), because part of the antiviral response is to induce apoptosis (1), and IRF-3 can participate in this induction independently of its role in the IFN response (37, 38). PARP cleavage was elevated following MG132 treatment (Fig. 2B, lanes 18–20), and this cleavage was reduced by the addition of Z-VAD-FMK (Fig. 2B, lanes 22–24). However, PARP cleavage was not elevated by the expression of vIRF-2 (Fig. 2B, compare PARP cleavage in the absence of vIRF-2, lanes 2 and 3, with that in its presence in lanes 8, 9, 15, and 16), indicating the viral protein is not pro-apoptotic, as suggested previously (39). Further evidence excluding vIRF-2 from a pro-apoptotic role was obtained by quantifying apoptosis in transfected cells by flow cytometry (supplemental Fig. S3).
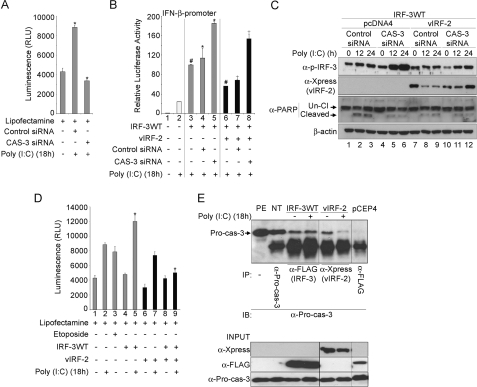
The Accelerated Decay of Activated IRF-3WT by vIRF-2 Depends on Caspase-3 Activity
Because Z-VAD-FMK is a pan-caspase inhibitor, the identity of the caspase most likely involved in IRF-3 decay was sought. Studies with caspase-specific inhibitors indicated that IRF-3 decay was reduced when caspase-3 activity was inhibited. Indeed, when functional levels of caspase-3 were knocked down by >50% with specific siRNA (Fig. 3A), IRF-3WT-mediated transactivation of the IFN-β promoter in reporter gene assays was increased significantly in the presence of vIRF-2. Thus, under these circumstances, inhibition of IRF-3 transactivation by vIRF-2 was significantly mitigated (Fig. 3B, compare the normalized IFN-β promoter activity in bars #7 and #8). However, mitigation was not complete, as compared with the level of promoter activity achieved with caspase-3 siRNA alone (Fig. 3B, compare the normalized IFN-β promoter activity in bars #8 and #5), suggesting vIRF-2 inhibits IRF-3 activity by another, caspase-3-independent, mechanism. Furthermore, IRF-3 transactivation of the IFN-β promoter, even in the absence of vIRF-2, was increased significantly in the presence of caspase-3 siRNA compared with control siRNA (Fig. 3B, compare bars #5 and #4, respectively). These data confirm those of Fig. 2B: that IRF-3WT turnover was reduced in the presence of Z-VAD-FMK, even in the absence of vIRF-2 (Fig. 2B, top panel, compare the level of IRF-3 in the absence of Z-VAD-FMK in lanes 1–3 with the level in its presence in lanes 4–6; see also supplemental Fig. S2B(i)). They therefore provide evidence in support of a cellular mechanism in which caspase-3 participates to turnover IRF-3 following activation of the antiviral response by poly(I:C) transfection. vIRF-2 accelerates this process.
FIGURE 3.
The accelerated decay of activated wild-type IRF-3 by vIRF-2 depends on caspase-3 activity. A, siRNA knockdown of caspase-3. Functional levels of caspase-3 protein were analyzed with the Caspase-Glo®-3/7 assay kit (Promega). HEK293 cells were transfected with either Lit28i Polylinker ShortCut® siRNA mix (Control siRNA) or caspase-3 ShortCut® siRNA (CAS-3 siRNA) for 24 h. They were then transfected with poly(I:C) for 18 h before harvesting for caspase activity assay. Activity assays were repeated at least twice, and average values (± S.D.) are presented from one representative experiment (*, p < 0.05, Student's t test). B, knockdown of caspase-3 elevates IFN-β promoter-driven reporter gene activity. HEK293 cells were transiently co-transfected with the full-length IFN-β promoter reporter plasmid (p125-luc) and an expression vector of FLAG-tagged IRF-3 wild-type (IRF-3WT), or Xpress-tagged vIRF-2-expression vector (vIRF-2), as indicated. The pRLSV40 plasmid constitutively expressing Renilla luciferase was added as an internal control to which firefly luciferase levels were normalized. Co-transfection with control (Control siRNA) and caspase-3 (CAS-3) siRNA was performed as indicated. Twenty-four hours after plasmid transfection, the cells were transfected with poly(I:C) (10 μg/ml), and luciferase activity was measured 18 h later. Average values (± S.D.) are presented from one representative experiment of at least three performed independently (*, #, and + indicate p < 0.05, Student's t test). Gray bars, cells expressing ectopic IRF-3; black bars, cells co-transfected to express ectopic vIRF-2. C, knockdown of caspase-3 elevates phospho-IRF-3 levels. HEK293 cells were transiently co-transfected with an expression vector for FLAG-tagged IRF-3 wild-type (IRF-3WT), and Xpress-tagged vIRF-2 (vIRF-2) or empty vector parental plasmid (pcDNA4), as indicated. Co-transfection with control (Control siRNA) and caspase-3 (CAS-3) siRNA was performed as indicated. Twenty-four hours after plasmid transfection, the cells were transfected with poly(I:C) (10 μg/ml). Protein extracts were prepared at the indicated times thereafter and analyzed by immunoblot with the following antibodies: anti-PARP, anti-Xpress (vIRF-2), anti-phospho-IRF-3 (α-p-IRF-3; Ser-396) and anti-β-actin. The anti-PARP antibody recognizes uncleaved PARP (Un-Cl, 116 kDa) and one cleaved fragment (Cleaved, 83 kDa). This experiment is representative of more than three performed independently. D, expression of vIRF-2 influences caspase-3/-7 activity. HEK293 cells were transiently transfected with an expression vector for FLAG-tagged IRF-3 wild type (IRF-3WT), and/or Xpress-tagged vIRF-2 (vIRF-2), as indicated. Twenty-four hours after plasmid transfection, the cells were treated with etoposide (50 μm) or DMSO and 30 min later transfected with poly(I:C) (10 μg/ml). Caspase-3/-7 activities were measured 18 h later. Average values (± S.D.) are presented from one representative experiment of three performed independently (*, p < 0.05, Student's t test). Black bars, cells co-transfected to express ectopic vIRF-2. E, pro-caspase-3 interacts with IRF-3 and vIRF-2. Upper panel: co-immunoprecipitation studies were performed with lysates of HEK293 cells that had been transfected with an expression vector for FLAG-tagged IRF-3 wild-type (IRF-3WT), or Xpress-tagged vIRF-2 (vIRF-2), as indicated. Twenty-four hours after plasmid transfection, the cells were transfected with poly(I:C) (10 μg/ml), and immunoprecipitation assays were performed 18 h later. Wild-type IRF-3 was immunoprecipitated with anti-FLAG polyclonal antibody and vIRF-2 with anti-Xpress monoclonal antibody. Immunoprecipitates and input extracts (lower panel) were separated by SDS-PAGE before immunoblotting with anti-Pro-caspase-3 antibody (α-Pro-cas-3). PE, protein extract; NT, non-transfected cells. Plasmid pCEP-4 expresses a negative control FLAG-tagged protein that did not immunoprecipitate pro-caspase-3. Note that all seven lanes were separated on the same gel and transferred to the same membrane, but because the vIRF-2 lanes were not contiguous with the others, intervening, irrelevant lanes were removed from the image. IP, antibody used for immunoprecipitation; IB, antibody used for immunoblot. Right panel, Input: these lanes demonstrate the expression levels for vIRF-2, IRF-3 and pro-caspase-3 in the lysates before immunoprecipitation.
Elevated IRF-3WT transactivation of the IFN-β promoter in the presence of caspase-3 siRNA (Fig. 3B, bar #5) was mirrored by the relative accumulation of phosphorylated IRF-3 under the same conditions (Fig. 3C, top panel, compare the phospho-IRF-3 level in the presence of caspase-3 siRNA, lanes 4–6 with that in the presence of control siRNA, lanes 1–3; see also supplemental Fig. S4(i)). Likewise, phospho-IRF-3 levels accumulated in the presence of vIRF-2 and caspase-3 siRNA when compared with vIRF-2 and control siRNA (Fig. 3C, top panel, compare the phospho-IRF-3 level in the presence of caspase-3 siRNA and vIRF-2, lanes 10–12 with that in the presence of vIRF-2 and control siRNA, lanes 7–9; see also supplemental Fig. S4(ii)). This increase in phospho-IRF-3 levels resulting from caspase-3 siRNA treatment in the presence of vIRF-2 was not as exaggerated as that in the absence because complete caspase-3 knockdown was not achieved and therefore active caspase-3 continued contributing to IRF-3 turnover in a mechanism accelerated by vIRF-2.
As expected, reduced cleavage of PARP was evident in the presence of caspase-3 siRNA (Fig. 3C, lanes 5 and 6), when compared with control siRNA (Fig. 3C, lanes 2 and 3). Cleaved PARP levels were also reduced in the presence of vIRF-2 compared with its absence (Fig. 3C, compare lanes 8 and 9 with lanes 2 and 3), regardless of the presence of caspase-3 siRNA. This finding is consistent with that from Fig. 2B (compare PARP cleavage in the absence of vIRF-2, lanes 2 and 3, with that in its presence in lanes 8, 9, 15, and 16) and suggests vIRF-2 might have either anti-apoptotic activity or sequester caspase-3 away from PARP, or both. Therefore, these data are also consistent with those indicating vIRF-2 is not pro-apoptotic (supplemental Fig. S3). Furthermore, vIRF-2 significantly reduced caspase-3/-7 activities following poly(I:C) treatment in the presence of IRF-3WT (Fig. 3D, bar #9), compared with the level in the absence of vIRF-2 (Fig. 3D, bar #5).
Thus, the data of Fig. 3 (B–D) suggest an interaction between activated IRF-3WT and caspase-3, and between vIRF-2 and caspase-3. To determine if this interaction is physical, pro-caspase-3 was immunoprecipitated and the presence of IRF-3WT and vIRF-2 determined by Western blot. Both proteins co-precipitated with pro-caspase-3. However, the level of vIRF-2 was reduced following poly(I:C) treatment (Fig. 3E), consistent with this viral protein being either a direct or indirect target of caspase-3 activity, as suggested from the data of Fig. 2B in which vIRF-2 accumulated in the presence of Z-VAD-FMK (Fig. 2B, compare vIRF-2 levels between lanes 7–9 in the absence of Z-VAD-FMK and lanes 10–12 in its presence; see also supplemental Fig. S2B(iv)).
IRF-3WT Accumulates in a Caspase-3-deficient Cell Line Unless Ectopic Caspase-3 Is Introduced, when vIRF-2 Accelerates IRF-3WT Decay
Because knockdown studies with siRNA did not completely abrogate caspase-3 activity, the transactivating function and fate of IRF-3WT were investigated in the human breast adenocarcinoma cell line, MCF-7, which is caspase-3-deficient (40). First, the full-length IFN-β reporter plasmid was found to be significantly activated by poly(I:C) transfection (Fig. 4A, compare bars #1 and #2), confirming the function of the antiviral response signaling pathway in this cell line. This activation was increased significantly by ectopic expression of IRF-3WT (Fig. 4A, bar #3). The introduction of caspase-3 into these cells reduced IRF-3WT transactivation of the IFN-β promoter significantly (Fig. 4A, compare bars #3 and #4), to that level resulting from vIRF-2 expression (Fig. 4A, bar #5). The expression of caspase-3 and vIRF-2 together synergistically reduced IRF-3WT transactivation (Fig. 4A, bar #6). The inhibition of IRF-3WT transactivation by vIRF-2, despite the absence of caspase-3 (Fig. 4A, bar #5) indicates that the viral protein also inhibits IRF-3 activity in a caspase-3-independent manner, as suggested previously (Fig. 3B).
