Abstract
A role for Wiskott-Aldrich syndrome protein (WASP) in chemotaxis to various agents has been demonstrated in monocyte-derived cell types. Although WASP has been shown to be activated by multiple mechanisms in vitro, it is unclear how WASP is regulated in vivo. A WASP biosensor (WASPbs), which uses intramolecular fluorescence resonance energy transfer to report WASP activation in vivo, was constructed, and following transfection of macrophages, activation of WASPbs upon treatment with colony-stimulating factor-1 (CSF-1) was detected globally as early as 30 s and remained localized to protrusive regions at later time points. Similar results were obtained when endogenous WASP activation was determined using conformation-sensitive antibodies. In vivo CSF-1-induced WASP activation was fully Cdc42-dependent. Activation of WASP in response to treatment with CSF-1 was also shown to be phosphatidylinositol 3-kinase-dependent. However, treatment with the Src family kinase inhibitors PP2 or SU6656 or disruption of the major tyrosine phosphorylation site of WASPbs (Y291F mutation) did not reduce the level of CSF-1-induced WASP activation. Our results indicate that WASP activation downstream of CSF-1R is phosphatidylinositol 3-kinase- and Cdc42-dependent consistent with an involvement of these molecules in macrophage migration. However, although tyrosine phosphorylation of WASP has been proposed to stimulate WASP activity, we found no evidence to indicate that this occurs in vivo.
Macrophages, terminally differentiated cells of the mononuclear phagocytic lineage, are found throughout the body and play important roles in normal tissue development and immune defense. However, in certain circumstances, excessive recruitment of macrophages has been shown to participate in the progression of several diseases, inflammatory (rheumatoid arthritis) or metabolic (atherosclerosis), as well as in tumor progression (1–3). Importantly expression of colony-stimulating factor-1 (CSF-1),4 the most pleiotropic macrophage growth factor, has been correlated with the progression of these disease states (for a review, see Ref. 4). Inhibition of undesirable macrophage recruitment to specific sites in response to CSF-1 is therefore an attractive goal for therapies (5).
In addition to stimulating survival, proliferation, and differentiation of monocytes and macrophages, CSF-1 is also a potent chemotactic factor inducing the migration of these cell types (for a review, see Ref. 4). CSF-1 stimulation leads to the rapid production of F-actin-rich protrusions and the spreading and migration of macrophages (4). All CSF-1 effects are mediated through its tyrosine kinase receptor (CSF-1R), which upon activation leads to phosphorylation of tyrosine residues in a number of signaling molecules. Downstream molecules essential for macrophage migration in response to CSF-1 include phosphatidylinositol 3-kinase (PI3K) isoforms β and δ (6, 7). PI3K may potentially regulate migration through the activation of guanine nucleotide exchange factor activity to Rac1 and Cdc42, which are required for CSF-1-elicited protrusions (8, 9) and chemotaxis (10). The major means by which Rac and Cdc42 regulate the Arp2/3 complex is through the Wiskott-Aldrich syndrome protein/Wiskott-Aldrich syndrome verprolin-homologous (WASP/WAVE) family of proteins (11). A Rac1-IRSp53-Abi1-WAVE2 complex has been shown to mediate CSF-1-induced macrophage motility (12, 13), and a unique role for WASP in macrophage chemotaxis to CSF-1, formylmethionylleucylphenylalanine, MCP-1, and MIP-1α has been demonstrated (14, 15). WASP is a hematopoietic cell-specific regulator of Arp2/3-dependent actin remodeling. The catalytically active domain of WASP lies in its C terminus, which is conserved among all WASP/WAVE proteins and contains a VCA (verprolin homology, cofilin-like, and acidic region) domain capable of activating the Arp2/3 complex. The other domains found in WASP can regulate, directly or indirectly, the activity of its VCA domain (for a review, see Ref. 16). Both WASP and N-WASP bind activated Cdc42 through their GTPase-binding domain, which is believed to cause a structural transition that results in dissociation of the intramolecular contacts leaving the VCA domain accessible for Arp2/3 binding (17, 18). In addition, biochemical studies have revealed that several signaling molecules, including WASP-interacting SH3 protein, WASP-interacting protein, Grb2, phosphoinositides, and Src family kinases, activate N-WASP (for reviews, see Refs. 16 and 19). Phosphorylation of WASP has also been proposed to activate Arp2/3-mediated actin polymerization in vitro (20–22).
Recently different probes have been developed that detect a conformational change in N-WASP and therefore reflect its activation (23–25). Using either a fluorescence resonance energy transfer (FRET)-based biosensor that detects a conformational change in N-WASP (23, 24) or antibodies that can only bind to the open conformation of N-WASP (25), N-WASP has been shown to be activated in response to epidermal growth factor in HEK293 cells and in MTLn3 carcinoma cells. This activity has been temporally localized to subcellular compartments important for carcinoma cell chemotaxis and invasion (24). We have adapted these approaches to explore the signal transduction pathways responsible for the activation of WASP in vivo.
EXPERIMENTAL PROCEDURES
Constructs
The WASP biosensor (WASPbs) construct was generated according to a previously described procedure (24). Briefly human WASP was used as a template for PCR amplification and insertion into the ECFP-EYFP-biosensor vector (24). Point mutations were introduced using the QuikChange kit from Stratagene (La Jolla, CA) according to the manufacturer's instructions.
