Abstract
Endothelial lipase (EL) is a member of a subfamily of lipases that act on triglycerides and phospholipids in plasma lipoproteins, which also includes lipoprotein lipase and hepatic lipase. EL has a tropism for high density lipoprotein, and its level of phospholipase activity is similar to its level of triglyceride lipase activity. Inhibition or loss-of-function of EL in mice results in an increase in high density lipoprotein cholesterol, making it a potential therapeutic target. Although hepatic lipase and lipoprotein lipase have been shown to function as homodimers, the active form of EL is not known. In these studies, the size and conformation of the active form of EL were determined. Immunoprecipitation experiments suggested oligomerization. Ultracentrifugation experiments showed that the active form of EL had a molecular weight higher than the molecular weight of a simple monomer but less than a dimer. A construct encoding a covalent head-to-tail homodimer of EL (EL-EL) was expressed and had similar lipolytic activity to EL. The functional molecular weights determined by radiation inactivation were similar for EL and the covalent homodimer EL-EL. We previously showed that EL could be cleaved by proprotein convertases, such as PC5, resulting in loss of activity. In cells overexpressing PC5, the covalent homodimeric EL-EL appeared to be more stable, with reduced cleavage and conserved lipolytic activity. A comparative model obtained using other lipase structures suggests a structure for the head-to-tail EL homodimer that is consistent with the experimental findings. These data confirm the hypothesis that EL is active as a homodimer in head-to-tail conformation.
Three members of the triglyceride lipase family, lipoprotein lipase (LPL),3 hepatic lipase (HL), and endothelial lipase (EL), contribute to lipoprotein catabolism in the plasma compartment. They are all secreted proteins that bind to heparan sulfate proteoglycans on the luminal side of endothelial cells where they interact with their lipoprotein substrates. They have different specificities for lipoproteins, and all hydrolyze triglycerides and phosphatidylcholine at the sn-1 position, albeit with widely differing efficiencies (1). The preferred lipoprotein substrates for LPL are the triglyceride-rich lipoproteins, chylomicrons, and very low density lipoproteins; the triglyceride lipase activity of LPL is more than 100-fold greater than its phospholipase activity. The primary lipoprotein substrates for HL are chylomicron remnants, intermediate density lipoproteins, and large triglyceridase-enriched HDL; its triglyceride lipase activity is about 20-fold higher than its phospholipase activity. EL is much more active on HDL, and its phospholipase activity is quite similar to its triglyceride lipase activity.
These three lipolytic enzymes share a number of structural features. By analogy to the crystal structure of pancreatic lipase (2), another member of the triglyceride lipase family, each has a clearly defined N-terminal and C-terminal structural domain, joined by a hinge region. They are all serine esterases with a catalytic triad of serine, aspartic acid, and histidine located in the N-terminal domain. The catalytic triad is covered by a lid domain that contributes to the preference for either triglyceride or phospholipid substrates (3–6). The C-terminal domain contributes to lipid binding and determines the preferences for binding to lipoproteins (7–12). The N-terminal portion of the EL molecule contains the active site of the enzyme. However, when this portion of the molecule is expressed without the C-terminal domain, it lacks enzymatic activity against phospholipid (13), triglyceride, and the more soluble micellar substrate tributyrin.4 Thus, the presence of the C-terminal domain is necessary for activity. All three enzymes are glycosylated to varying degrees, and two of the glycosylation sites (one each in the N- and C-terminal domains) are common to all three enzymes (14, 15). All three enzymes are also heparin-binding proteins, and regions that contribute to heparin binding are found in both the N- and C-terminal domains (16–18).
EL and, to a lesser extent, LPL are subject to proteolytic cleavage by proprotein convertases at a prototypical RXKR site in the hinge region, but HL lacks the site and is not cleaved (19, 20). The catalytically active forms of both LPL and HL have been shown to be homodimers (18, 21–27), and in the case of LPL, the orientation of the subunits of the dimer has been shown to be head-to-tail (28–30). The present study tested the hypothesis that EL also functions as a head-to-tail dimer.
EXPERIMENTAL PROCEDURES
Construction of a Head-to-Tail Dimer of EL (EL-EL)
We used PCR to engineer a head-to-tail dimer of two monomeric subunits of EL (EL-EL) in an approach similar to that of Wong et al. (28) for LPL using the same short hinge of 8 amino acids (GSIEGRLE) to create the head-to-tail dimer of EL (Fig. 1).
FIGURE 1.
Schematic diagram of EL-EL construct. Shown is a schematic diagram representing EL-EL, the homodimer of EL in a head-to-tail conformation. A short hinge (linker) of 8 amino acid residues (GSIEGRLE) joined the two monomeric subunits of EL using overlap extension PCR. EL-EL contained sequences coding for the signal peptide (SP) of EL, the mature EL protein, the 8 amino acid-peptide linker containing a factor Xa site, and another mature EL protein. Each monomeric subunit of mature EL protein has an N-terminal domain (N) and a C-terminal domain (C). To allow comparison of the levels of expression of the dimeric construct (EL-EL) and wild-type EL, we constructed C-terminal myc-His-tagged proteins.
Two cDNAs of human EL were amplified separately by PCR. Each cDNA contained engineered restriction sites to facilitate assembly and subcloning into the expression plasmid pcDNA3.1/myc-His(−)A (Invitrogen). The sense primer of the first cDNA of EL (5′-ACGTTCTAGAGGCAGGATGAGCAACTCCG-3′) contained an XbaI restriction site and the first 13 coding nucleotides of the EL cDNA. The antisense primer of the first cDNA of EL (5′-CGCTCTCGAGACGACCTTCGATGGATCCGGGAAGCTCCACAGTGGGAC-3′) contained the last 20 coding nucleotides of the EL cDNA, a factor Xa site (coding for 8 amino acids, GSIEGRLE), and an XhoI restriction site. The sense primer of the second cDNA of EL (5′-GCGTCTCGAGAGCCCCGTACCTTTTGGTC-3′) contained an XhoI restriction site and the first 19 coding nucleotides of the mature EL protein. The antisense primer of the second cDNA of EL (5′-TGACAAGCTTTCAGGGAAGCTCCACAGTGGGAC-3′) contained the last 23 coding nucleotides of the EL cDNA and a HindIII restriction site. These two PCR products were purified, digested using XbaI, XhoI, and HindIII restriction enzymes, and then ligated using T4 ligase from a rapid ligation kit (Roche Applied Science). The final product contained sequences coding for the signal peptide of EL, the mature EL protein, an 8-amino acid-peptide linker containing a factor Xa site, and another mature EL protein (Fig. 1). To allow comparison of the levels of expression of the dimeric construct (EL-EL) and wild-type EL, we constructed C-terminal myc-His-tagged proteins by inserting the full-length cDNA into pcDNA3.1/myc-His(−)A plasmid expression vector (Invitrogen). The construct was sequenced to confirm accuracy.