FIGURE 4.
IRF-3WT accumulates in a caspase-3-deficient cell line unless ectopic caspase-3 is introduced, when vIRF-2 accelerates the decay. A, IFN-β promoter-driven reporter gene activity is elevated in the absence of caspase-3. Caspase-3-deficient MCF-7 cells were transiently co-transfected with the full-length IFN-β promoter reporter plasmid (p125-luc) and an expression vector for FLAG-tagged IRF-3 wild type (IRF-3WT), or Xpress-tagged vIRF-2 (vIRF-2) or pcDNA3/caspase-3, a caspase-3 expression vector (CAS-3), as indicated in the figure. The pRLSV40 plasmid constitutively expressing Renilla luciferase was added as an internal control to which firefly luciferase levels were normalized. Twenty-four hours after plasmid transfection, the cells were transfected with poly(I:C) (10 μg/ml), and luciferase activity was measured 18 h later. Average values (± S.D.) are presented from one representative experiment (●, *, #, and + p < 0.05, Student's t test) of at least three performed independently. White bars, cells lacking ectopic IRF-3 or vIRF-2; gray bars, cells expressing ectopic IRF-3; black bars, cells co-transfected to express ectopic vIRF-2. B, IRF-3WT accumulates in a caspase-3-deficient cell line. MCF-7 cells were transiently co-transfected with an expression vector for FLAG-tagged IRF-3 wild-type (IRF-3WT), or Xpress-tagged vIRF-2 (vIRF-2) or pcDNA3/Caspase-3, a caspase-3 expression vector (CAS-3) or the empty vector pcDNA4, as indicated in the figure. Protein extracts were analyzed by immunoblot. C, vIRF-2 sequesters caspase-3 activity. Caspase-3/7 activity was measured in MCF-7 cells transfected with plasmids described above, as indicated in the figure. Twenty-four hours after plasmid transfection, the cells were transfected with poly(I:C) (10 μg/ml). Caspase-3/-7 activities were measured 18 h later. Average values (±S.D.) are presented from one representative experiment of three performed independently (* and #, p < 0.05, Student's t test). Black bars, cells co-transfected to express ectopic vIRF-2.
These effects on IRF-3WT transactivation of the IFN-β promoter were mirrored directly by the levels of IRF-3WT (Fig. 4B). In particular, within 24 h of poly(I:C) transfection of MCF-7 cells already transfected with the caspase-3 expression vector, IRF-3WT levels were reduced by ∼60% (Fig. 4B, lane 6), compared with the levels in the absence of caspase-3 (Fig. 4B, lane 3). However, co-transfecting the vIRF-2 and caspase-3 expression vectors resulted in IRF-3WT levels becoming undetectable within 24 h of poly(I:C) treatment (Fig. 4B, lane 12).
The coincident transfection of the vIRF-2 and caspase-3 expression vectors also reduced the levels of both proteins substantially (Fig. 4B, compare vIRF-2 and pro-caspase-3 levels, in lanes 10–12, with the level of vIRF-2 without caspase-3, lanes 7–9, and pro-caspase-3 levels without vIRF-2, lanes 4–6). The data suggest a multiprotein complex forms between minimally vIRF-2, caspase-3, and IRF-3, leading to a reduction in the levels of each of them.
vIRF-2 significantly inhibited caspase-3 activity against the Z-DEVD-aminoluciferin substrate (Fig. 4C, compare bars #6 and #10), providing further evidence against a pro-apoptotic role for vIRF-2. Moreover, reduced caspase-3/-7 activity in the presence of vIRF-2 (Fig. 4C, bars #9 and #10) is consistent with the reduced levels of caspase-3 in the presence of this viral protein (Fig. 4B, lanes 10–12). In these studies, the enzyme activities measured in the absence of caspase-3 (Fig. 4C, bars #1, #2, and #4) represent basal caspase-7 activities and therefore enable the contribution of ectopic caspase-3 activity to be determined (Fig. 4C, bars #3, #6, #9, and #10).
Intracellular Localization of vIRF-2, IRF-3WT, and Caspase-3
Because IRF-3 stability in the presence of vIRF-2 protein expression was higher in caspase-3-deficient MCF-7 cells than in HEK293 cells, the distribution of these proteins in the breast adenocarcinoma cell line was investigated (Fig. 5). The studies were performed 16 h following poly(I:C) transfection, because after longer times in the presence of caspase-3, IRF-3 levels become undetectable (see Fig. 4B). The vIRF-2 protein assumed a diffuse cytoplasmic and nuclear distribution, with some concentration around the inner nuclear membrane (Fig. 5A). Upon activation of the antiviral response by poly(I:C) transfection, this distribution became more punctate with expression emphasized in the cytoplasm and not the nucleus (Fig. 5A). Conversely, IRF-3 redistributed to both the nuclear and cytoplasmic compartments from a predominantly cytoplasmic location upon poly(I:C) transfection (Fig. 5B), as described previously (23). When vIRF-2 and IRF-3 were co-expressed, IRF-3 was sufficiently abundant to be detected readily, consistent with the Western blot data (Fig. 4B, lanes 7–9), but its redistribution to the nucleus was minimized (Fig. 5C). These data are consistent with caspase-3-independent inhibition of IRF-3 transactivation by vIRF-2, as suggested previously (Figs. 3B and 4A). Both vIRF-2 and IRF-3, when expressed independently in MCF-7 cells, were sufficiently abundant to be identified as colocalizing with ectopic caspase-3, predominantly in the cytoplasm (Fig. 5, D and E, respectively).
FIGURE 5.
