Abstract
The 3,4-dihydroxyphenyl-l-alanine (Dopa)-containing proteins of mussel byssus play a critical role in wet adhesion and have inspired versatile new synthetic strategies for adhesives and coatings. Apparently, however, not all mussel adhesive proteins are beholden to Dopa chemistry. The cDNA-deduced sequence of Pvfp-1, a highly aromatic and redox active byssal coating protein in the green mussel Perna viridis, suggests that Dopa may be replaced by a post-translational modification of tryptophan. The N-terminal tryptophan-rich domain of Pvfp-1 contains 42 decapeptide repeats with the consensus sequences ATPKPW1TAW2K and APPPAW1TAW2K. A small collagen domain (18 Gly-X-Y repeats) is also present. Tandem mass spectrometry of isolated tryptic decapeptides has detected both C2-hexosylated tryptophan (W1) and C2-hexosylated hydroxytryptophan (W2), the latter of which is redox active. The UV absorbance spectrum of W2 is consistent with 7-hydroxytryptophan, which represents an intriguing new theme for bioinspired opportunistic wet adhesion.
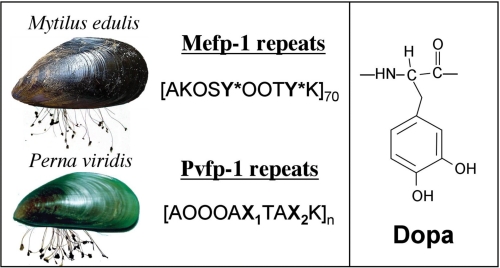
The amino acid 3,4-dihydroxyphenyl-l-alanine (Dopa)3 occurs in many proteins of the mussel holdfast or byssus (1, 2) and has recently been incorporated into mussel-inspired synthetic polymers with versatile adhesive consequences (3–7). One byssal protein in particular, mussel foot protein-1 (Mfp-1), has been investigated from over 15 mussel byssi, where it protectively coats compliant collagen-like proteins in the thread core (8–11). Mfp-1s typically contain 10–15 mol % Dopa in a highly conserved repeating peptide structure (8). In the blue mussel Mytilus edulis fp-1 (Mefp-1), for example, the consensus decapeptide AKPSYPPTYK is repeated over 70 times in tandem, and much of the tyrosine (Y) is converted to Dopa (Y*) (Fig. 1). In stark contrast to this, only trace levels of Dopa were detected in Pvfp-1 from the green mussel Perna viridis (Linnaeus 1758) (12), a notoriously invasive fouling species originally from the Indo-Pacific region (13). Because P. viridis fp-1 (Pvfp-1) and its homologue, Mefp-1, are both strongly aromatic, quinogenic, and composed of highly polar decapeptide repeats (12), identifying the Dopa-mimetic substitutes in Pvfp-1 has been a matter of considerable interest.
FIGURE 1.
The common blue mussel M. edulis and green mussel P. viridis are shown with attached byssal threads. Distinct consensus decapeptide repeat sequences are associated with the repeat domains in fp-1 foot proteins of the two species. The amino acid Dopa (Y*, right panel) is prominent in mefp-1 repeats but absent from Pvfp-1. O denotes trans-4-hydroxyprolines. Residues denoted as X1 and X2 are shown by this study to be derived from tryptophan.
MATERIALS AND METHODS
Protein Isolation from Mussel Feet
Green mussels (P. viridis) were collected from Tampa harbor in Florida. The feet were severed from about 100 freshly shucked mussels and stored at −80 °C. Protein extraction from mussel feet was adapted from a previous report (12). Frozen mussel feet (in lots of 10 g of wet weight) were thawed and depigmented by scraping with a scalpel. The depigmented feet were homogenized in a glass tissue grinder with 50 ml of 5% acetic acid and two protease inhibitors (pepstatin A and leupeptin, both 30 mm). The homogenate was centrifuged (15,000 × g, 4 °C, 30 min), and the recovered supernatant (S1) was chilled in an ice bath. 70% perchloric acid was added dropwise with stirring to a final perchloric acid concentration of 1.4% (v/v). The mixture was further stirred and centrifuged (15,000 × g, 4 °C) for 30 min. The supernatant (S2) was collected, dialyzed in 500 volumes of 5% acetic acid for 12 h at 4 °C, freeze-dried at −80 °C, and resuspended in 0.2 ml of 5% acetic acid.