Cell Culture and Transfections
HEK293 cells and sublines stably expressing profurin or PC5a were maintained in Dulbecco's modified Eagle's medium, 10% fetal bovine serum, and 1× antibiotic/antimycotic supplement (Invitrogen). The cells were transfected with EL constructs using LipofectamineTM reagent (Invitrogen). Twenty four h after transfection, the medium was replaced with Dulbecco's modified Eagle's medium containing 10 units/ml heparin (Sigma) and incubated for another 24 h. Thirty min prior to harvesting this conditioned medium, more heparin was added to the medium, bringing the final concentration of heparin to 20 units/ml. Conditioned media were collected, subjected to low speed centrifugation, and frozen in aliquots at −80 °C. For some experiments where high enzyme activity was required (sucrose gradients and radiation inactivation), the conditioned medium was concentrated 5-fold using an Amicon ultracentrifugal filtration device (30,000 Mr cutoff) (Millipore).
Western Blotting
The levels of protein expression of wild-type EL and EL-EL were estimated by Western blot. For each experiment, we used conditioned medium from cells transfected to express green fluorescent protein (GFP) as a negative control. Ten μl of conditioned media were resolved on 10% BisTris or 7% Tris acetate SDS-PAGE (Invitrogen) and transferred to Hybond ECL nitrocellulose membrane (Amersham Biosciences). Proteins were detected using either a polyclonal rabbit anti-human EL primary antibody specific for a peptide in the N-terminal region or a monoclonal mouse anti-Myc primary antibody (clone 9E10) and a horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch).
Co-immunoprecipitation
Conditioned media from HEK293 cells transfected with GFP and co-transfected with EL-myc-His and EL-FLAG or a 1:1 mixture of media from HEK293 cells, which had been transfected with EL-myc-His or EL-FLAG, were incubated with mouse (or rabbit) anti-Myc (Abcam) or mouse anti-FLAG IgG (Sigma) at 4 °C in the presence of 0.1% Triton X-100 (Fisher) for 2 h. Protein G magnetic beads (New England Biolabs) were added to each sample, and samples were incubated overnight at 4 °C. Supernatant and pellet were separated after centrifuging for 10 min at 10,000 × g, 4 °C. Pellets were washed three times with phosphate-buffered saline and eluted from the beads with 4× loading buffer and 10× dithiothreitol (Invitrogen). Western blot analysis of samples was performed using a rabbit anti-EL antibody or a rabbit anti-Myc (Biovision) or a mouse anti-FLAG antibody (HRP conjugate, Sigma).
Sucrose Gradient Ultracentrifugation
Sucrose gradients were prepared as described (27). Briefly, 5–20% sucrose gradients containing 50 mm ammonium hydroxide, pH 8.1, were prepared. Glucose-6-phosphate dehydrogenase (G6PDH) (110 kDa) was added to each sample to serve as an internal standard. Cytochrome c (12.5 kDa), ovalbumin (45 kDa), malate dehydrogenase (74 kDa), and catalase (240 kDa) were centrifuged in two separate tubes and served as external standards. After centrifugation (22 h at 200,000 × g at 4 °C), 600-μl fractions were collected and assayed for enzyme activity and protein concentration. The protein concentration was determined in each fraction of external standards using a commercially available kit (BCA protein assay kit, Pierce).
Radiation Inactivation
Conditioned medium containing partially cleaved myc-His-tagged EL was prepared by transfection in HEK293 cells as described above. Media containing either uncleaved myc-His-tagged wild-type EL or uncleaved myc-His-tagged EL-EL dimer were prepared in a similar manner by transfection in HEK293 cells that stably express profurin. Apotransferrin (2 mg/ml, Sigma) and G6PDH (4 units/ml, Sigma) were added to the media. Samples of these media (0.55 ml) were placed in glass vials, quick frozen, sealed, and stored at −80 °C. Vials were shipped and returned on dry ice. Irradiations were performed with a linear electron accelerator at the Armed Forces Radiobiology Research Institute, Bethesda, or at the National Institute of Standards and Technology, Gaithersburg, MD. Frozen samples were irradiated at −135 °C with a beam of 13-MeV electrons as described (31). Two aliquots not irradiated served as the zero dose controls. Radiation dose was determined with thermoluminescent or alanine dosimeters. Irradiated frozen samples were stored at −80 °C until assayed for triglyceride lipase activity. The activity (A) at each dose was divided by the activity at zero dose (A0) to determine the fractional residual activity at each dose. For each enzyme preparation, the natural logarithm of the fraction remaining was plotted as a function of radiation dose. The slopes of these lines were determined by linear regression (GraphPad Prizm) and were used to calculate molecular mass as described previously (31). The slopes for the inactivation of G6PDH were identical in all sample sets.
Triglyceride Lipase and Phospholipase Assays
Emulsions of triolein or dipalmitoylphosphatidylcholine were used to measure triglyceride lipase or phospholipase activity, respectively (1, 32, 33). For the triglyceride lipase assay, the emulsion contained triolein and egg phosphatidylcholine containing glycerol tri[9,10-3H]oleate stabilized with glycerol. For the phospholipase assay, a similar glycerol-stabilized emulsion was used that contained radiolabeled phospholipids ([14C]dipalmitoylphosphatidylcholine) and cholesteryl oleate as the neutral lipid core. Samples were incubated for 15 min at 37 °C. All enzyme activities are reported as nanomoles of free fatty acid liberated/h-ml conditioned medium as source of enzyme. All assays were done in triplicate.