IRF-3WT, vIRF-2, and caspase-3 co-localize in the cytoplasm. A, vIRF-2 redistributes from the nucleus and the cytoplasm (panel: vIRF-2) to the cytoplasm following activation of the antiviral response by poly(I:C) transfection (panel: vIRF-2+IC). MCF-7 cells were transfected with the expression vector for Xpress-tagged vIRF-2. After 24 h they either were (+IC) or were not transfected with poly(I:C), fixed, and stained a further 16 h later. B, IRF-3 redistributes from the cytoplasm (panel: IRF-3) to the nucleus and the cytoplasm following activation of the antiviral response by poly(I:C) transfection (panel: IRF-3+IC). MCF-7 cells were transfected with the expression vector for FLAG-tagged IRF-3. After 24 h they either were (+IC) or were not transfected with poly(I:C), fixed, and stained a further 16 h later. C, vIRF-2 and IRF-3 co-localize in the cytoplasm. MCF-7 cells were co-transfected with the expression vectors for Xpress-tagged vIRF-2 and FLAG-tagged IRF-3. After 24 h they were (+IC) or were not transfected with poly(I:C) and fixed and stained a further 16 h later. D, vIRF-2 and caspase-3 co-localize in the cytoplasm. MCF-7 cells were co-transfected with the expression vectors for Xpress-tagged vIRF-2 and caspase-3 (CAS-3). After 24 h they were (+IC) or were not transfected with poly(I:C) and fixed and stained a further 16 h later. E, IRF-3 and caspase-3 co-localize in the cytoplasm. MCF-7 cells were cotransfected with the expression vectors for FLAG-tagged IRF-3 and caspase-3 (CAS-3). After 24 h they were (+IC) or were not transfected with poly(I:C) and fixed and stained a further 16 h later. F, vIRF-2, IRF-3, and caspase-3 co-localize in the cytoplasm. MCF-7 cells were co-transfected with the expression vectors for Xpress-tagged vIRF-2, FLAG-tagged IRF-3, and caspase-3 (CAS-3). After 24 h they were (+IC) or were not transfected with poly(I:C) and fixed and stained a further 16 h later. A region of co-localization in the merge panel (white arrow) was magnified, and the images are presented in the bottom row of panels.
When MCF-7 cells were co-transfected with vIRF-2, IRF-3 and caspase-3 expression vectors, the proteins they encode co-localized in the cytoplasm (Fig. 5F). Poly(I:C) transfection reduced IRF-3 protein to barely detectable levels, consistent with the Western blot data (Fig. 4B). Nevertheless, cytoplasmic co-localization of vIRF-2, IRF-3, and caspase-3 could still be determined in discrete regions of the cytoplasm (Fig. 5F, bottom row of magnified images) when sufficient IRF-3 levels were detectable.
Endogenous IRF-3 Is Affected by vIRF-2 in the Same Way as Ectopic IRF-3WT
The studies to date were performed with overexpressed, epitope-tagged ectopic IRF-3. We therefore examined the fate of endogenous IRF-3, to determine if it is regulated by vIRF-2 in the same way as ectopic IRF-3WT.
First, activation of the full-length IFN-β promoter by endogenous IRF-3 in response to poly(I:C) transfection was verified (Fig. 6A, compare bars #1 and #2). This activation was increased significantly following treatment with the pan caspase inhibitor Z-VAD-FMK (Fig. 6A, compare bars #2 and #3). These data are consistent with those of Fig. 2B, showing elevated levels of IRF-3WT in the presence of this inhibitor. The presence of vIRF-2 significantly inhibited IFN-β promoter activity (Fig. 6A, compare bars #2 and #4), by >50%, consistent with the inhibitory activity of vIRF-2 on ectopic IRF-3WT (Fig. 1A). Treatment with Z-VAD-FMK in the presence of vIRF-2 completely restored IFN-β promoter activity to the levels seen in the absence of vIRF-2 (Fig. 6A, compare bars #3 and #5). These data are consistent with the increase in ectopic IRF-3WT levels in the presence of vIRF-2 and Z-VAD-FMK (Fig. 2B). Because IFN-β promoter activity is a surrogate marker for IRF-3 function, we then quantified active endogenous IRF-3 binding to its cognate oligonucleotide by ELISA, under similar experimental conditions (Fig. 6B). For these experiments an engineered derivative of HEK293 cells was studied, clone #3–9, in which all cells can be induced to express vIRF-2 (data not shown). Thus, this clonal cell line was stably transfected with a doxycycline-inducible vIRF-2 expression cassette (see “Experimental Procedures”) and compared with isogenic empty vector (EV) cells. Both cell lines were treated with doxycycline to mitigate any effects of leaky vIRF-2 expression in the absence of doxycycline treatment. Measuring endogenous IRF-3 activity in clone #3–9 cells generated similar data to those obtained for IFN-β promoter activity in transiently transfected HEK293 cells (Fig. 6A): vIRF-2 expression significantly inhibited endogenous IRF-3 activity by ∼50% (Fig. 6B, for DOX+DMSO-treated cells, compare IRF-3 activity in EV cells at 12 h with that in clone #3–9 cells). This inhibition was significantly repressed by the presence of Z-VAD-FMK (Fig. 6B, compare IRF-3 activity at 12 h in DOX+DMSO-treated clone #3–9 cells with that of DOX+Z-VAD-FMK-treated clone #3–9 cells). Moreover, the level of IRF-3 activity at 12 h in DOX+DMSO-treated EV cells is comparable to that in DOX+Z-VAD-FMK-treated clone #3–9 cells, suggesting the caspase inhibitor can almost completely restore endogenous IRF-3 activity in the presence of vIRF-2. Consistent with these data, dimeric IRF-3 levels in clone #3–9 cells increased with Z-VAD-FMK treatment (Fig. 6C, upper panel, compare the levels of dimeric (D) IRF-3 in lanes 11 and 12 with those in lanes 8 and 9).
FIGURE 6.