P. viridis foot protein-1 (Pvfp-1) was purified in three stages: gel filtration, reversed phase HPLC, and finally, again, gel filtration. Initial isolation was achieved by gel filtration chromatography on a Shodex KW-803 column (5 μm, 8 × 300 mm). The column was equilibrated and eluted with 5% acetic acid at a flow rate of 0.2 ml/min. A maximum volume of 200 μl of S2 was loaded onto the Shodex column/run. The eluted volume was monitored at 280 nm, and the fractions under the Pvfp-1 peak were pooled (1–1.5 ml) and further resolved by C8 HPLC (Brownlee Aquapore RP-300, 7 μm, 4.6 × 250 mm) at a flow rate of 1 ml/min with an acetonitrile gradient described by Ref. 12. The collected fractions were assayed by redox cycling after acid urea gel electrophoresis (12) and amino acid analysis as described below to identify Pvfp-1 containing fractions. Pvfp-1 fractions were pooled and lyophilized at −80 °C. After resuspending in 200 μl of 5% acetic acid, Pvfp-1-positive fractions were run once more on Shodex KW-803 (above conditions). Fractions eluting between 33 and 43 min were redox active and had amino acid compositions consistent with previous studies (12). Purified Pvfp-1 fractions were recovered by freeze-drying. The N terminus of purified Pvfp-1 was sequenced by automated Edman degradation on a microsequencer (model 2090; Porton Instruments, Tarzana, CA) with a modified gradient program (14).
Mass Spectrometry
The mass of intact Pvfp-1 was determined by matrix-assisted laser desorption and ionization with time-of-flight (MALDI-TOF) mass spectrometry (Voyager DE, AB Biosystems, Foster City, CA) in the positive ion mode with delayed extraction. The MALDI matrix was prepared by dissolving sinapinic acid (10 mg/ml) in 30 vol % acetonitrile. Purified Pvfp-1 was dissolved in this matrix solution to give a final concentration between 1 and 10 pmol/μl. About 1 μl of sample was then spotted to the target plate and allowed to dry under vacuum (500 microns). Sample spots were irradiated at 337 nm using an N2 laser (LSI, Inc., Cambridge, MA) with a pulse width of 8 ns and a frequency of 5 Hz. Soft ionization generated singly, doubly, and triply protonated ions of the Pvfp-1 protein variants. The singly and doubly protonated ions of bovine serum albumin were used as molecular mass calibrants at m/z 66,430 and 33,215, respectively).
Cloning and Sequencing Pvfp-1 cDNA
To obtain the partial cDNA sequence of Pvfp-1, total RNA was first isolated from the phenol gland at the tip of P. viridis foot tissue using an RNase plant mini kit from Qiagen. A single freshly dissected foot was pulverized under liquid nitrogen using a mortar and pestle, after which the mini kit protocol was adopted verbatim. Following that, the first strand cDNA was synthesized using Superscript II reverse transcriptase at 50 °C with an Adapter primer, 5′-GGCCACGCGTCG ACTAGTACT(T)16-3′ (Invitrogen). The product of the reverse-transcribed reaction was used as a template for subsequent PCRs.
Assuming repetition of the known trypsinized internal peptide sequence APPPAX1TAX2K (where X denotes an unknown amino acid residues) from a previous study (12), a degenerate oligonucleotide (sense, 5′-AARGCNCCNCCNCCNGC-3′, which corresponds to the sequence KAPPPA) was designed and combined with an abridged universal amplification primer (antisense, 5′-GGCCACGCGTCGACTAGTAC-3′; Invitrogen) to amplify the 3′ end of Pvfp-1 from reverse transcriptase templates. The cloned 3′ cDNA end provided the C-terminal sequence of Pvfp-1 along with some 3′-untranslated region.
Another degenerate oligonucleotide (sense, 5′-GCNGTNTAYCAYCCNCCNTC-3′) was designed on the basis of N-terminal sequence of Pvfp-1 (AVYHPPS) and coupled with a gene-specific reverse primer (antisense, 5′-GGTTTGCCATATCCACCATAACC-3′ corresponding to GYGGYGK) cloned above to amplify the 5′ end cDNA of Pvfp-1.
Once the cDNA sequences encoding the mature Pvfp-1s were cloned, a GeneRacer kit (Invitrogen) was used to obtain the 5′ end untranslated region of the cDNA of Pvfp-1 from full-length transcripts by 5′ rapid amplification to cDNA ends. PCR was performed with gene-specific primer (antisense, 5′-GCTTTCCATGCAGTCCATGCAGGTGGATG-3′ (corresponding to HPPAWTAWK) coupled with a GeneRacer 5′ sense primer from Invitrogen.