Other Enzyme Assays
Malate dehydrogenase activity was measured by incubating 2 μl of sample with 294 μl of substrate made of 367.4 μm oxaloacetate and 204 μm NADH in 40 mm Tris-H2SO4, pH 7.4, at 25 °C. The decrease in absorbance was monitored at 340 nm. G6PDH activity was measured by incubating 5 μl of sample in a final volume of 200 μl containing 50 mm Tris-HCl, pH 7.8, 3 mm MgCl2, 0.2 mm NADP, and 3.3 mm glucose 6-phosphate for 15 min at 25 °C and monitoring the increase in absorbance at 340 nm.
Comparative Protein Structure Modeling Methods
The human EL models were generated using the molecular modeling suite MODELLER version 8v1 (Accelrys Discovery Studio, San Diego) and evaluated with MODELLER versions 8v1 and 9v1 (34–37). As input for the modeling algorithm, the human EL protein sequence, NCBI protein data base accession code NP_006024.1, was aligned to the following lipase structures obtained from the Protein Data Bank: Homo sapiens pancreatic lipase and Sus scrofa co-lipase complex (PDB accession code 1N8S), H. sapiens pancreatic lipase and S. scrofa co-lipase complex inhibited by undecane phosphonate methyl ester (PDB accession code 1LPB), and Equus caballus pancreatic lipase (PDB accession code 1HPL). Multiple sequence alignments with the human EL protein sequence and those of the aforementioned three protein structures were generated by ClustalW (EMBL-EBI) using default parameter settings (38). The human EL sequence has 27% identity to human pancreatic lipase and 25% identity to horse pancreatic lipase. These identical residues located throughout the crystal structures of the pancreatic lipases served as anchors to guide the modeling of human endothelial lipase. The EL sequence contains 37 residues at the C terminus that were not used in the modeling process because there is no significant sequence homology to the other template lipase structures in this region. The human EL models were evaluated using the discrete optimized protein energy algorithms of MODELLER version 9v1 (39). To investigate the model structure in the context of a homodimer, the best scoring human endothelial lipase monomer models were structurally aligned to each monomer of the horse pancreatic lipase homodimer using MacPyMOL version 0.96.
RESULTS
EL Is a Dimer That Forms Intracellularly
To understand the intermolecular interaction of EL, we used two EL constructs, one encoding a FLAG tag and the other a myc-His tag at the C terminus. Neither tag had any effect on EL lipolytic activity (data not shown). Constructs encoding EL-myc-His and EL-FLAG were co-transfected in HEK293 cells. Conditioned medium was harvested and subjected to immunoprecipitation using an anti-Myc antibody to precipitate any Myc-containing proteins. The precipitated proteins were examined by Western blotting using an anti-FLAG antibody as well as an anti-EL antibody specific for a peptide in the N-terminal domain. No immunoreactive bands were detected in conditioned medium from cells transfected with a construct encoding GFP. As shown in Fig. 2, when constructs encoding EL-myc-His and EL-FLAG were co-transfected in HEK293 cells, the anti-FLAG antibody detected FLAG-tagged EL in the material that had been immunoprecipitated with anti-Myc antibody, indicating that there was interaction between EL-FLAG and EL-myc-His proteins, consistent with dimer formation. When conditioned medium containing EL-FLAG was mixed with conditioned medium containing EL-myc-His and subsequently immunoprecipitated, no interaction was detected, suggesting that the association between monomeric units occurs intracellularly and that the association between subunits is very strong.
FIGURE 2.
EL protein forms immunoprecipitable complex during secretion but not after secretion. HEK293 conditioned media were prepared as follows. Lanes 1 and 4 are from cells expressing GFP; lanes 2 and 5 are mixtures of media from cells separately transfected with either EL-myc-His or EL-FLAG; lanes 3, 6, and 7 are media from cells co-transfected with EL-myc-His and EL-FLAG. Western blots (WB) show supernatants (lanes 1–3) and pellets (lanes 4–6) after immunoprecipitation (IP) with mouse anti-Myc from samples as indicated. Lane 7 shows the pellet after immunoprecipitation with mouse anti-FLAG from conditioned media from HEK293 cells co-transfected with EL-myc-His and EL-FLAG. These Western blots used either an antibody against an EL peptide, FLAG tag, or Myc tag for detection. F indicates the full-length monomer size of EL; N indicates the N-terminal domain of EL, and C indicates the C-terminal domain of EL.
Active EL Is Larger than a Monomer and Consistent with Partially Cleaved Dimer
Sucrose gradient centrifugation of conditioned medium from cells expressing wild-type EL was used to determine the size of the functional unit of EL. The major peak of the triglyceride lipase activity of EL appeared as a distinct species somewhat smaller than G6PDH (which has a molecular weight of 110,000) as shown in Fig. 3A. A linear relationship was observed when the molecular weights of standard proteins versus sucrose concentrations were plotted (Fig. 3B). The relative molecular weight of the EL functional unit was 94,500, considerably smaller than a dimer of full-length monomers (136 kDa) and larger than a monomer (68 kDa). This functional size is most consistent with partially cleaved forms where part of the cleaved monomer remains associated with an uncleaved monomer (Fig. 4A).
FIGURE 3.
A, sucrose gradient centrifugation of wild-type EL. After 5–20% sucrose gradient ultracentrifugation, samples were collected. Each fraction was tested for TG lipase activity (♦) and G6PDH activity (■). The percentage of sucrose was also measured in each fraction collected. B, molecular weight of wild-type EL determined by ultracentrifugation. The molecular standards used in this experiment were cytochrome c (12.5 kDa), ovalbumin (45 kDa), malate dehydrogenase (74 kDa), G6PDH (114 kDa), and catalase (240 kDa). After centrifugation (22 h at 200,000 × g at 4 °C), fractions were collected and assayed for enzyme activity and protein concentration. There is a linear relationship between the molecular weights of standard proteins and the sucrose percentage.
FIGURE 4.