Cells and Transfections
COS cells and HEK293 cells (ATCC, Manassas, VA) were cultured according to their specifications. RAW/LR5 cells were derived from the murine monocyte/macrophage RAW 264.7 cell line (8) and were grown in RPMI 1640 medium (Mediatech, Inc.) containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Sigma). All cells were maintained at 37 °C in a 5% CO2 incubator. Transient transfections were performed using the SuperFect reagent from Qiagen according to the manufacturer's instructions. Reduction of Cdc42 expression was achieved through the retroviral infection of RAW/LR5 cells with short hairpin RNAs directed against the Cdc42 mRNA using pSUPER.reto.puro plasmids (Oligoengine, Seattle, WA).5
Cell Stimulation and Immunofluorescence Staining
For all experiments involving a CSF-1 stimulation cells were treated according to Cox et al. (8). 20 ng/ml murine recombinant CSF-1 (R&D Systems, Minneapolis, MN) was added or not to the cells for the indicated times at 37 °C. Following fixation in 3.7% formaldehyde and permeabilization in 0.2% Triton X-100 cells were stained using either WASP/N-WASP conformation-sensitive antibody (CSA) (25) or anti-Myc antibodies (Roche Applied Science) followed by incubation with labeled secondary antibodies and Alexa Fluor 568 phalloidin (Molecular Probes, Invitrogen). Mean fluorescence intensity of entire cells was measured at 20× and plotted versus time after CSF-1 addition.
Microscopy and FRET Analysis
Spectral analysis of WASPbs expressing HEK293 cells (see Fig. 1B) was done using a Leica TCS SP2 AOBS confocal microscope with a 63×, numerical aperture 1.4 objective (Leica Microsystems, Exton, PA). Cells were illuminated with a 405 nm laser, and fluorescence was detected from 445 (440–450) to 595 nm (590–600) with a 10-nm-wide slit stepping up the spectrum. The intensity of the whole image was plotted versus the wavelength to get the fluorescence spectra and normalized to the signal at 495 nm.
FIGURE 1.
Characterization of the WASPbs. A, schematic of the WASPbs where, upon illumination of CFP, FRET can occur from CFP to YFP in the closed conformation of the construct. GBD, GTPase-binding domain. B, after cotransfection of HEK293 cells with the WASPbs and either constitutively active (Q61L; black squares) or dominant-negative (N17; white dots) Cdc42, fluorescence spectra of transfected cells following excitation of CFP were measured by confocal microscopy. Quantification of fluorescence spectra was performed by plotting the intensity of the whole image versus the wavelength. In the presence of constitutively active Cdc42 a decrease in the YFP peak (515–525 nm) was observed indicating a loss of FRET upon WASPbs activation. C, COS cells were cotransfected with the WASPbs and constitutively active (Q61L) Cdc42 and then incubated in either DMSO (black bar) or 5 μm wiskostatin (WISK; white bar) for 1 h. Following fixation, WASP activity was deter mined as the ratio of the donor fluorescence over FRET (see “Experimental Procedures” for a detailed description). Data from one representative experiment, reported as donor/FRET, are expressed as mean ± S.E. (error bars) (n = 17 and 19 cells, respectively; *, p = 0.007). AU, arbitrary units.
FRET image sequences of WASPbs-expressing cells were obtained essentially as described in Lorenz et al. (24). Acquisition was performed with IP Lab v3.51 (Scanalytics Inc.), and FRET analyses were performed with IP Lab v3.51 and with ImageJ (W. S. Rasband, ImageJ, National Institutes of Health, Bethesda, MD, 1997–2006). For ratiometric FRET analysis, after background subtraction the total cellular donor fluorescence intensity was divided by the total FRET fluorescence intensity. Only cells expressing low levels of the WASPbs, as measured by the acceptor fluorescence intensity, were analyzed because overexpression of WASP induced artifacts similar to those reported for cells overexpressing N-WASP (24). Absolute FRET values were variable between experiments due to varying illumination intensity and exposure conditions. Therefore we only compared paired conditions within the same experiment to avoid instrument or other variability that was not related to specific regulation of WASP activity. To compensate for this, for each experiment, each condition was compared with the unstimulated condition of the same experiment and expressed as a percentage. The percentage of control values from at least three independent experiments were subsequently averaged together. Results were then reported as donor/FRET values for individual experiments or as percent change compared with the basal (or unstimulated) controls when multiple experiments were compared.
Immunoprecipitation and Western Blotting
RAW/LR5 cells (see Fig. 5A) were pretreated with 10 μm PP2 (Calbiochem, EMD Bioscience) or with DMSO (vehicle) for 1 h at 37 °C before being subjected to CSF-1 stimulation as described above. COS cells transfected with the indicated WASPbs constructs with or without Myc-Cdc42Q61L (see Fig. 6B) or RAW/LR5 cells transfected with Myc-WASP (see Fig. 5A) were incubated in the presence or absence of 12 μm pervanadate for 30 min at 37 °C according to Cory et al. (21). Cells were lysed in ice-cold lysis buffer (1% Triton X-100, 25 mm Tris, 137 mm NaCl, 2 mm EDTA, 1 mm orthovanadate, 1 mm benzamidine, 10 μg/ml aprotinin, and 10 μg/ml leupeptin, pH 7.4). Immunoprecipitations were carried out by incubating the cleared cell lysates at 4 °C with the appropriate antibody prebound to protein A/G-agarose beads (Santa Cruz Biotechnology). Samples were resolved by SDS-PAGE, transferred onto polyvinylidene difluoride membranes (Immobilon-P, Millipore) followed by incubation with the indicated primary antibodies and secondary antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA). Signals were detected using the Super Signal West Pico chemiluminescent substrate from Pierce, and images were acquired and analyzed using a Kodak Image Station 440. Antibodies used for these applications were as follows: anti-phosphotyrosine (Tyr(P)-99, Santa Cruz Biotechnology), anti-phospho-Hck (Tyr-411, Santa Cruz Biotechnology), anti-green fluorescent protein or anti-Myc (Roche Applied Science), anti-β-actin (AC-15, Sigma), and anti-WASP (B19, Santa Cruz Biotechnology).
FIGURE 5.