Generally, PCR was carried out in 25 μl of 1× Buffer B (Fisher) and 5 pmol of each primer, 5 μmol of each dNTP, 1 μl of first strand reaction, and 2.5 units of Taq DNA polymerase (Fisher) for 35 cycles on a Robocycler (Stratagene). Each cycle consisted of 30 s at 94 °C, 30 s at 50 °C, and 1 min at 72 °C, with a final extension of 15 min. The PCR products were subjected to 1% agarose gel electrophoresis, purified, and cloned into a PCR TA vector (TOPO TA cloning kit; Invitrogen) and transformed into competent “Top10” cells (Invitrogen) for amplification, purification, and sequencing.
Byssal Thread Analysis
Fifteen to twenty threads were severed from the proximal end of each byssal stem, washed in 500 volumes of MilliQ water, and blotted dry. The dry threads were weighed and hydrolyzed by one of two methods: 6 m HCl for all amino acids except for Trp and 4 m NaOH for Trp (15). Acid hydrolysis was done with reusable hydrolysis vials (Pierce) at 110 °C for 24 h and flash evaporated under high vacuum, whereas NaOH hydrolysates were titrated to pH 3–4 with glacial acetic acid. Purification of Trp-derived amino acids was achieved by C-18 HPLC (Brownlee Aquapore AOD-300, 7 μm, 4.6 × 250 mm) over an acetonitrile gradient of 0–10%. The eluate was monitored continuously at 220 and 280 nm, and all of those peaks absorbing at both wavelengths were collected for analysis by electrospray ionization and tandem mass spectrometry.
For routine amino acid analysis, the purified Pvfp-1s were hydrolyzed in one of three ways: 1) 4 m methanesulfonic acid with 0.5% 3-methyl-indole (16), 2) 6 m HCl with 5% phenol in vacuo at 110 °C for 24 h, or 3) 4 m NaOH in vacuo at 110 °C for 24 h (15). The first method was dropped because of extremely low tryptophan derivative recovery. The HCl hydrolysate was flash evaporated at 50 °C under vacuum and shaken to dryness with ≤0.5 ml of MilliQ water followed by methanol; NaOH hydrolysates were titrated to pH 4. Amino acid analysis was performed according to conditions described earlier with a Beckman System 6300 auto analyzer (14) on which an authentic C2-(α-d-mannopyranosyl)-tryptophan (C-ManTrp) standard (17) eluted just after Met with a run time of 31.5 min.
Trp-containing Peptides
Preparation of tryptic peptides from Pvfp-1, including purification by C-18 HPLC (Brownlee, A300, 4.6 × 260 mm) was done exactly as described by Ohkawa et al. (12). Proteolysis was stopped by acidification to pH 4 with glacial acetic acid before HPLC. Three peak fractions corresponding to peptides d, e, and j described by Ohkawa were selected for analysis by electrospray ionization mass spectrometry followed by tandem mass spectrometry with collision-induced decomposition using the PE Sciex QStar quadrupole/time-of-flight tandem mass spectrometer (PerkinElmer Life Science) in the University of California at Santa Barbara Mass Spectrometry Facility. Interpretation of fragments produced from the peptides by collision-induced decomposition was assisted by comparison with an authentic C-ManTrp standard and by mock fragmentation of various model sequences using Protein Prospector, version 5.1.4, on ExPASy Tools.
UV-visible Spectra of Indole Derivatives
Chemical modifications based on 1-nitroso-2-naphthylation (18) and nitration (19) were performed as described. The former reacts only with the 5-hydroxy indole isomer, and the latter, which was developed for catechol detection, gave a bright yellow color for Pvfp-1 and its tryptic peptides. UV-visible spectra were obtained for 0.1 mm solutions of l-tryptophan (Sigma), C2-mannosyltryptophan (donated by S. Manabe), peptide e, and four hydroxyindoles (4-hydroxyindole (Acrõs Organics), 5-hydroxytryptophan (Sigma), 6-hydroxyindole (Oakwood Products, Columbia, SC), and 7-hydroytryptophan (SynChem OHG, Felsberg, Germany)) using an HP model 8453 UV-visible scanning spectrophotometer with UV-visible ChemStation (Rev 06.03). All of the solutions were buffered with 5% (v/v) acetic acid and scanned between 235 and 335 nm in masked 40-μl quartz cells (Starna, Atascadero, CA).