Endogenous IRF-3 shares the same fate as ectopic IRF-3. A, vIRF-2 inhibition of the transcriptional activity of the full-length IFN-β promoter (p125-luc) reporter vector driven by endogenous IRF-3 is repressed by the caspase inhibitor Z-VAD-FMK. HEK293 cells were transiently co-transfected with p125-luc and the Xpress epitope-tagged vIRF-2-expressing plasmid, pcDNA4/vIRF-2 (vIRF-2; black bars), or the empty parental plasmid backbone, pcDNA4 (light bars). The pRLSV40 plasmid constitutively expressing Renilla luciferase was added as an internal control to which firefly luciferase levels were normalized. Twenty-four hours later, cells were treated for 30 min with either DMSO (as the vehicle negative control) or Z-VAD-FMK (10 μm) and then transfected with poly(I:C) (10 μg/ml). Luciferase activity was measured 12 h later. The data represent the mean (± S.D.) from one representative experiment of three performed independently (*, #, and +, p < 0.005, Student's t test). B, vIRF-2 inhibition of endogenous IRF-3 activity is repressed by Z-VAD-FMK (Z-VAD) treatment. The clone #3–9 cell line (#3–9) harboring doxycycline-inducible vIRF-2 and the negative control empty vector cell line (EV) were both treated with doxycycline (DOX) for 24 h. They were then treated with either DMSO (as the vehicle negative control) or Z-VAD-FMK (Z-VAD; 10 μm) and transfected with poly(I:C) (10 μg/ml) for the times indicated. IRF-3 activity was determined with the TransAMTM IRF-3 ELISA (Active Motif). The data represent the mean (± S.D.) of two independent experiments (* and #, p < 0.005, Student's t test). C, vIRF-2-accelerated decay of endogenous IRF-3 is repressed by Z-VAD-FMK (Z-VAD) treatment. This experiment was performed essentially as described for Fig. 6B, with the exception that IRF-3 levels were measured by Western blot. The clone #3–9 cell line (#3–9) harboring doxycycline-inducible vIRF-2 and the negative control empty vector cell line (EV) were both treated with doxycycline (DOX) for 24 h. They were then treated with either DMSO (as the vehicle negative control) or Z-VAD-FMK (10 μm) and transfected with poly(I:C) (10 μg/ml) for the times indicated. Immunoblotting was performed on 10 μg of lysates separated either by native PAGE (upper panel) or denaturing PAGE (lower three panels). Native PAGE enabled IRF-3 dimeric (D) and monomeric (M) forms to be detected with anti-IRF-3 antibody. Aliquots of the same lysates were analyzed by denaturing PAGE to quantify total IRF-3, c-Myc-tagged vIRF-2, and β-actin. D, vIRF-2 inhibition of the transcriptional activity of the full-length IFN-β promoter (p125-luc) reporter vector driven by endogenous IRF-3 is repressed by caspase-3 siRNA. HEK293 cells were transiently co-transfected with p125-luc and the Xpress epitope-tagged vIRF-2-expressing plasmid, pcDNA4/vIRF-2 (vIRF-2; black bars), or the empty parental plasmid backbone, pcDNA4 (light bars). The pRLSV40 plasmid constitutively expressing Renilla luciferase was added as an internal control to which firefly luciferase levels were normalized. At the same time, either siRNA for caspase-3 (CAS-3 siRNA) or negative control scrambled siRNA (Control siRNA) was transfected. Twenty-four hours later, cells were transfected with poly(I:C) (10 μg/ml), and luciferase activity was measured 12 h later. The data represent the mean (± S.D.) from one representative experiment of three performed independently (* and #, p < 0.005; +, p < 0.05, Student's t test). E, measuring endogenous IRF-3 activity in the presence of vIRF-2 and MG132. The clone #3–9 cell line (#3–9) harboring doxycycline-inducible vIRF-2 and the negative control empty vector cell line (EV) were treated with doxycycline (DOX) for 24 h. They were then treated with either DMSO (as the vehicle negative control) or MG132 (10 μm) and transfected with poly(I:C) (10 μg/ml) for the times indicated. IRF-3 activity was determined with the TransAMTM IRF-3 ELISA (Active Motif). The data represent the mean (± S.D.) of two independent experiments (*, #, and +, p < 0.005, Student's t test).
To verify the effects of Z-VAD-FMK on endogenous IRF-3 involved caspase-3, the effect of caspase-3 siRNA on endogenous IRF-3 was determined. Essentially, the experiment presented in Fig. 3B was repeated, but IFN-β promoter activity was measured. As expected, IFN-β promoter activity was inhibited significantly by the presence of vIRF-2 (Fig. 6D, compare bars #2 and #4). However, this inhibition was significantly repressed by caspase-3 siRNA (Fig. 6D, compare bars #4 and #5). Thus, as for IRF-3WT (Fig. 3B), inhibition of endogenous IRF-3 transactivation by vIRF-2 was significantly mitigated by down-regulating caspase-3. Mitigation was not complete though, as measured by comparing the level of promoter activity achieved with caspase-3 siRNA alone (Fig. 6D, compare the IFN-β promoter activity in bars #3 and #5), suggesting vIRF-2 might also inhibit endogenous IRF-3 activity by a caspase-3-independent mechanism. Furthermore, even in the absence of vIRF-2, there is modest, but significantly increased, IFN-β promoter activity in the presence of caspase-3 siRNA compared with control siRNA (Fig. 6D, compare bars #2 and #3). This pattern is consistent with that seen for ectopic IRF-3WT (Fig. 3B), suggesting caspase-3 contributes to the negative regulation of IRF-3 activity even in the absence of vIRF-2.