Although Hck is activated in response to CSF-1, WASP activation downstream of CSF-1 is Src family kinase (SFK)-independent. A, RAW/LR5 cells were treated for various times with CSF-1 prior to lysis and immunoprecipitation (IP) of phosphotyrosine (PY)-containing proteins as described under “Experimental Procedures.” Immunoprecipitates were then subjected to Western blotting using a phosphospecific Hck (P-Hck) antibody that detects only the activated form of Hck (upper panel). Actin content of the whole cell lysate is shown as a loading control. Cells pretreated with DMSO (control) or 10 μm PP2 were incubated with CSF-1 for the indicated times prior to lysis, and Hck activation was determined as above (middle panel). RAW/LR5 cells expressing Myc-WASP were preincubated with 10 μm PP2 and then treated with 12 μm pervanadate (PV) for 30 min where indicated. After cell lysis, WASP was immunoprecipitated using Myc antibodies followed by Western blotting for either phosphotyrosine or WASP (lower panel). B, RAW/LR5 cells expressing the WASPbs were pretreated with DMSO (control) or 10 μm PP2 prior to CSF-1 stimulation for 30 s. After fixation WASP activation was determined and is represented as percent change compared with unstimulated DMSO-treated cells (n ≥ 4 independent experiments; ±S.E. (error bars)). C, RAW/LR5 cells were pretreated with either DMSO, 10 μm PP2, 10 μm SU6656, or 100 μm LY294002 (LY) and then stimulated with CSF-1 for 30 s prior to fixation. Cells were subsequently stained with CSA (n = 5 independent experiments; ±S.E.). *, statistically significant difference when compared with unstimulated control cells.
FIGURE 6.
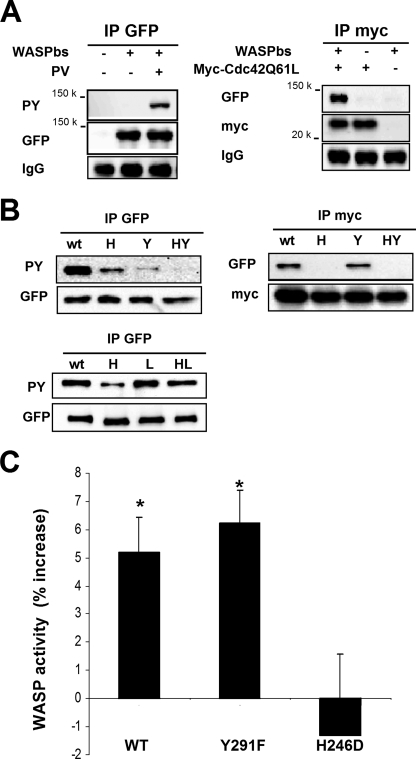
WASP activation downstream of CSF-1 is independent of WASP phosphorylation. A, WASPbs-expressing RAW/LR5 cells were treated or not with 12 μm pervanadate (PV) for 30 min (upper left panel) or cotransfected with Myc-Cdc42Q61L (upper right panel). After cell lysis, WASPbs or Cdc42Q61L was immunoprecipitated (IP) using green fluorescent protein (GFP) or Myc tag antibodies, respectively. Samples were subjected to Western blotting with the indicated antibodies. B, similar experiments were performed to compare WASPbs wild type (wt), H246D (H), Y291F (Y), H246D/Y291F (HY), L270P (L), and L270P/H246D (LH) mutants in their ability to bind Cdc42Q61L and to be tyrosine-phosphorylated. The indicated versions of the WASPbs were expressed in COS-7 cells, which were either subsequently treated with pervanadate (upper and lower left panels) or cotransfected with Cdc42Q61L (upper right panel). C, RAW/LR5 cells expressing WASPbs wild type (WT), Y291F, or H246D were subjected to CSF-1 stimulation for 30 s. After fixation WASP activation was determined and is represented as percent change compared with their respective unstimulated conditions (n ≥ 4 independent experiments; ±S.E. (error bars)). *, statistically significant difference when compared with unstimulated control cells. PY, phosphotyrosine.
Data Analysis
All data are represented as the mean, and error bars represent the S.E. Significance was analyzed using paired Student's t test, and differences between two means with a p value <0.05 were considered significant.
RESULTS
Characterization of the WASP Biosensor
To specifically address how WASP activity is regulated in vivo we have constructed a FRET-based WASPbs. Based on the known autoinhibitory conformation of both WASP and N-WASP, the WASPbs was designed to exhibit intramolecular FRET between cyan fluorescent protein (CFP; donor) and yellow fluorescent protein (YFP; acceptor) fused to the N or C termini of full-length WASP, respectively (see Fig. 1A); it is similar to the previously described N-WASP biosensor (24). Activation of WASP induces a conformational change that would result in increasing the CFP-YFP distance leading to a decreased FRET signal (Fig. 1A). Validating the functionality of the biosensor, HEK293 cells expressing the WASPbs alone (data not shown) or in the presence of dominant-negative Cdc42 (Cdc42N17) exhibited two emission peaks at 475 (CFP) and 525 nm (YFP) after excitation of CFP (Fig. 1B). This observation is consistent with WASP being present in a closed conformation and permitting FRET. Moreover in the presence of constitutively active Cdc42 (Cdc42Q61L) a reduction in the YFP peak at 525 nm was observed suggesting that a structural change had occurred upon Cdc42Q61L binding (Fig. 1B). To confirm that a decrease in FRET was indicative of a conformational change in the WASPbs, experiments were performed using the WASP inhibitor wiskostatin that binds to and stabilizes the closed, autoinhibited conformation of WASP (26). Following fixation of COS-7 cells cotransfected with WASPbs and Cdc42Q61L, WASP activity was determined as the loss of FRET and quantified as a ratio of the donor emission divided by the FRET emission (see “Experimental Procedures” for more details). Pretreatment of the cells with 5 μm wiskostatin inhibited the activation of the WASPbs induced by Cdc42Q61L (Fig. 1C) indicating that changes in FRET observed in the presence of Cdc42Q61L reflect changes in WASPbs conformation.