A, cleavage products of EL monomer and EL dimer. A schematic diagram is shown of all the potential intermediate forms of EL monomer and EL dimer induced by the proteolytic cleavage of EL by proprotein convertases. All the possible forms varying from uncleaved to partially cleaved and fully cleaved in the case of EL monomer and EL dimer are represented. The relative molecular mass of each of these forms in kDa is shown. B, Western blot of EL and EL-EL detected by an antibody against Myc tag. Expression in HEK293 conditioned media of Myc-tagged EL and EL-EL was detected by Western blot using a mouse anti-Myc monoclonal antibody and HRP-conjugated secondary antibody. Schematic diagrams of the different forms of EL detected by the anti-Myc antibody are shown. C, Western blot of EL and EL-EL detected by an antibody against EL. Expression in HEK293 conditioned media of Myc-tagged EL and EL-EL was detected by Western blot using a rabbit anti-human EL polyclonal antibody and HRP-conjugated secondary antibody. Schematic diagrams of the different forms of EL as detected by the anti-EL antibody are shown.
A Covalent Head-to-Tail EL Dimer Is Highly Active
We engineered a construct encoding a covalent head-to-tail dimer of two monomeric subunits of EL (EL-EL) joined by a short hinge of eight amino acid residues (GSIEGRLE) using an approach similar to that of Wong et al. (28) for LPL. The final cDNA product contained sequences coding for the signal peptide of EL, the mature EL protein, the 8-amino acid peptide linker followed by another mature EL protein, and a C-terminal myc-His tag (Fig. 1). The expression of the dimeric construct (EL-EL) was compared with wild-type EL, which was also myc-His-tagged. Western blots of conditioned media of transiently transfected HEK293 cells using an antibody against the Myc tag as primary antibody (Fig. 4B) showed a band corresponding to a full-length monomer (68 kDa) in the case of wild-type EL and a dimer (136 kDa) in the case of EL-EL, as well as the cleavage products that are myc-His-tagged. Although dimeric forms are not always present in Western blots for wild-type EL, a dimeric form and partially cleaved dimer are also visible in the wild-type EL lane. In the Western blot of the same media using an antibody against EL as primary antibody (Fig. 4C), we observed the full-length monomer of wild-type EL (68 kDa) and the full-length dimer of EL-EL (136 kDa), and as expected some cleavage products (20). Interestingly, in the case of EL-EL we detected cleavage products of intermediate size between a monomer and a dimer corresponding to a monomer plus the N-terminal domain (108 kDa) and a monomer plus the C-terminal domain (96 kDa).
We compared the enzymatic activity of wild-type EL and EL-EL using lipid emulsions of triglyceride and phospholipid to measure triglyceride lipase and phospholipase activity, respectively. The EL-EL dimer showed both triglyceride lipase and phospholipase activities. For each transfection, we consistently measured more lipase activity (both triglyceride lipase and phospholipase) in EL-EL despite the protein band in the respective Western blot often appearing of similar or lesser intensity than the band of the full-length wild-type EL (Fig. 4B). The ratio of the triglyceride lipase activity to phospholipase activity (triglyceridase/phospholipase ratio) reflects the substrate specificity of the enzyme and was not significantly different for EL-EL (1.46 ± 0.57) than for EL (1.56 ± 0.24). Thus the EL protein in a dimeric head-to-tail conformation (EL-EL) was active both as a triglyceride lipase and phospholipase. These data are consistent with a model that EL is active as dimer in a head-to-tail conformation.
Intermediate Oligomeric Forms of EL Are Enzymatically Active
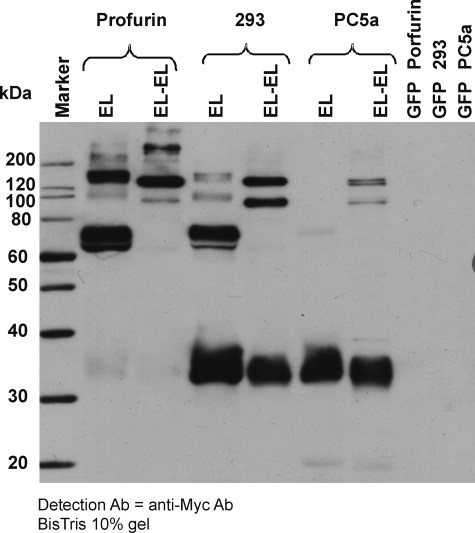
EL is active as a dimer in a head-to-tail conformation, but other intermediate forms could potentially also be active provided that an intact active site is present and preserved. To test that hypothesis, we expressed wild-type EL and EL-EL in HEK293 cells under conditions promoting different levels of cleavage. We used HEK293 cells stably expressing profurin, which has been reported to prevent EL cleavage, or expressing PC5a which has been reported to fully cleave EL (20). Fig. 5 shows that indeed more full-length EL and EL-EL dimer were present in the media of cells expressing profurin, whereas wild-type EL and EL-EL dimer were more heavily cleaved in the presence of PC5a expression. Interestingly, EL-EL appeared more resistant to cleavage in HEK293 cells and in the absence or presence of PC5a in comparison with wild-type EL. These data support the concept that the oligomeric state of EL is dimeric and that the linker hinge in the EL-EL protein stabilizes the enzyme, making it less prone to inactivating enzymatic cleavage by enzymes of the proprotein convertase family such as PC5a. The linker hinge also ensures that the N-terminal and C-terminal domains remain attached and presumably preserve an active site.
FIGURE 5.
Western blots of EL and EL-EL under conditions promoting different levels of cleavage. The level of protein expression in HEK293 conditioned media was detected by Western blot using a mouse anti-Myc monoclonal antibody (Ab) and HRP-conjugated secondary antibody. GFP (as negative control), EL, and EL-EL were expressed in HEK293 cells under conditions promoting different levels of cleavage. HEK293 cells stably expressing profurin prevented EL cleavage, and HEK293 cells stably expressing PC5a promote the cleavage of EL.
We measured the triglyceride lipase and phospholipase activity of wild-type EL and EL-EL in these three experimental conditions (Table 1). In the case of EL, both the triglyceride lipase and phospholipase activity of wild-type EL decreased as the amount of cleavage increased (Fig. 5 and Table 1). Similar results were observed in the case of EL-EL, where both the triglyceride lipase and phospholipase activity of EL-EL decreased as the amount of cleavage increased (Fig. 5 and Table 1). EL-EL differed from wild-type EL only in the extent of the effect. Indeed, EL-EL only partially lost activity in the presence of PC5a, whereas wild-type EL was more drastically affected. Our activity data (triglyceride lipase and phospholipase) associated with the mass indicated that as less protein was cleaved in the case of EL-EL compared with wild-type EL, more lipolytic enzymatic activity remained (Fig. 5 and Table 1). These data indicated that not only is EL active as a homodimer but that the cleavage of EL results in a loss of activity and the partially or wholly cleaved forms of EL are less active or inactive.