Finally, we verified that inhibition of the proteasome does not obviate vIRF-2-mediated inhibition of endogenous IRF-3 transactivation, as suggested from our studies of ectopic IRF-3WT (Fig. 2A). Again IRF-3 activity was quantified by functional ELISA (Fig. 6E). First, when comparing clone #3–9 and EV cells treated with doxycycline, IRF-3 activity was suppressed by ∼50% in the vIRF-2 expressing cells (Fig. 6E, compare IRF-3 activity at 12 h from EV cells treated with DOX+DMSO with that of clone #3–9 cells treated with DOX+DMSO), as described above (Fig. 6B). This vIRF-2 inhibition of IRF-3 activity was not reversed by MG132 treatment. Rather, inhibition was increased significantly (Fig. 6E, compare IRF-3 activity at 12 h from clone #3–9 cells treated with DOX+DMSO with that of clone #3–9 cells treated with DOX+MG132). This effect of MG132 in the presence of vIRF-2 is consistent with the suggestion from the data of Fig. 2A that vIRF-2 might recruit to IRF-3 either a protein or a protein complex responsible for IRF-3 decay that is normally targeted for degradation by the proteasome, but is therefore stabilized by MG132.
Moreover, treating EV cells with MG132 increased IRF-3 activity significantly, again consistent with the known proteasome involvement in normal IRF-3 turnover (Fig. 6E, compare IRF-3 activity at 12 h from EV cells treated with DOX+DMSO with that of EV cells treated with DOX+MG132).
DISCUSSION
The present study of the function of the KSHV vIRF-2 protein, in addition to elucidating the molecular mechanisms of a viral immune modulator, has identified a previously unrecognized cellular mechanism of negative feedback control of the IFN antiviral response. Our data reveal that in a cell in which this response is invoked by transfection of synthetic double-stranded RNA, a proteolytic process that involves caspase-3 activity depletes fully activated IRF-3. The studies with MCF-7 cells (Figs. 4 and 5) provide conclusive genetic proof for a mechanism of IRF-3 turnover involving caspase-3. This mechanism contributes to IRF-3 destabilization, along with the activity of the proteasome, to negatively regulate the antiviral response.
KSHV is unique among viruses that infect humans in that it encodes a family of four vIRFs (41). Sequence comparisons indicate they share homology with cellular IRFs, from which they are likely to have been derived (17). The only other virus known to encode vIRFs is the KSHV-related non-human rhesus rhadinovirus (42, 43). However, all four of the KSHV vIRFs function in different ways, and vIRF-2 is not the only KSHV protein capable of suppressing the IFN antiviral response (see Ref. 16).
By accelerating the caspase-3-dependent process of IRF-3 turnover, the KSHV vIRF-2 protein functions as a catalyst in this IRF-3-degradative process, inhibiting IRF-3 transactivation and the concomitant antiviral response. The inhibition of IRF-3WT transactivation by vIRF-2, in MCF-7 cells in the absence of ectopic caspase-3 expression (Fig. 4A) and also in HEK293 cells during siRNA knockdown of caspase-3 (Fig. 3B), indicates that the viral protein can also repress IRF-3 activity independently of caspase-3; this observation was confirmed for endogenous IRF-3 (Fig. 6D). Confocal microscopy analyses suggest this caspase-3-independent activity may be due to vIRF-2 preventing IRF-3 redistribution to the nucleus (Fig. 5C). Indeed, vIRF-2 is likely multifunctional, as suggested from studies in the Pitha laboratory on the N-terminal exon-encoded protein (18, 19) that we refer to as vIRF-2×1 (17), to distinguish it from the two-exon-encoded full-length vIRF-2 protein of our present and previous (20) studies. Transactivation of the IFNA4 promoter upon Newcastle disease virus infection was inhibited by vIRF-2, likely through interacting with IRF-1 and post-translationally activated IRF-3, and/or the p300/CBP transcriptional coactivator (18). vIRF-2×1 also physically interacts with double-stranded RNA-activated protein kinase R (19). This interaction suppresses protein kinase R function, for example, reducing inhibition of eukaryotic translational initiation factor 2α activity by protein kinase R-mediated phosphorylation. Moreover, the interaction of vIRF-2 with IRF-1 identified by Burysek et al. (18) appears to be pleiotropic. Kirchhoff et al. identified positive regulatory IRF-1-dependent domains in the promoter of CD95 (Fas) ligand and recognized that vIRF-2 repressed transactivation of this promoter, perhaps through IRF-1 and NF-κB inhibition, in turn reducing activation-induced cell death (39).
A mutant of IRF-3, IRF-3(5A), which homodimerizes upon phosphorylation of the serine residues in C-terminal site 1, but lacks phosphorylation targets in site 2 (Fig. 1A, lower panel), is still depleted in the absence of vIRF-2, but accumulates in its presence (Fig. 1E). Because IRF-3(2A) and IRF-3(2A5A) are incapable of homodimerization (27) and do not interact with vIRF-2 (Fig. 1D), we infer that dimerization is necessary for IRF-3 interaction with the viral protein. However, homodimerization and vIRF-2 interaction are insufficient for vIRF-2-accelerated decay of IRF-3, because IRF-3(5A) remains abundant in the presence of the viral protein (Figs. 1E and 2A). Thus, authentic phosphorylation of IRF-3 at both C-terminal sites is required for IRF-3 turnover involving caspase-3. These data suggest the caspase-3-dependent mechanism only operates on fully phosphorylated and functional IRF-3 that has undergone complete conformational rearrangement and is competent for transcriptional activation of cognate genes; constitutively expressed, non-activated IRF-3 is disregarded.
Increasingly, the paradigm that caspases function only in a pro-apoptotic manner is being dispelled. Active caspases operate in multiple cellular processes, through both proteolytic and non-proteolytic activities, without activating apoptosis (see Refs. 30–32). These processes include cell differentiation, proliferation, and activation of inflammatory cytokines (44). Caspase-3 has non-apoptotic functions in various cell types (33, 34, 45–47), and transcription factors represent a class of non-apoptotic caspase targets (31), e.g. NFATc2 is cleaved by caspase-3 in apoptotic and non-apoptotic T cells (48).