RESULTS
A previous investigation by Ohkawa et al. (12) reported the following attributes for Pvfp-1: 1) an apparent mass of 89 kDa based on SDS-PAGE, 2) marked quinone-like redox cycling activity without Dopa, 3) significant carbohydrate content, particularly with respect to mannose, N-acetylglucosamine and fucose, and 4) an Edman-derived primary sequence dominated by two closely related consensus repeats: AOOOAX1TAX2K and APOKOX1TAX2K, in which O denotes trans-4-hydroxyproline. The X1 and/or X2 positions were consistent with the presence of an aromatic amino acid but could not be reconciled with any known modification of Tyr or Dopa.
In the present investigation, purified Pvfp-1 was shown to consist of at least two variants (supplemental Fig. S1) with masses of 50 and 64 kDa observed by MALDI-TOF mass spectrometry (supplemental Fig. S2). The N-terminal sequence of the isolated protein was found to be (A)VY*HPPSX1TAX2IAOK (supplemental Fig. S1), where X1 and X2 represent Edman cycles devoid of detectable phenylthiohydantoin derivatives. Y* denotes Dopa and was the only Dopa detected in Pvfp-1.
To further explore the chemistry of X1 and X2, we first deduced the complete sequence of Pvfp-1 from its corresponding cDNA prepared by standard cloning procedures. Two Pvfp-1 variants, probably related by alternative splicing, were found with the shorter differing from the longer by exactly 15 decapeptide repeats. In the longer variant 1 (calculated mass, 61 kDa), APPPAWTAWK and ATPKPWTAWK consensus repeats occur 14 and 29 times, respectively (Fig. 2 and supplemental Fig. S3). Other intriguing features in Pvfp-1 are a distinct collagen-like sequence with 18 tripeptide Gly-X-Y repeats, followed by a short interval (14 amino acids long), before returning to a Trp-rich C-terminal sequence. It must therefore be concluded that in Pvfp-1 some modification of tryptophan, not tyrosine, provides the basis for the unknown aromatic amino acids in the decapeptide repeats.
FIGURE 2.
Complete protein sequence of Pvfp-1 variant 1 deduced from cDNA. The signal peptide is italicized, and the mature N terminus is indicated by arrows. Three directly sequenced peptide sequences are underlined: the N terminus (solid line), decapeptide repeats (peptides e and d, dashed line), and collagen (peptide j, dotted line). The single Dopa (Y) residue is circled. The variant 2 sequence is in the supporting data. The GenBankTM accession numbers for variants 1 and 2 are AAY46226.1 and AAY46227.1.
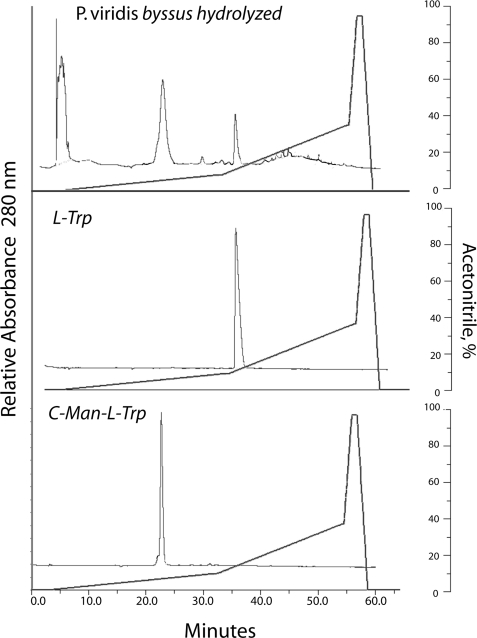
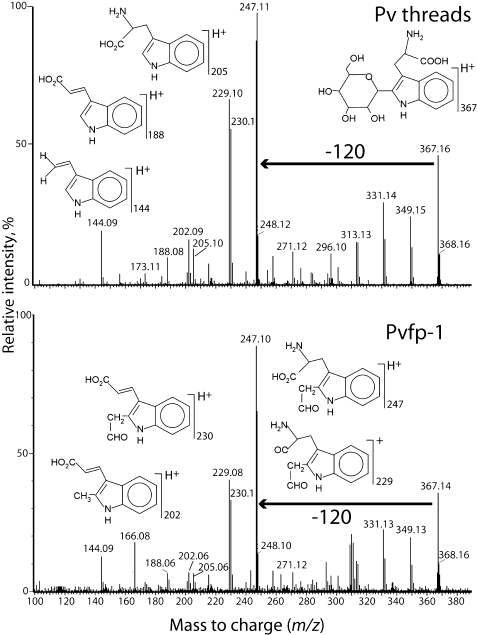