TABLE 1.
Triglyceride lipase and phospholipase activities of EL and EL-EL under conditions promoting different levels of cleavage
Wild-type EL and EL-EL were expressed in HEK293 cells under control conditions and under conditions promoting different levels of cleavage. HEK293 cells stably expressing profurin prevent EL cleavage, and HEK293 cells stably expressing PC5a promote the cleavage of EL. The triglyceride lipase activity (TG activity) and phospholipase activity (PL activity) of each conditioned medium were expressed in nanomoles of free fatty acid product (P) formed/h-ml of conditioned media as source of lipase. The activity values reported in this table are the mean values and the standard deviations (mean ± S.D., n = 3). The ratio of the triglyceride lipase to phospholipase activity (TG/PL ratio) represent the substrate specificity of the enzyme. The last column represents the mean values and the mean ± S.D. of the triglyceride/phospholipase ratios across independent experiments (n = 5 for HEK 293 cell media and n = 4 each for cell media from HEK293 cells expressing profurin or PC5a). Statistical analysis was run using an unpaired t test to determine whether the mean of the triglyceride/phospholipase ratios were significantly different (p < 0.05). The triglyceride/phospholipase ratios of EL-EL and wild-type EL were not statistically different experimentally whatever the cell types used for the transfection.
| 293 cells | Lipases | TG activity | PL activity | TG/PL ratio (mean ± S.D.) |
|---|---|---|---|---|
| nmol P/h/ml | nmol P/h/ml | |||
| PC5a | EL | 172.5 ± 8.4 | 92.7 ± 7.4 | 1.92 ± 0.12 |
| EL-EL | 425.5 ± 8.6 | 173.1 ± 10.6 | 1.56 ± 0.62 | |
| EL | 265.9 ± 3.8 | 172.4 ± 1.1 | 1.56 ± 0.24 | |
| EL-EL | 484.9 ± 20.2 | 226.0 ± 11.3 | 1.46 ± 0.57 | |
| Profurin | EL | 445.2 ± 10.5 | 306.5 ± 79.1 | 1.72 ± 0.24 |
| EL-EL | 673.5 ± 22.1 | 313.1 ± 4.5 | 1.79 ± 0.58 |
Comparison of the Molecular Mass of Uncleaved Wild-type EL and Uncleaved Dimeric EL
The functional size of EL was examined using radiation inactivation. Conditioned medium was prepared from cells expressing myc-His-tagged wild-type EL that was partially cleaved. The same construct was transfected in cells expressing profurin to produce EL that was uncleaved, as was the myc-His-tagged head-to-tail construct EL-EL, to produce the covalently linked dimer, also uncleaved. As shown in Fig. 6, all three preparations gave single exponential decays as a function of radiation dose. Unlike the sucrose density analysis, radiation inactivation is not influenced by glycosylation. The inactivation is a function only of the protein component of the enzyme. The predicted protein mass of a dimer of wild-type EL is 109,201 Da and of the dimer with the covalent linker is 110,061 Da. Both of the uncleaved preparations gave highly similar decay functions, which indicated a functional molecular mass of protein of 103.3 kDa for the uncleaved wild-type EL and 108.5 kDa for the uncleaved dimer that contains the additional 8-amino acid linker. Thus we can conclude that EL functions as a head-to-tail homodimer. The wild-type EL that had undergone cleavage also gave a single exponential decay as a function of radiation dose that was clearly different from the other two. The functional molecular mass of protein calculated for that preparation was 79 kDa. The predicted molecular masses of the protein components of partially cleaved forms are 90,629 and 82,629 Da, respectively (these are the 108- and 96-kDa forms in Fig. 4A with carbohydrate). These data indicate that, after cleavage, one of the domains can dissociate from the intact monomer, whereas the other remains with the monomer and the molecule remains active.
FIGURE 6.
Radiation inactivation of EL and EL-EL. The loss of triglyceride lipase activity of EL as a function of radiation dose (megarads) is shown. Three sources of EL were studied as follows: conditioned medium from cells expressing myc-His-tagged wild-type EL that was partially cleaved (◇, wt(cleaved)), conditioned medium from cells expressing profurin to produce EL that was uncleaved (Δ, wt(uncleaved)), and conditioned medium from cells expressing profurin to produce EL-EL that was uncleaved (□, dimer(uncleaved)).
Structural Modeling of EL
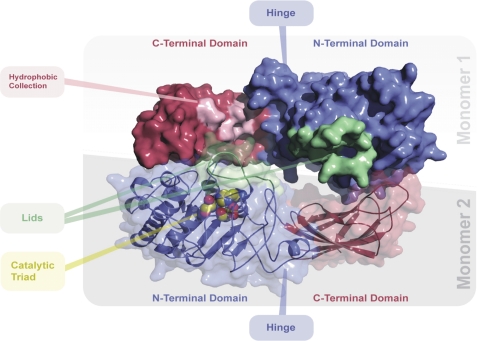
Comparative models of EL were created computationally to assess the feasibility of head-to-tail homodimer formation with EL and the possible impact of such a configuration on activity. Presently, no high resolution structures of EL have been determined, in part due to the marginal stability of the EL protein under laboratory conditions. Therefore, comparative modeling was employed to infer likely structures of the protein. Previously published structures of pancreatic lipases from human and horse were used as structural templates in conjunction with multiple sequence alignments of lipase sequences from several different species to generate three-dimensional models of human EL monomers. After model refinements using the MODELLER suite, the best scoring EL monomer models were aligned to the structure of the horse pancreatic lipase, which crystallized as a head-to-tail dimer (40). The EL dimer model (Fig. 7) aligns well to each monomer within the horse pancreatic lipase dimer, and the overall positions of the N terminus, activation lid, hinge, and C-terminal domains are similar to those of horse pancreatic lipase. The EL dimer model contains no steric clashes between monomers at the dimer interface. Within the EL dimer model, the N-terminal catalytic triad is in proximity (within 30 Å) to the C-terminal domain of the other monomer.