The type I IFN-driven antiviral response can inhibit cell growth by blocking proliferation and inducing apoptosis, and it can enhance the adaptive immune response (see Ref. 1). Recent reports suggest IRF-3 itself is pro-apoptotic, providing cell death as a strategy to inhibit persistent virus infection (37). The mechanism may depend on the induction of expression of the pro-apoptotic protein, Noxa, because IRF-3 transactivates the gene encoding this protein (38). Our work has revealed caspase-3 inhibits IRF-3 by contributing to its depletion (Fig. 3) and concomitant functional inactivation (Fig. 6) and that vIRF-2 inhibits caspase-3/-7 function, as measured by cleavage of the luminogenic substrate Z-DEVD-aminoluciferin (Fig. 4C). These data suggest that vIRF-2 might sequester caspase-3 to augment IRF-3 decay, either directly or through recruitment of a protease that is in turn activated by caspase-3 and then degrades the vIRF-2/caspase-3 complex. This possibility is also supported by the data of Figs. 1E and 2A, in which the decay of incompletely activated IRF-3(5A) is inhibited by the presence of vIRF-2. We interpret this observation as IRF-3 being recruited by vIRF-2 to a caspase-3-containing complex, but the conformation of IRF-3(5A) is not amenable to its turnover by the complex. Certainly, in the context of the KSHV-infected cell, vIRF-2 expression kinetics are consistent with such a role: Krishnan et al. described vIRF-2 expression within 2 h of de novo KSHV infection (49).
Hence, vIRF-2 catalyzes caspase-3-mediated decay of IRF-3, functioning as a putative platform to recruit IRF-3 and caspase-3 and directing the proteolytic enzyme to initiate IRF-3 degradation. Whether caspase-3 acts either directly on IRF-3 to effect its depletion or, more likely, activates a proteolytic enzyme that then acts on IRF-3 remains to be established. Even if, in the KSHV-infected cell, the augmentation of this IRF-3 decay through sequestration of caspase-3 by vIRF-2 resulted in apoptosis, a panoply of anti-apoptotic KSHV factors, including vIRF-2 itself (39), would be expected to counteract it and prevent progression (16). Indeed, different KSHV proteins repress other aspects of the type I IFN-driven antiviral response. For example, the KSHV K3 and K5 proteins down-regulate cell surface expression of major histocompatibility class I molecules by augmenting their endocytosis (50–52; see Ref. 53).
In conclusion, our work reveals a cellular process for terminating IRF-3 transactivation, not previously appreciated, and a new mechanism of virus evasion of the IFN response.
Supplementary Material
Acknowledgments
We thank John Hiscott (Lady Davis Institute for Medical Research, Canada), Reiner U. Jäanicke (University of Düsseldorf, Germany), Takashi Fujita (Kyoto University, Japan), and Gemma Kelly, Claire Shannon-Lowe, and Jianmin Zuo (University of Birmingham) for generously providing reagents. Martin Rowe kindly provided reagents and helpful comments on the manuscript and reagents. The authors acknowledge the helpful suggestions of Roger Grand.
This work was supported by the Medical Research Council (Grant G0400408) and Cancer Research UK (Grant C7934) (to D. J. B.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- IFN
- interferon
- IRF
- IFN regulatory factor
- KSHV
- Kaposi sarcoma-associated herpesvirus
- KS
- Kaposi sarcoma
- vIRF
- viral IRF
- PARP
- poly(adenosine diphosphate-ribose) polymerase
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone
- EV
- empty vector
- siRNA
- small interference RNA
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Randall R. E., Goodbourn S. (2008) J. Gen. Virol. 89,1–47 [DOI] [PubMed] [Google Scholar]
- 2.Servant M. J., ten Oever B., LePage C., Conti L., Gessani S., Julkunen I., Lin R., Hiscott J. (2001) J. Biol. Chem. 276,355–363 [DOI] [PubMed] [Google Scholar]
- 3.Smith E. J., Marié I., Prakash A., García-Sastre A., Levy D. E. (2001) J. Biol. Chem. 276,8951–8957 [DOI] [PubMed] [Google Scholar]
- 4.Panne D., Maniatis T., Harrison S. C. (2007) Cell 129,1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dragan A. I., Carrillo R., Gerasimova T. I., Privalov P. L. (2008) J. Mol. Biol. 384,335–348 [DOI] [PubMed] [Google Scholar]
- 6.Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. (2003) Science 300,1148–1151 [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4,491–496 [DOI] [PubMed] [Google Scholar]
- 8.Sarkar S. N., Peters K. L., Elco C. P., Sakamoto S., Pal S., Sen G. C. (2004) Nat. Struct. Mol. Biol. 11,1060–1067 [DOI] [PubMed] [Google Scholar]
- 9.Bibeau-Poirier A., Gravel S. P., Clément J. F., Rolland S., Rodier G., Coulombe P., Hiscott J., Grandvaux N., Meloche S., Servant M. J. (2006) J. Immunol. 177,5059–5067 [DOI] [PubMed] [Google Scholar]
- 10.Hiscott J. (2007) J. Biol. Chem. 282,15325–15329 [DOI] [PubMed] [Google Scholar]
- 11.Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K. P., Yamaoka S. (2006) Nat. Immunol. 7,598–605 [DOI] [PubMed] [Google Scholar]
- 12.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418,646–650 [DOI] [PubMed] [Google Scholar]
- 13.Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003) Cell 113,803–809 [DOI] [PubMed] [Google Scholar]
- 14.Andrejeva J., Childs K. S., Young D. F., Carlos T. S., Stock N., Goodbourn S., Randall R. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101,17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkin D. M. (2006) Int. J. Cancer 118,3030–3044 [DOI] [PubMed] [Google Scholar]