Murine monocyte/macrophage cells, RAW/LR5 (8), were transfected with the WASPbs and then stimulated with CSF-1 for various times before fixation. As shown in Fig. 2A, WASP activation was detected globally as early as 30 s following CSF-1 addition and appeared to be localized to protrusive regions at later times. Quantification of whole cell donor/FRET signals indicated a significant peak in WASP activity at 30 s after CSF-1 addition (Fig. 2, B and C). These results were confirmed using fluorescent lifetime imaging microscopy as an alternative method (not shown). The biosensor had a FRET efficiency in the inactive conformation of ∼19%, similar to the FRET efficiency observed for the highly structural homologue N-WASPbs (24), and 5% after activation, corresponding to an estimated dye-to-dye distance of 6.4 and 8.2 nm, respectively (data not shown).
FIGURE 2.
WASP is activated rapidly following CSF-1 addition. A, RAW/LR5 cells transfected with the WASPbs were fixed, and WASP activity was determined as the ratio of donor/FRET emission. Global transition of the WASPbs from an inactive (blue color) to active (green and red colors) state was visualized by microscopy, whereas hot spots of WASP activity were found in discreet subcellular regions only (red signals). Both phase (upper panels) and ratio images (lower panels) are shown for the indicated times. The inset is a higher magnification of the boxed region showing that WASP activation is enriched in CSF-1-induced protrusions. Bar, 10 μm. B, WASP activity of cells treated with or without CSF-1 for 30 s was quantified and reported as the donor/FRET ratio and expressed as mean ± S.E. (error bars) (n = 39 and 35 cells, respectively, from one representative experiment). *, p < 1 × 10−7. C, quantification of WASP activity was determined by measuring donor/FRET signals (n ≥ 3 independent experiments; n ≥ 30 cells per experiment). Data (mean ± S.E.) are represented as percent change compared with unstimulated WASPbs-expressing cells. ch., channel.
To ensure that the WASPbs was reflecting the activation of endogenous WASP in RAW/LR5 cells, a CSA was used as an alternative approach to determine the activation state of endogenous WASP. This antibody was generated against a region only accessible when the protein is in an opened, active conformation (25). Consistent with a highly conserved GTPase-binding domain between N-WASP and WASP, CSA was able to detect WASP by Western blot in RAW/LR5 (data not shown). However, because it has been reported that a small amount of N-WASP is also expressed in macrophages (27) the affinity of the CSA for WASP and N-WASP was determined by examining lysates of COS cells transfected with either the WASPbs used in this study or the previously reported N-WASP biosensor (24). The affinity of CSA for either WASP or N-WASP was determined as a ratio of CSA staining intensity of WASP/N-WASP following normalization for the expression of the constructs using anti-green fluorescent protein (supplemental Fig. 1). The 5-fold higher affinity of the CSA for WASP and the fact that WASP is expressed at 15 times the level of N-WASP in RAW/LR5 cells6 suggest that this reagent can be used to determine the activation state of endogenous WASP. When used in immunofluorescence experiments on fixed cells, CSA detected an increase of WASP activity 30 s after CSF-1 addition with hot spots of active WASP observed in CSF-1-elicited protrusions (Fig. 3A). Quantification of stained cells indicated a significant increase in fluorescence intensity 30 s following CSF-1 addition that decreased to base-line levels by 5 min (Fig. 3B), thus paralleling the rapid and transient CSF-1-induced activation of WASP as measured with the FRET biosensor.
FIGURE 3.
Endogenous WASP is activated following CSF-1 addition with kinetics similar to that of the WASPbs. A, RAW/LR5 cells were stimulated with CSF-1 for various times prior to fixation and staining with CSA and phalloidin. Representative images are shown for either unstimulated (upper panels) or CSF-1-treated conditions (lower panels). Note the colocalization of CSA and F-actin in protrusions following CSF-1 addition as indicated by white arrows. Bar, 10 μm. B, percent increase in CSA intensity per cell is reported as average ± S.E. (error bars) (n = 6 independent experiments).
Cdc42 Dependence of WASP Activation in Response to CSF-1
Activated Cdc42 has been shown to bind to and activate WASP and N-WASP in vitro (18). Therefore the involvement of Cdc42 in WASP activation in vivo was assessed. Cotransfection of RAW/LR5 cells with WASPbs and Cdc42N17 resulted in a decrease in WASP activity compared with cells expressing the WASPbs alone (Fig. 4A). This is consistent with an interpretation that despite serum starvation there always was some constitutive level of WASP activity in resting cells. Importantly treatment with CSF-1 was unable to rescue the inhibition of WASP activity in this situation indicating that CSF-1 signaling to WASP cannot bypass Cdc42 in these cells (Fig. 4A). Conversely RAW/LR5 cells showed an enhanced WASP activity when cotransfected with WASPbs and constitutively active Cdc42 (Cdc42V12) compared with WASPbs alone-expressing cells. In addition, CSF-1 stimulation did not potentiate the Cdc42V12-dependent increase in WASPbs activity (Fig. 4A), suggesting that the activation of WASPbs was already maximal. Similar results were also obtained with Cdc42Q61L (data not shown). To confirm a specific role for Cdc42 in WASP activation the ability of CSF-1 to induce WASP activation in cells with reduced endogenous levels of Cdc42 was determined. Cdc42 levels were stably reduced by greater than 70% through expression of a short hairpin RNA sequence targeting Cdc42.5 There was no significant increase in WASP activation following CSF-1 addition as determined using the CSA in cells with reduced levels of Cdc42 (Fig. 4B). Control infected cells showed a level of WASP activation in response to CSF-1 similar to that of non-infected cells (compare Figs. 4B and 3B). Altogether these data clearly demonstrate that WASP activation in response to CSF-1 is Cdc42-dependent.
FIGURE 4.