Because tryptophan is unstable to hydrolysis by HCl, purified Pvfp-1 and P. viridis byssal threads were hydrolyzed with 4 m NaOH. After hydrolysis, component aromatic amino acids were chromatographically separated by C-18 HPLC (Fig. 3) and analyzed by mass spectrometry to help deduce their relationship to tryptophan. Two ions with masses of 367 and 529 Da eluting between 22 and 24 min were detected; the latter decomposed readily to 367 through a loss of 162 Da (hexose) (supplemental Fig. S4), suggesting a cleavage that is typical of O- and N-linked hexoses (20, 21). The 367-Da ion and its fragmentation, most notably the 120-Da loss to m/z 247, are identical with standard C2-[α-d-mannopyranosyl]-tryptophan (22, 23) (Fig. 4). The hexose linked to Trp in Pvfp-1 is likely to be mannose, but tandem mass spectrometry by itself is unable to distinguish different hexoses. Because C-ManTrp elutes just after Met in amino acid analysis, this method was used to estimate a C2-mannosyltryptophan content of 1–2 dry weight % in byssal threads and 3–4 dry weight % in Pvfp-1 following base hydrolysis (supplemental Table S1). Only trace levels of Trp could be detected in Pvfp-1; thus the Trp in byssus must come from other proteins.
FIGURE 3.
C-18 HPLC of C2-hexosyl Trp from NaOH hydrolyzed P. viridis byssal threads (top panel), standard l-Trp (middle panel), and standard C2-mannosyltryptophan (bottom panel). Fractions at 22–25 min were sampled by electrospray ionization mass spectrometry and collision-induced decomposition. Standard 7-hydroxytryptophan, which does not survive NaOH hydrolysis, eluted at 17 min.
FIGURE 4.
Mass spectrometric analysis of the m/z 367 peak obtained from C-18 HPLC of NaOH hydrolyzed P. viridis threads (top panel) and purified Pvfp-1 (bottom panel). The loss of 120 Da is a signature for C-hexosylation.
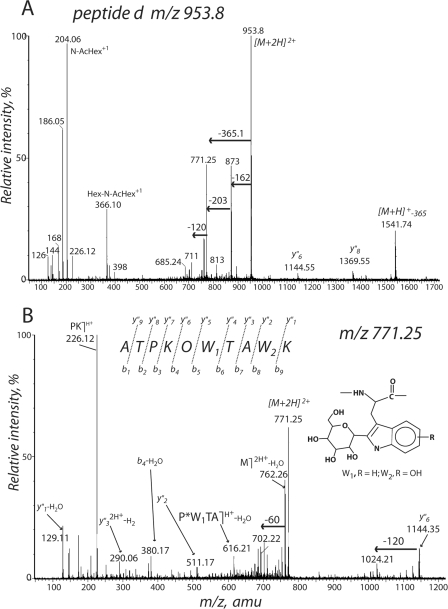
To detect modified tryptophans more directly in the decapeptide sequences of Pvfp-1, we used tandem mass spectrometry with collision-induced decomposition of three peptides following trypsin digestion of Pvfp-1 (supplemental Fig. S5). Peptides d and e were selected because 75% of their sequences had already been established by Edman chemistry (12). The doubly charged parent ion (M+2H)2+ (m/z 953.8) for the tryptic decapeptide d readily decomposes to another ion (M+2H)2+ (m/z 771.25) after a neutral loss of 162 to m/z 873 (hexose) followed by another neutral loss of 204 to m/z 771 (N-acetyl hexose) (Fig. 5A). The oxonium ion of hexosyl-N-acetylhexose is evident at m/z 366. The 771.25 peak loses water to become m/z 762, which then undergoes the 120 Da (60 × 2) neutral loss typical of C2-mannosylation; y″6 or OW1TAW2K (m/z 1144) does the same. The internal fragment, OW1TA (m/z 616.1), indicates a 366-Da mass for X1 (Fig. 5B). Notably, the 511.1 (y″2) and 290 (y″32+-2H) ions corresponding to W2K and AW2K are larger than the comparable W1-containing fragments by 16 Da. C-ManTrp + 16 is consistent with C2-hexosylated hydroxytryptophan (W2K) and the quinoid counterpart of C2-hexosylated hydroxytryptophan (AW2K) (Fig. 5B). A complete annotation of fragment masses is given in supplemental Fig. S6.