FIGURE 7.
Molecular model of the EL dimer. Top view of the human endothelial lipase dimer model is shown. The N-terminal “head” domains are shaded in blue; the C-terminal “tail” domains are shaded in red. The activation lid is colored green. The three catalytic residues, serine 169, aspartic acid 193, and histidine 274 are rendered in a space-filling format and located under the activation lid. A collection of exposed hydrophobic residues, Met-351, Leu-382, Pro-383, Ile-386, Val-387, Phe-398, Leu-399, Val-400, Pro-437, Pro-440, and Gly-441, is highlighted in pink on monomer 1. Monomer 2 is rendered so as to reveal the N- and C-terminal domains, hinge, and secondary structure.
DISCUSSION
LPL, HL, and EL are members of the triglyceride lipase gene subfamily involved in plasma lipoprotein metabolism. EL has a particular tropism for HDL and modulates HDL metabolism in mice and in humans (41, 42). To date, only the three-dimensional structure pancreatic lipase has been resolved by x-ray crystallography (2). Based on the high amino acid homology and the conservation of the catalytic triad, LPL, HL, and EL are assumed to present three-dimensional structures similar to pancreatic lipase. Both HL and LPL have been shown to be active in a homodimeric form, using methods varying from ultracentrifugation, gel filtration, antibody inhibition, to radiation inactivation (18, 21, 23, 24, 26–28, 43). Mutants of LPL engineered by molecular biology have shown that LPL is active as a homodimer in a head-to-tail conformation (28–30).
The purpose of this work was to determine the active form of EL. Initial experiments of immunoprecipitation of media from co-transfection of constructs encoding EL-myc and EL-FLAG showed that proteins immunoprecipitated with anti-Myc antibody contained both Myc-tagged EL and FLAG-tagged EL. Thus, there was an interaction between EL-FLAG and EL-myc proteins, consistent with an oligomerization of EL. However, when conditioned media from cells expressing either EL-myc or EL-FLAG were incubated together prior to immunoprecipitation, no evidence of interaction was observed. This suggests that dimerization occurs intracellularly, before secretion, and that there is a high affinity, stable interaction between subunits.
Ultracentrifugation studies of other triglyceride lipase family members suggested that the plasma compartment lipases are dimers. Human HL was shown to have a molecular mass of 121 kDa (43), and similar studies of rat HL showed 113 kDa (26). Together, these data suggested that HL exists as a dimer. Analysis of the sedimentation coefficient of bovine LPL yielded a molecular weight of 96,900, whereas the molecular weight of the monomer was 48,300 (21). Thus the native LPL enzyme appears to be a dimer of presumably identical subunits. Similarly, the bovine LPL activity profiles from sedimentation equilibrium as well as from gel filtration indicated that activity was associated almost exclusively with the dimer fraction (24). Sucrose density gradient centrifugation was also used to determine that monomeric LPL refolded in the presence of calcium becomes dimeric (44).
A complicating issue in determining the functional size of catalytically active EL relates to its susceptibility to proteolytic cleavage by proprotein convertases (19, 20). One can predict that, upon cleavage of a dimeric uncleaved EL, a number of possible forms might be obtained (Fig. 4A) and that some of these partially cleaved forms could potentially retain catalytic activity. The lack of enzymatic activity of the N-terminal portion of EL (14) indicates that an interaction with the C-terminal domain is essential to activity. Even if EL is active as a dimer in a head-to-tail conformation, other intermediate forms could potentially also be active as long as an intact active site is present and the contribution of the C-terminal domain is preserved. Using classic sucrose density ultracentrifugation methods, we compared the mobility of wild-type EL to that of several proteins of known molecular masses and calculated that the molecular mass of wild-type EL was 94.5 kDa. As this preparation of EL was made in HEK293 cells, which also secrete proprotein convertases, EL in the preparation was at least partially cleaved. The estimated molecular mass is consistent with either of the partially cleaved forms (Fig. 4A) where an intact N-terminal or C-terminal domain remains associated with an intact monomer. These data suggest that one or both of these partially cleaved forms is active.
We then prepared EL in a head-to-tail homodimer using methods similar to those used by Wong et al. (28), wherein we made a construct encoding such a dimer covalently linked by an 8-amino acid hinge region (Fig. 1). The construct also contained a myc-His tag to allow estimation of expression. The dimer construct expressed nearly as well as the wild type. However, the triglyceride and phospholipase activities of the dimer preparations were consistently higher that those of the wild type. The ratios of triglyceride to phospholipase activities were essentially unchanged, indicating that the covalent linkage of the dimer had no effect on substrate specificity. To determine the effect of cleavage on activity, we compared activity from wild type to that of the dimer in conditions of normal, high, or low activity of proprotein convertases. As shown in Table 1, activity was highest in the absence of cleavage for both wild-type and dimeric EL and lowest for both in the presence of the highest degree of cleavage. Interestingly, the dimeric form appeared to be more resistant to loss of activity than the wild type. This may be due to a resistance to cleavage by proprotein convertases or to the fact that the covalent hinge between the C terminus of the original first EL monomer keeps the N terminus of the second EL monomer attached so that an active molecule is retained. Even with a high degree of cleavage (Fig. 5) of EL prepared in cells expressing PC5, robustly detectable activity remains (Table 1), suggesting that fully cleaved N- and C-terminal fragments can remain together and maintain an active site.
The functional size of EL was also examined using radiation inactivation to determine the size of the active unit. The radiation inactivation method was developed to determine the size of enzymes (45) and has been previously used to determine the functional molecular mass of both LPL (23, 46) and HL (25, 44). For bovine milk LPL, the combined data of radiation inactivation under a number of different conditions yielded a functional size of 72 kDa for the unglycosylated protein, which is close to that expected for a dimer, 77 kDa (23). For rat LPL the functional molecular mass was calculated to be 127 kDa, again the unglycosylated size (46), consistent with a dimer. Thus the smallest unit required for enzyme function constitutes a dimer in the case of LPL. There is some discrepancy in the literature with regard to the functional unit of HL. When recombinant human HL was subjected to radiation inactivation, the size of functional HL was calculated to be 109 kDa (unglycosylated), the size of a homodimer (43). An examination of rat HL from liver, adrenal gland, and ovaries showed that the liver enzyme had a functional molecular mass of 63 kDa (monomeric), but the enzymes from the adrenals and ovaries both showed functional molecular masses of 117 kDa (dimeric) (26). Unlike the sucrose density analysis, radiation inactivation is not influenced by glycosylation and might therefore be a more accurate method in the case of glycosylated protein such as EL. Another advantage to the radiation inactivation is that it is a method for determining the functional size of a protein without the need of prior purification. Uncleaved preparations of wild-type EL and EL-EL gave similar functional molecular mass estimates, consistent with the concept that EL is active as a homodimer and are in agreement with the data previously generated for LPL (23, 46) and HL (25, 43). Interestingly, cleavage of only one “side” of the homodimer results in an intermediate size form (Fig. 4A) that is still active, although less so than the full homodimer.