WASP activation downstream of CSF-1 is Cdc42- and PI3K-dependent. A, RAW/LR5 cells were transfected with the WASPbs alone or with either constitutively active (V12) or dominant-negative (N17) Cdc42. Cells were left unstimulated (white bars) or subjected to CSF-1 stimulation for 30 s (black bars). Quantification of WASP activity was determined and is represented as percent change compared with unstimulated cells expressing the WASPbs alone. A minimum of 30 cells were imaged for each condition per experiment (n ≥ 3 independent experiments; ±S.E. (error bars)). B, cells infected with non-interfering short hairpin RNA (Ctrl sh) or cells with reduced levels of Cdc42 (Cdc42 sh#1) were treated with CSF-1 for 30 s followed by staining with CSA. Percent increase in CSA intensity per cell is reported as average ± S.E. (n = 3 independent experiments; ≥100 cells per experiment). *, p < 0.05. C, RAW/LR5 cells expressing the WASPbs alone or with constitutively active Cdc42 (V12) were pretreated with DMSO (control) or 100 nm wortmannin (WM) prior to CSF-1 stimulation for 30 s. Data are represented as percent change compared with unstimulated DMSO-treated cells. A minimum of 20 cells were imaged for each condition per experiment and averaged (n ≥ 4 independent experiments; ±S.E.). *, statistically significant difference when compared with the condition without wortmannin.
Requirement for PI3K in WASP Activation
It has been speculated that following treatment of monocytes/macrophages with CSF-1 PI3K activates guanine nucleotide exchange factors that in turn activate the Rho GTPases Rac and Cdc42 that are required for chemotaxis (28). Because Cdc42 binding appeared to be essential for WASP activity in RAW/LR5 cells, we then asked whether its putative activator PI3K was indeed controlling WASP activity in our system. To address this question, cells expressing the WASPbs were preincubated in the presence or absence of PI3K inhibitors, wortmannin, or LY294002 before CSF-1 stimulation for 30 s, and the WASP activation was measured. PI3K inhibition completely blocked WASP activation induced by CSF-1 as measured with either the WASPbs (Fig. 4C) or CSA (Fig. 5C). Expression of Cdc42V12 in WASPbs-expressing cells was able to bypass the requirement of PI3K for WASP activation in response to CSF-1 (Fig. 4B) consistent with a situation where PI3K is upstream of Cdc42, which itself is required for the activation of WASP.
Tyrosine Phosphorylation and WASP Activation
Recent reports have suggested that there are also Cdc42-independent mechanisms of WASP activation. Tyrosine phosphorylation of N-WASP or WASP or introduction of a phosphomimicking mutation has been shown to stimulate their activities in vitro (21, 29). The hematopoietic cell-specific Src family kinase Hck has been proposed to phosphorylate and activate WASP in macrophages (21). Immunoprecipitation experiments were performed to identify activated Hck in the phosphotyrosine-containing fraction of RAW/LR5 cell lysates after addition of CSF-1. Hck was activated after addition of CSF-1 with kinetics similar to that of WASP activation (Fig. 5A). As a control, we verified that the CSF-1-induced increase in Hck tyrosine phosphorylation was abolished when RAW/LR5 cells were pretreated with the Src family kinase inhibitor PP2 (Fig. 5A, middle panel). In addition, tyrosine phosphorylation of Myc-tagged WASP expressed in RAW/LR5 cells was shown to be fully dependent on Src family kinase activity (Fig. 5A, lower panel). Then to determine whether Src family kinases were required for WASP activation in vivo, cells expressing the WASPbs were pretreated with PP2, and the ability of CSF-1 to activate WASP was analyzed. At concentrations of PP2 that effectively inhibited CSF-1-induced Hck activation, PP2 had no effect on either the basal level or the CSF-1-induced activation of WASP as shown in Fig. 5B. In addition, there was no significant difference in the intensity of staining using the CSA between CSF-1-stimulated cells in the presence or absence of PP2 or SU6656, a structurally unrelated pan-Src family kinase inhibitor (Fig. 5C). Altogether these data clearly demonstrate that Src family kinases are not required for WASP activation in response to CSF-1.
Because several other tyrosine kinases have been shown to have the ability to phosphorylate WASP the major tyrosine-phosphorylated site of WASP (21) was mutated to phenylalanine (Y291F) in the biosensor to assess the functional relevance of phosphorylation in WASP activity in vivo. In addition and to confirm our results on the role of Cdc42, the Cdc42 binding site (30) of the WASPbs was mutated alone (H246D) or in combination with the tyrosine residue (H246D/Y291F). The effects of these mutations on either Cdc42 binding or phosphorylation of WASP were tested in COS-7 cells expressing the different WASPbs constructs by co-expressing Cdc42Q61L and/or treating the cells with pervanadate to induce global tyrosine phosphorylation (21) prior to immunoprecipitation of the biosensor or Cdc42Q61L (Fig. 6A). Verifying the efficacy of the GTPase-binding domain mutation, WASPbs H246D was incapable of binding Cdc42Q61L. Meanwhile the Y291F mutation had no effect on the ability of the WASPbs to bind Cdc42 (Fig. 6B). Confirming the status of this tyrosine residue as a major target for kinases in WASP, the Y291F mutation resulted in massively impaired pervanadate-induced phosphorylation, and this residual phosphorylation was abrogated in the double mutant (H246D/Y291F) (Fig. 6B). These results indicated that the biosensor mutations had the desired effect of blocking either Cdc42 binding and/or phosphorylation, and therefore these probes were suitable for the analysis of the role of Cdc42 and tyrosine phosphorylation in WASP activation in response to CSF-1 stimulation.
It should be noted that there was an apparent reduction in the level of pervanadate-induced tyrosine phosphorylation in the H246D mutant (Fig. 6B), and the presence of Cdc42 enhanced the level of WASP tyrosine phosphorylation induced by pervanadate (data not shown). Others have reported similar results on the facilitating role of Cdc42 in the tyrosine phosphorylation of N-WASP in vitro by relieving the autoinhibition and allowing N-WASP to be accessible for subsequent phosphorylation (22, 31). To address this possibility a constitutively open conformation of the WASPbs was generated by the replacement of leucine 270 with a proline residue (32). Interestingly the open conformation of WASPbs (L270P) had an ability to be phosphorylated that was retained in the absence of Cdc42 binding (L270P/H246D) (Fig. 6B). These results are consistent with a role for Cdc42 in facilitating WASP phosphorylation by inducing an open conformation.