FIGURE 5.
Tryptic peptide d (parent ions [M+2H]2+m/z 953 and 771) following collision-induced decomposition. O, trans-4-hydroxyproline; W1, C2-hexosylTrp; W2, C2-hexosylhydroxyTrp. See supporting data for complete fragment annotation.
Mannose is plausible as the C2-hexose given that WX1X2W is a well accepted sequence signature for C2-mannosylation of tryptophan in proteins (20,24–26). The precise ring assignment of the hydroxyl group in hydroxytryptophan (OHTrp) is not possible from electrospray ionization tandem mass spectrometry, but a phenyl ring placement is consistent with the loss of two hydrogens to form a doubly protonated quinoid ion at m/z 290 and by the strong quinone-like redox cycling activity in Pvfp-1 and tryptic decapeptides (12).
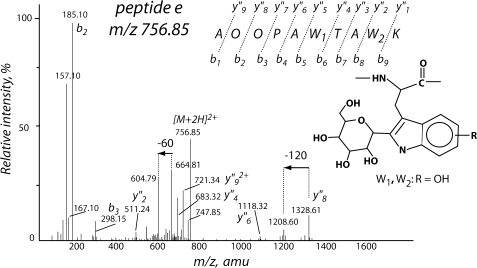
Peptide e (m/z 757) is less glycosylated than peptide d but shows some trends similar to those in the 771 ion during fragmentation (Fig. 6). The y″8 ion (OPAW1TAW2K) at m/z 1328 shows a neutral loss of 120 Da. The fragment ions implicate both W1 and W2 as C2-[α-d-mannopyranosyl]-hydroxytryptophan; the y″2 (W2K) and y″4 ions (TAW2K) are particularly suggestive of W2 (Fig. 6; complete fragment annotation in supplemental Fig. S7).
FIGURE 6.
Tryptic peptide e (parent ion [M+2H]2+m/z 757) following collision-induced decomposition. O denotes trans-4-hydroxyproline; W1 and W2 both are consistent with C2-hexosylhydroxyTrp. See supporting data for complete fragment annotation.
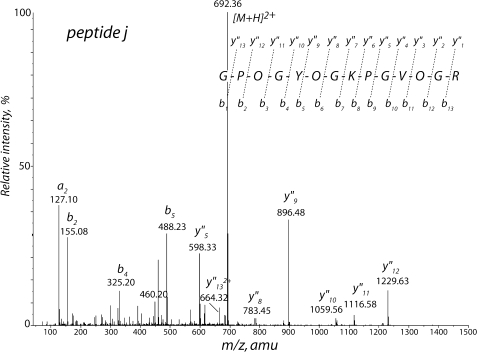
The fragmentation of peptide j (Fig. 7 and supplemental Fig. S8) was undertaken to corroborate the odd collagen-like primary structure predicted by the cDNA-deduced Pvfp-1 sequence (Fig. 2). Peptide j is 14 residues long, and its sequence corresponds to residues 496–509 with Gly at every third residue and three of the five prolines converted to trans-4-hydroxyproline.
FIGURE 7.
Tryptic peptide j (parent ion [M+2H]2+m/z 692) after collision-induced decomposition. O denotes trans-4-hydroxyproline. See supporting data for complete fragment annotation.
Neither hydroxytryptophan nor its C-hexosylated forms were detected after methansulfonic acid or NaOH hydrolysis of the byssus or Pvfp-1. We observed that whereas standard 5- or 7-hydroxytryptophan were stable to hydrolysis in methansulfonic acid, standard C-ManTrp was not. OHTrp standards hydrolyzed in the presence of sugars were not detected by amino acid composition. C2-[α-d-mannopyranosyl]-hydroxytryptophan would thus be unlikely to survive methansulfonic acid. OHTrp recovery following alkaline hydrolysis is prevented by the formation of quinonimine at alkaline pH (27).