In an elegant model of LPL (47), a head-to-tail conformation was proposed where the monomers associate side-by-side. This conformation has enough space for the lid to move freely to enable a conformational change upon substrate binding (47). Based on this, we developed a similar model for EL (Fig. 7). Our EL dimer model contains no steric clashes between monomers at the dimer interface. Within this model, the N-terminal catalytic triad of one-half of the dimer is in close proximity to the C-terminal domain of the other half. This model suggests how the C-terminal domain could affect substrate specificity by playing a role in positioning the substrate in proximity to the catalytic triad of the other half of the dimer across the dimer interface. In contrast, in a monomer model, the C-terminal domain is almost twice as far from the catalytic triad (∼60 Å). Indeed in the EL dimer model, a hydrophobic collection of amino acids (Met-351, Leu-382, Pro-383, Ile-386, Val-387, Phe-398, Leu-399, Val-400, Pro-437, Pro-440, and Gly-441) in the C-terminal domain is in proximity to the catalytic triad (under the activation lid) of the N-terminal domain of the other monomer (Fig. 7). Furthermore, there is ample volume for the activation lids of each monomer to open into the middle part of the dimer interface, and therefore head-to-tail packing could help stabilize the opening of the lid through ionic or hydrophobic interactions on the surface of the opposing monomer, similar to the observations of Kobayashi et al. (29) with their computational model of LPL. These observations, in conjunction with the biochemical activity data, suggest that head-to-tail dimerization of EL may help position the C-terminal domain, important for substrate specificity, in close proximity to the catalytic region of the enzyme while in turn stabilizing the movement of the activation lid.
In conclusion, HL and LPL have been shown using a variety of methods to be active in a homodimeric form and, at least for LPL, in a head-to-tail conformation. We comprehensively studied EL functional size using a variety of similar methods, including ultracentrifugation, radiation inactivation, covalent EL homodimer, immunoprecipitation, and molecular modeling. Our data are wholly consistent with a model in which EL oligomerizes into a homodimer that is active in a head-to-tail conformation. Only the full length of EL is present on the cell surface (data not shown), and it is conceivable that dimerization of EL allows its optimal activity against and interaction with its substrates, HDL, on the endothelial surface. Furthermore, EL is proteolytically cleaved in the hinge region by proprotein convertases, resulting in partially cleaved dimeric forms of EL that retain some activity and could have particular physiologic roles. Because EL is a pharmacologic target for inhibition as a strategy to raise HDL cholesterol levels, detailed structural information regarding the active forms of EL could be important to the optimal design of EL inhibitors.
Acknowledgments
We are indebted to Valeska Redon, Theo Hill, and Debra Cromley for their excellent technical assistance and John (Ioannis) Stylianou for help on the figures.
This work was supported, in whole or in part, by National Institutes of Health Grant HL55323 from the NHLBI (to D. J. R.).
N. Griffon, J. M. Glick, and D. J. Rader, unpublished observations.
- LPL
- lipoprotein lipase
- EL
- endothelial lipase
- HL
- hepatic lipase
- HDL
- high density lipoprotein
- GFP
- green fluorescent protein
- G6PDH
- glucose-6-phosphate dehydrogenase
- HRP
- horseradish peroxidase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank.
REFERENCES
- 1.McCoy M. G., Sun G. S., Marchadier D., Maugeais C., Glick J. M., Rader D. J. (2002) J. Lipid Res. 43,921–929 [PubMed] [Google Scholar]
- 2.Winkler F. K., D'Arcy A., Hunziker W. (1990) Nature 343,771–774 [DOI] [PubMed] [Google Scholar]
- 3.Dugi K. A., Dichek H. L., Talley G. D., Brewer H. B., Jr., Santamarina-Fojo S. (1992) J. Biol. Chem. 267,25086–25091 [PubMed] [Google Scholar]
- 4.Dugi K. A., Dichek H. L., Santamarina-Fojo S. (1995) J. Biol. Chem. 270,25396–25401 [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi J., Applebaum-Bowden D., Dugi K. A., Brown D. R., Kashyap V. S., Parrott C., Duarte C., Maeda N., Santamarina-Fojo S. (1996) J. Biol. Chem. 271,26296–26301 [PubMed] [Google Scholar]
- 6.Griffon N., Budreck E. C., Long C. J., Broedl U. C., Marchadier D. H., Glick J. M., Rader D. J. (2006) J. Lipid Res. 47,1803–1811 [DOI] [PubMed] [Google Scholar]
- 7.Wong H., Davis R. C., Nikazy J., Seebart K. E., Schotz M. C. (1991) Proc. Natl. Acad. Sci. U.S.A. 88,11290–11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis R. C., Wong H., Nikazy J., Wang K., Han Q., Schotz M. C. (1992) J. Biol. Chem. 267,21499–21504 [PubMed] [Google Scholar]
- 9.Dichek H. L., Parrott C., Ronan R., Brunzell J. D., Brewer H. B., Jr., Santamarina-Fojo S. (1993) J. Lipid Res. 34,1393–1440 [PubMed] [Google Scholar]