The different versions of WASPbs were then transfected in RAW/LR5 cells prior to stimulation with CSF-1 for 30 s, fixation, and measurement of WASP activation as described above. Expression levels, assessed by YFP fluorescence intensity, were similar for all WASPbs constructs. An increase in WASPbs Y291F activity following stimulation was observed and was equivalent to the obtained increase of the unaltered WASPbs (Fig. 6B). Consistent with this result, WASPbs Y291F activity was also present in CSF-1-elicited protrusions at later times, and induction of tyrosine phosphorylation with pervanadate in WASPbs-expressing cells had no effect on the level of WASP activity compared with untreated cells (data not shown). Also expression of a phosphomimetic version of WASPbs (Y291E) did not show an enhanced basal level of WASP activity as compared with WASPbs wild type or Y291F (supplemental Fig. 2). Conversely CSF-1-induced activation of WASP was abolished when using the WASPbs H246D mutant (Fig. 6B) confirming the results obtained using Cdc42N17 (Fig. 4). Similar results were obtained using the double WASPbs mutant (H246D/Y291F) (data not shown). Overall the data obtained with the mutated biosensors were consistent with those obtained with the Src family kinase inhibitors (PP2 and SU6656) or with co-expression of the Cdc42 mutants and demonstrated that activation of WASP in response to CSF-1 was independent of tyrosine phosphorylation but tightly regulated by Cdc42.
DISCUSSION
WASP, a known regulator of Arp2/3-dependent actin assembly, is specifically expressed in hematopoietic cells where it controls chemotaxis among other processes. Although the functional importance of WASP is indisputable, the relevant signaling pathways triggered by physiological stimulants and leading to its activation are not unambiguously established as many studies are essentially based on biochemical in vitro assays. To address these issues, we constructed and characterized a novel FRET-based biosensor to report WASP activity in vivo. This report describes for the first time its utilization to study how WASP is regulated in a monocyte/macrophage cell line responding to a physiological stimulant, namely CSF-1. The biological relevance of the biosensor is further supported by the fact that both WASPbs and endogenous WASP were activated in response to CSF-1 with similar kinetics.
Previous studies have shown that WASP isolated from neutrophils was able to bind purified activated Cdc42, and overexpression of WASP in normal rat kidney and porcine aortic endothelial cells is able to induce actin assembly in a Cdc42-dependent manner, suggesting that WASP is a downstream effector of Cdc42 (33). Cdc42 is required for protrusions induced by the chemoattractant formylmethionylleucylphenylalanine in neutrophils through activation of the Arp2/3 complex (34) potentially through WASP. However, the relevance of the Cdc42-WASP axis in the control of actin polymerization has been questioned by several studies as overexpression of a mutated WASP that cannot bind Cdc42 still induces actin polymerization in COS cells (30), and more importantly, expression of this mutant in WASP-deficient T cells is capable of rescuing the actin defects due to WASP deficiency to the same extent as expression of wild-type WASP (35). The reasons for these discrepancies are unclear, but it should be noted that all these reports do not directly evaluate the activation state of WASP but solely rely on actin polymerization as a consequence of WASP activation. An important advantage of the biosensor is that it directly reports WASP activation instead of a downstream outcome that can result from additional causes and allows us to dissect the signal transduction pathways connecting activation of the CSF1-R to activation of WASP. Following CSF-1 stimulation, PI3K is required for activation of Cdc42 leading to its subsequent binding to and activation of WASP, strongly supporting the initial speculation that the role of PI3K in CSF-1-induced chemotaxis is mediated by Rho GTPases (28).
Several studies have suggested that tyrosine phosphorylation of WASP may participate in the initiation or the duration of WASP activity. Others have shown that WASP phosphorylation depends on the presence of Cdc42 as it is believed that Cdc42 binding is required to relieve the autoinhibition of WASP to allow it to be accessible for subsequent phosphorylation (22, 31, 36). Our data are consistent with this latter proposition because invalidation of the Cdc42 binding site of WASP (H246D mutation) not only inhibited its interaction with Cdc42 but also reduced the level of WASP phosphorylation in response to pervanadate (Fig. 6). In addition, the presence of constitutively active Cdc42 enhanced the level of WASP tyrosine phosphorylation induced by pervanadate (data not shown), and WASPbs phosphorylation occurred in the absence of Cdc42 binding when a mutation was introduced that maintains WASP in an open conformation (Fig. 6).
Although our results show that tyrosine phosphorylation was clearly not essential for WASP activation, a more subtle role for WASP phosphorylation cannot be ruled out. As discussed above, phosphorylated WASP may remain activated for longer periods of time (22, 31). In addition, the subcellular localization of N-WASP to specific compartments in NIH 3T3 cells appears to be affected by the phosphorylation status of its tyrosine 256 (37). However, experiments looking at areas of protrusions and comparing the status of wild-type WASPbs and the Y291F mutant did not show any differences in localized WASP activation (data not shown). Alternatively stimulants other than CSF-1 may trigger different signaling cascades truly leading to functional WASP phosphorylation in macrophages. Although this study addressed in detail the role of two key mechanisms of WASP activation, Cdc42 binding and tyrosine phosphorylation, other levels of control have also been identified. The binding of SH3 domain-containing proteins, such as Nck, to the proline-rich region of WASP has been shown to induce WASP-dependent actin polymerization in vitro (38–40). Binding of phosphatidylinositol 4,5-bisphosphate to the basic region of N-WASP has been proposed to partially stimulate its actin polymerization activity synergistically with Cdc42 (39, 41). However, a recent biochemical study indicates that phosphoinositide regulation of WASP might differ significantly from that of N-WASP with phosphatidylinositol 4,5-bisphosphate alone having no effect on WASP activity and even being inhibitory with regard to Cdc42-induced stimulation of WASP (40). Very recently, WASP dimerization has been suggested to regulate its activity (42), although the existence of WASP dimers in cells under physiological conditions remains to be shown. However, the authors suggest that an initial change in conformation is necessary for dimerization to occur and that dimerization maintains WASP in an open conformation (42). Therefore, the WASPbs should prove to be useful to address these issues and to study other domains of WASP through a deletion and/or mutagenesis approach.