- 16.Aresté C., Blackbourn D. J. (2009) Trends Microbiol. 17,119–129 [DOI] [PubMed] [Google Scholar]
- 17.Cunningham C., Barnard S., Blackbourn D. J., Davison A. J. (2003) J. Gen. Virol. 84,1471–1483 [DOI] [PubMed] [Google Scholar]
- 18.Burysek L., Yeow W. S., Pitha P. M. (1999) J. Hum. Virol. 2,19–32 [PubMed] [Google Scholar]
- 19.Burýsek L., Pitha P. M. (2001) J. Virol. 75,2345–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuld S., Cunningham C., Klucher K., Davison A. J., Blackbourn D. J. (2006) J. Virol. 80,3092–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin R., Mamane Y., Hiscott J. (1999) Mol. Cell. Biol. 19,2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoneyama M., Suhara W., Fukuhara Y., Fukuda M., Nishida E., Fujita T. (1998) EMBO J. 17,1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamura T., Yoneyama M., Yamaguchi K., Suhara W., Mori W., Shiota K., Okabe Y., Namiki H., Fujita T. (2001) Genes Cells 6,375–388 [DOI] [PubMed] [Google Scholar]
- 24.Kelly G. L., Milner A. E., Tierney R. J., Croom-Carter D. S., Altmann M., Hammerschmidt W., Bell A. I., Rickinson A. B. (2005) J. Virol. 79,10709–10717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panne D., McWhirter S. M., Maniatis T., Harrison S. C. (2007) J. Biol. Chem. 282,22816–22822 [DOI] [PubMed] [Google Scholar]
- 26.Qin B. Y., Liu C., Lam S. S., Srinath H., Delston R., Correia J. J., Derynck R., Lin K. (2003) Nat. Struct. Biol. 10,913–921 [DOI] [PubMed] [Google Scholar]
- 27.Mori M., Yoneyama M., Ito T., Takahashi K., Inagaki F., Fujita T. (2004) J. Biol. Chem. 279,9698–9702 [DOI] [PubMed] [Google Scholar]
- 28.Hilton L., Moganeradj K., Zhang G., Chen Y. H., Randall R. E., McCauley J. W., Goodbourn S. (2006) J. Virol. 80,11723–11732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saira K., Zhou Y., Jones C. (2007) J. Virol. 81,3077–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham M. C., Shaham S. (2004) Trends Cell Biol. 14,184–193 [DOI] [PubMed] [Google Scholar]
- 31.Lamkanfi M., Festjens N., Declercq W., Vanden Berghe T., Vandenabeele P. (2007) Cell Death Differ 14,44–55 [DOI] [PubMed] [Google Scholar]
- 32.Li J., Yuan J. (2008) Oncogene 27,6194–6206 [DOI] [PubMed] [Google Scholar]
- 33.Fujita J., Crane A. M., Souza M. K., Dejosez M., Kyba M., Flavell R. A., Thomson J. A., Zwaka T. P. (2008) Cell Stem Cell 2,595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janzen V., Fleming H. E., Riedt T., Karlsson G., Riese M. J., Lo Celso C., Reynolds G., Milne C. D., Paige C. J., Karlsson S., Woo M., Scadden D. T. (2008) Cell Stem Cell 2,584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi K., Kawai T., Kumar H., Sato S., Yonehara S., Akira S. (2006) J. Immunol. 176,4520–4524 [DOI] [PubMed] [Google Scholar]
- 36.Lakhani S. A., Masud A., Kuida K., Porter G. A., Jr., Booth C. J., Mehal W. Z., Inayat I., Flavell R. A. (2006) Science 311,847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters K., Chattopadhyay S., Sen G. C. (2008) J. Virol. 82,3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goubau D., Romieu-Mourez R., Solis M., Hernandez E., Mesplède T., Lin R., Leaman D., Hiscott J. (2009) Eur. J. Immunol. 39,527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirchhoff S., Sebens T., Baumann S., Krueger A., Zawatzky R., Li-Weber M., Meinl E., Neipel F., Fleckenstein B., Krammer P. H. (2002) J. Immunol. 168,1226–1234 [DOI] [PubMed] [Google Scholar]
- 40.Jänicke R. U., Sprengart M. L., Wati M. R., Porter A. G. (1998) J. Biol. Chem. 273,9357–9360 [DOI] [PubMed] [Google Scholar]
- 41.Russo J. J., Bohenzky R. A., Chien M. C., Chen J., Yan M., Maddalena D., Parry J. P., Peruzzi D., Edelman I. S., Chang Y., Moore P. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93,14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Searles R. P., Bergquam E. P., Axthelm M. K., Wong S. W. (1999) J. Virol. 73,3040–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander L., Denekamp L., Knapp A., Auerbach M. R., Damania B., Desrosiers R. C. (2000) J. Virol. 74,3388–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller M., Rüegg A., Werner S., Beer H. D. (2008) Cell 132,818–831 [DOI] [PubMed] [Google Scholar]
- 45.Wilhelm S., Wagner H., Häcker G. (1998) Eur. J. Immunol. 28,891–900 [DOI] [PubMed] [Google Scholar]
- 46.Rosado J. A., Lopez J. J., Gomez-Arteta E., Redondo P. C., Salido G. M., Pariente J. A. (2006) J. Cell. Physiol. 209,142–152 [DOI] [PubMed] [Google Scholar]
- 47.Acarin L., Villapol S., Faiz M., Rohn T. T., Castellano B., González B. (2007) Glia 55,954–965 [DOI] [PubMed] [Google Scholar]
- 48.Wu W., Misra R. S., Russell J. Q., Flavell R. A., Rincón M., Budd R. C. (2006) J. Biol. Chem. 281,10682–10690 [DOI] [PubMed] [Google Scholar]
- 49.Krishnan H. H., Naranatt P. P., Smith M. S., Zeng L., Bloomer C., Chandran B. (2004) J. Virol. 78,3601–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coscoy L., Ganem D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97,8051–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishido S., Wang C., Lee B. S., Cohen G. B., Jung J. U. (2000) J. Virol. 74,5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson P. G., Efstathiou S., Doherty P. C., Lehner P. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97,8455–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rezaee S. A., Cunningham C., Davison A. J., Blackbourn D. J. (2006) J. Gen. Virol. 87,1781–1804 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.