- 10.Lookene A., Groot N. B., Kastelein J. J., Olivecrona G., Bruin T. (1997) J. Biol. Chem. 272,766–772 [DOI] [PubMed] [Google Scholar]
- 11.Keiper T., Schneider J. G., Dugi K. A. (2001) J. Lipid Res. 42,1180–1186 [PubMed] [Google Scholar]
- 12.Broedl U. C., Jin W., Fuki I. V., Glick J. M., Rader D. J. (2004) FASEB J. 18,1891–1893 [DOI] [PubMed] [Google Scholar]
- 13.Gauster M., Hrzenjak A., Schick K., Frank S. (2005) J. Lipid Res. 46,977–987 [DOI] [PubMed] [Google Scholar]
- 14.Miller G. C., Long C. J., Bojilova E. D., Marchadier D., Badellino K. O., Blanchard N., Fuki I. V., Glick J. M., Rader D. J. (2004) J. Lipid Res. 45,2080–2087 [DOI] [PubMed] [Google Scholar]
- 15.Brown R. J., Miller G. C., Griffon N., Long C. J., Rader D. J. (2007) J. Lipid Res. 48,1132–1139 [DOI] [PubMed] [Google Scholar]
- 16.Hill J. S., Yang D., Nikazy J., Curtiss L. K., Sparrow J. T., Wong H. (1998) J. Biol. Chem. 273,30979–30984 [DOI] [PubMed] [Google Scholar]
- 17.Lutz E. P., Merkel M., Kako Y., Melford K., Radner H., Breslow J. L., Bensadoun A., Goldberg I. J. (2001) J. Clin. Invest. 107,1183–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sendak R. A., Bensadoun A. (1998) J. Lipid Res. 39,1310–1315 [PubMed] [Google Scholar]
- 19.Jin W., Fuki I. V., Seidah N. G., Benjannet S., Glick J. M., Rader D. J. (2005) J. Biol. Chem. 280,36551–36559 [DOI] [PubMed] [Google Scholar]
- 20.Jin W., Wang X., Millar J. S., Quertermous T., Rothblat G. H., Glick J. M., Rader D. J. (2007) Cell Metab. 6,129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iverius P. H., Ostlund-Lindqvist A. M. (1976) J. Biol. Chem. 251,7791–7795 [PubMed] [Google Scholar]
- 22.Bengtsson G., Olivecrona T. (1981) Eur. J. Biochem. 113,547–554 [DOI] [PubMed] [Google Scholar]
- 23.Olivecrona T., Bengtsson-Olivecrona G., Osborne J. C., Jr., Kempner E. S. (1985) J. Biol. Chem. 260,6888–6891 [PubMed] [Google Scholar]
- 24.Osborne J. C., Jr., Bengtsson-Olivecrona G., Lee N. S., Olivecrona T. (1985) Biochemistry 24,5606–5611 [DOI] [PubMed] [Google Scholar]
- 25.Schoonderwoerd K., Hom M. L., Luthjens L. H., Vieira van Bruggen D., Jansen H. (1996) Biochem. J. 318,463–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berryman D. E., Mulero J. J., Hughes L. B., Brasaemle D. L., Bensadoun A. (1998) Biochim. Biophys. Acta 1382,217–229 [DOI] [PubMed] [Google Scholar]
- 27.Ben-Zeev O., Doolittle M. H. (1999) Methods Mol. Biol. 109,257–266 [DOI] [PubMed] [Google Scholar]
- 28.Wong H., Yang D., Hill J. S., Davis R. C., Nikazy J., Schotz M. C. (1997) Proc. Natl. Acad. Sci. U.S.A. 94,5594–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi Y., Nakajima T., Inoue I. (2002) Eur. J. Biochem. 269,4701–4710 [DOI] [PubMed] [Google Scholar]
- 30.Lutz E. P., Kako Y., Yagyu H., Heeren J., Marks S., Wright T., Melford K., Ben-Zeev O., Radner H., Merkel M., Bensadoun A., Wong H., Goldberg I. J. (2004) J. Biol. Chem. 279,238–244 [DOI] [PubMed] [Google Scholar]
- 31.Harmon J. T., Nielsen T. B., Kempner E. S. (1985) Methods Enzymol. 117,65–94 [DOI] [PubMed] [Google Scholar]
- 32.Nilsson-Ehle P., Schotz M. C. (1976) J. Lipid Res. 17,536–541 [PubMed] [Google Scholar]
- 33.Belfrage P., Vaughan M. (1969) J. Lipid Res. 10,341–344 [PubMed] [Google Scholar]
- 34.Fiser A., Do R. K., Sali A. (2000) Protein Sci. 9,1753–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiser A., Sali A. (2003) Bioinformatics 19,2500–2501 [DOI] [PubMed] [Google Scholar]
- 36.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29,291–325 [DOI] [PubMed] [Google Scholar]
- 37.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234,779–815 [DOI] [PubMed] [Google Scholar]
- 38.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22,4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen M. Y., Sali A. (2006) Protein Sci. 15,2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bourne Y., Martinez C., Kerfelec B., Lombardo D., Chapus C., Cambillau C. (1994) J. Mol. Biol. 238,709–732 [DOI] [PubMed] [Google Scholar]
- 41.Jaye M., Lynch K. J., Krawiec J., Marchadier D., Maugeais C., Doan K., South V., Amin D., Perrone M., Rader D. J. (1999) Nat. Genet. 21,424–428 [DOI] [PubMed] [Google Scholar]
- 42.Badellino K. O., Rader D. J. (2004) Curr. Opin. Cardiol. 19,392–395 [DOI] [PubMed] [Google Scholar]
- 43.Hill J. S., Davis R. C., Yang D., Wen J., Philo J. S., Poon P. H., Phillips M. L., Kempner E. S., Wong H. (1996) J. Biol. Chem. 271,22931–22936 [DOI] [PubMed] [Google Scholar]
- 44.Zhang L., Lookene A., Wu G., Olivecrona G. (2005) J. Biol. Chem. 280,42580–42591 [DOI] [PubMed] [Google Scholar]
- 45.Kempner E. S., Schlegel W. (1979) Anal. Biochem. 92,2–10 [DOI] [PubMed] [Google Scholar]
- 46.Garfinkel A. S., Kempner E. S., Ben-Zeev O., Nikazy J., James S. J., Schotz M. C. (1983) J. Lipid Res. 24,775–780 [PubMed] [Google Scholar]
- 47.van Tilbeurgh H., Roussel A., Lalouel J. M., Cambillau C. (1994) J. Biol. Chem. 269,4626–4633 [PubMed] [Google Scholar]