In addition to being required for chemotaxis WASP and N-WASP are also required for proper exocytosis (43, 44) and endocytosis (45, 46) and for the formation of invasive structures such as podosomes (47–49) and invadopodia (50). An increasing body of evidence shows that, in several cancers, recruitment of large numbers of macrophages to primary tumors can significantly contribute to their progression to an invasive, metastatic phenotype through a paracrine interaction (51). Interestingly the concomitant formation of WASP-dependent podosomes in macrophages and N-WASP-dependent invadopodia in tumor cells appears to be an important feature of tumor progression in vivo (52, 53). In all these crucial situations, the underlying molecular and cellular mechanisms are not fully understood. In future studies, the utilization of biosensors that can detect activation of WASP or N-WASP in time and space and in vivo in tumors should be a valuable tool to determine their exact contributions to these processes. As we have demonstrated here, specific alterations of these biosensors should also be a powerful way to assess the relevance of a variety of putative binding partners and signaling pathways. Overall because its biological significance is now being established, this novel and versatile technology will undoubtedly be central to a number of investigations in need of precise information on protein activity and regulation in vivo.
Supplementary Material
Acknowledgments
We are grateful to Shailesh Shenoy for help with imaging, and we thank members of the John Condeelis, Jeffrey Segall, and E. Richard Stanley laboratories for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 GM071828 (to D. C.), GM38511 (to J. C.), and P01 CA100324 (to M. L.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
A. Dovas, I.-C. Gevrey, A. Grossi, W. G. Abou-Kheir, and D. Cox, manuscript submitted.
B. M. Isaac, D. Ishihara, L. Nusblat, J. Condeelis, and D. Cox, manuscript in preparation.
- CSF-1
- colony-stimulating factor-1
- CFP
- cyan fluorescent protein
- FRET
- fluorescence resonance energy transfer
- PI3K
- phosphatidylinositol 3-kinase
- WASP
- Wiskott-Aldrich syndrome protein
- WASPbs
- WASP biosensor
- CSA
- WASP/N-WASP conformation-sensitive antibody
- YFP
- yellow fluorescent protein
- WAVE
- Wiskott-Aldrich syndrome verprolin-homologous
- VCA
- verprolin homology, cofilin-like, and acidic region
- N-WASP
- neuronal WASP
- SH3
- Src homology 3.
REFERENCES
- 1.Campbell I. K., Rich M. J., Bischof R. J., Hamilton J. A. (2000) J. Leukoc. Biol. 68,144–150 [PubMed] [Google Scholar]
- 2.Smith J. D., Trogan E., Ginsberg M., Grigaux C., Tian J., Miyata M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92,8264–8268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin E. Y., Nguyen A. V., Russell R. G., Pollard J. W. (2001) J. Exp. Med. 193,727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pixley F. J., Stanley E. R. (2004) Trends Cell Biol. 14,628–638 [DOI] [PubMed] [Google Scholar]
- 5.Aharinejad S., Sioud M., Lucas T., Abraham D. (2007) Methods Mol. Biol. 361,227–238 [DOI] [PubMed] [Google Scholar]
- 6.Vanhaesebroeck B., Jones G. E., Allen W. E., Zicha D., Hooshmand-Rad R., Sawyer C., Wells C., Waterfield M. D., Ridley A. J. (1999) Nat. Cell Biol. 1,69–71 [DOI] [PubMed] [Google Scholar]
- 7.Jones G. E., Prigmore E., Calvez R., Hogan C., Dunn G. A., Hirsch E., Wymann M. P., Ridley A. J. (2003) Exp. Cell Res. 290,120–131 [DOI] [PubMed] [Google Scholar]
- 8.Cox D., Chang P., Zhang Q., Reddy P. G., Bokoch G. M., Greenberg S. (1997) J. Exp. Med. 186,1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen W. E., Jones G. E., Pollard J. W., Ridley A. J. (1997) J. Cell Sci. 110,707–720 [DOI] [PubMed] [Google Scholar]
- 10.Allen W. E., Zicha D., Ridley A. J., Jones G. E. (1998) J. Cell Biol. 141,1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takenawa T., Miki H. (2001) J. Cell Sci. 114,1801–1809 [DOI] [PubMed] [Google Scholar]
- 12.Kheir W. A., Gevrey J. C., Yamaguchi H., Isaac B., Cox D. (2005) J. Cell Sci. 118,5369–5379 [DOI] [PubMed] [Google Scholar]
- 13.Abou-Kheir W., Isaac B., Yamaguchi H., Cox D. (2008) J. Cell Sci. 121,379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zicha D., Allen W. E., Brickell P. M., Kinnon C., Dunn G. A., Jones G. E., Thrasher A. J. (1998) Br. J. Haematol. 101,659–665 [DOI] [PubMed] [Google Scholar]
- 15.Badolato R., Sozzani S., Malacarne F., Bresciani S., Fiorini M., Borsatti A., Albertini A., Mantovani A., Ugazio A. G., Notarangelo L. D. (1998) J. Immunol. 161,1026–1033 [PubMed] [Google Scholar]
- 16.Takenawa T., Suetsugu S. (2007) Nat. Rev. Mol. Cell Biol. 8,37–48 [DOI] [PubMed] [Google Scholar]
- 17.Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K. (2000) Nature 404,151–158 [DOI] [PubMed] [Google Scholar]
- 18.Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. (1999) Cell 97,221–231 [DOI] [PubMed] [Google Scholar]
- 19.Miki H., Takenawa T. (2003) J. Biochem. 134,309–313 [DOI] [PubMed] [Google Scholar]
- 20.Cory G. O., Cramer R., Blanchoin L., Ridley A. J. (2003) Mol. Cell 11,1229–1239 [DOI] [PubMed] [Google Scholar]
- 21.Cory G. O., Garg R., Cramer R., Ridley A. J. (2002) J. Biol. Chem. 277,45115–45121 [DOI] [PubMed] [Google Scholar]
- 22.Torres E., Rosen M. K. (2003) Mol. Cell 11,1215–1227 [DOI] [PubMed] [Google Scholar]
- 23.Ward M. E., Wu J. Y., Rao Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101,970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz M., Yamaguchi H., Wang Y., Singer R. H., Condeelis J. (2004) Curr. Biol. 14,697–703 [DOI] [PubMed] [Google Scholar]
- 25.Sukumvanich P., DesMarais V., Sarmiento C. V., Wang Y., Ichetovkin I., Mouneimne G., Almo S., Condeelis J. (2004) Cell. Motil. Cytoskeleton 59,141–152 [DOI] [PubMed] [Google Scholar]
- 26.Peterson J. R., Bickford L. C., Morgan D., Kim A. S., Ouerfelli O., Kirschner M. W., Rosen M. K. (2004) Nat. Struct. Mol. Biol. 11,747–755 [DOI] [PubMed] [Google Scholar]
- 27.Stamm L. M., Pak M. A., Morisaki J. H., Snapper S. B., Rottner K., Lommel S., Brown E. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102,14837–14842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ridley A. J. (2001) FEBS Lett. 498,168–171 [DOI] [PubMed] [Google Scholar]
- 29.Suetsugu S., Hattori M., Miki H., Tezuka T., Yamamoto T., Mikoshiba K., Takenawa T. (2002) Dev. Cell 3,645–658 [DOI] [PubMed] [Google Scholar]
- 30.Kato M., Miki H., Imai K., Nonoyama S., Suzuki T., Sasakawa C., Takenawa T. (1999) J. Biol. Chem. 274,27225–27230 [DOI] [PubMed] [Google Scholar]
- 31.Torres E., Rosen M. K. (2006) J. Biol. Chem. 281,3513–3520 [DOI] [PubMed] [Google Scholar]
- 32.Devriendt K., Kim A. S., Mathijs G., Frints S. G., Schwartz M., Van Den Oord J. J., Verhoef G. E., Boogaerts M. A., Fryns J. P., You D., Rosen M. K., Vandenberghe P. (2001) Nat. Genet. 27,313–317 [DOI] [PubMed] [Google Scholar]
- 33.Symons M., Derry J. M., Karlak B., Jiang S., Lemahieu V., Mccormick F., Francke U., Abo A. (1996) Cell 84,723–734 [DOI] [PubMed] [Google Scholar]
- 34.Sun C. X., Magalhães M. A., Glogauer M. (2007) J. Cell Biol. 179,239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badour K., Zhang J., Shi F., Leng Y., Collins M., Siminovitch K. A. (2004) J. Exp. Med. 199,99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guinamard R., Aspenström P., Fougereau M., Chavrier P., Guillemot J. C. (1998) FEBS Lett. 434,431–436 [DOI] [PubMed] [Google Scholar]
- 37.Wu X., Suetsugu S., Cooper L. A., Takenawa T., Guan J. L. (2004) J. Biol. Chem. 279,9565–9576 [DOI] [PubMed] [Google Scholar]
- 38.Rivero-Lezcano O. M., Marcilla A., Sameshima J. H., Robbins K. C. (1995) Mol. Cell. Biol. 15,5725–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohatgi R., Nollau P., Ho H. Y., Kirschner M. W., Mayer B. J. (2001) J. Biol. Chem. 276,26448–26452 [DOI] [PubMed] [Google Scholar]
- 40.Tomasevic N., Jia Z., Russell A., Fujii T., Hartman J. J., Clancy S., Wang M., Beraud C., Wood K. W., Sakowicz R. (2007) Biochemistry 46,3494–3502 [DOI] [PubMed] [Google Scholar]
- 41.Prehoda K. E., Scott J. A., Mullins R. D., Lim W. A. (2000) Science 290,801–806 [DOI] [PubMed] [Google Scholar]
- 42.Padrick S. B., Cheng H. C., Ismail A. M., Panchal S. C., Doolittle L. K., Kim S., Skehan B. M., Umetani J., Brautigam C. A., Leong J. M., Rosen M. K. (2008) Mol. Cell 32,426–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pivniouk V. I., Snapper S. B., Kettner A., Alenius H., Laouini D., Falet H., Hartwig J., Alt F. W., Geha R. S. (2003) Int. Immunol. 15,1431–1440 [DOI] [PubMed] [Google Scholar]
- 44.Gasman S., Chasserot-Golaz S., Malacombe M., Way M., Bader M. F. (2004) Mol. Biol. Cell 15,520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merrifield C. J., Qualmann B., Kessels M. M., Almers W. (2004) Eur. J. Cell Biol. 83,13–18 [DOI] [PubMed] [Google Scholar]
- 46.Benesch S., Polo S., Lai F. P., Anderson K. I., Stradal T. E., Wehland J., Rottner K. (2005) J. Cell Sci. 118,3103–3115 [DOI] [PubMed] [Google Scholar]
- 47.Calle Y., Chou H. C., Thrasher A. J., Jones G. E. (2004) J. Pathol. 204,460–469 [DOI] [PubMed] [Google Scholar]
- 48.Jones G. E., Zicha D., Dunn G. A., Blundell M., Thrasher A. (2002) Int. J. Biochem. Cell Biol. 34,806–815 [DOI] [PubMed] [Google Scholar]
- 49.Linder S., Nelson D., Weiss M., Aepfelbacher M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96,9648–9653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., Condeelis J. (2005) J. Cell Biol. 168,441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Condeelis J., Pollard J. W. (2006) Cell 124,263–266 [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi H., Pixley F., Condeelis J. (2006) Eur. J. Cell Biol. 85,213–218 [DOI] [PubMed] [Google Scholar]
- 53.Wyckoff J., Wang W., Lin E. Y., Wang Y., Pixley F., Stanley E. R., Graf T., Pollard J. W., Segall J., Condeelis J. (2004) Cancer Res. 64,7022–7029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.