Tandem mass spectrometry supports the presence of OHTrp but is unable to specify the position of the hydroxy group. To further clarify this important point, Pvfp-1 and peptide e were reacted with nitrosonaphthol, which is specific for the 5-hydroxy position on the indole (18); both were negative. In addition, the UV absorbance spectrum of peptide e at pH 3.5 was compared with a series of related indole derivatives (28, 29). In contrast to l-tryptophan and C-ManTrp, peptide e has a λmax at 269 nm shifted to a lower wavelength (Fig. 8 and supplemental Fig. S9), and an extinction coefficient of 18,000 m−1 cm−1 at 269 nm that is consistent with the presence of two modified tryptophans enhanced by mannosylation (28). Of the four hydroxy-indole derivatives tested, only 7-hydroxytryptophan shared the same λmax values as peptide e. Indeed, the two spectra are nearly superimposable between 250 and 300 nm. Although future studies need to further characterize the tryptophan chemistry of peptide e by proton NMR and electrochemistry, the identification W2 as C2 hexosyl-7-hydroxy tryptophan in Pvfp-1 appears reasonable. A final test of hydroxy-indole reactivity with Arnow's reagent showed that only 7-hydroxy-tryptophan produced the same bright yellow product formed by Pvfp-1 (supplemental Fig. S9).
FIGURE 8.
Ultraviolet absorbance spectra of Pvfp-1-derived peptide e and model hydroxyindoles in 5% acetic acid with extinction coefficients at selected wavelengths. Top panel, 4-hydroxyindole. Second panel, 5-hydroxytryptophan. Third panel, 6-hydroxyindole. Bottom panel, 7-hydroxytryptophan. The spectrum of peptide e (broken line) is superimposed with all other spectra.
DISCUSSION
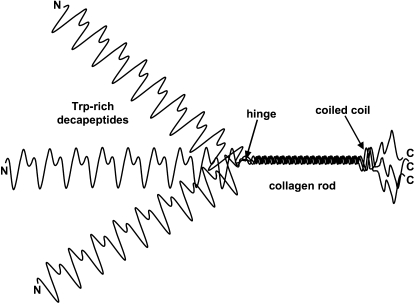
Pvfp-1 resembles other mussel coating proteins in its canonical, Pro- and Lys-rich repeats. It diverges significantly, however, in its reliance on modifications of tryptophan rather than tyrosine for its redox chemistry. Pvfp-1 consists of two closely related variants, the larger of which has predicted and observed masses of 61 and 64 kDa, respectively. Both variants show a highly repetitive sequence with four distinct domains: a Trp-rich decapeptide repeat (consensus: APPPAWTAWK and ATPKPWTAWK) domain largely in the N-terminal two-thirds, a collagen domain (Gly-X-Y repeats), a short hinge (residues 450–459), and a predicted, short coiled-coil region before returning to Trp-rich repeats at the C terminus (Fig. 9). The collagen domain is reminiscent of other collagen-like proteins such as the macrophage scavenger receptor (30), Torpedo acetylcholinesterase (31), and complement C1q (32), in which a small collagen domain directs trimer formation. The proposed trimeric structure remains speculative for Pvfp-1 because only the single chain mass has been determined with any accuracy by MALDI-TOF.
FIGURE 9.
Model of trimeric Pvfp-1 based on the trimerization imposed by formation of the collagen domain. Structure of the long N-terminal and short C-terminal repeat domains is not predictable at present. The coiled-coil region was predicted by Coils (window 14) in ExPAsy Tools.
Pvfp-1 exhibits extensive post-translational decoration. Pro is targeted for hydroxylation throughout both decapeptide consensus repeats and in the collagen domains; Thr-2 in ATPKPWTAWK appears to be O-glycosylated with O-(N-acetyl)-hexosyl-hexose, which agrees with the high levels of N-acetyl-glucosamine reported earlier (12) but differs from the previous designation of Pro for position 2 in peptide e. Possibly, the phenylthiohydantoin derivative of glycosylated Thr has the same elution time as Pro-phenylthiohydantoin on C-18 HPLC. Notably, Trp undergoes C2-hexosylation (probably by mannose) and hydroxylation. Whereas W1 and W2 are always hexosylated, W2 seems favored for hydroxylation. Peptides d and e may be representative of other decapeptides in Pvfp-1 with respect to C-hexosylation and hydroxylation. At this stage, however, aside from ample evidence for 120-Da mass losses, i.e. C-hexosyl-Trp in other peptides, collision-induced decomposition of many peptides remains too complex for confident interpretation. Pvfp-1 has only a single Dopa located near the N terminus.
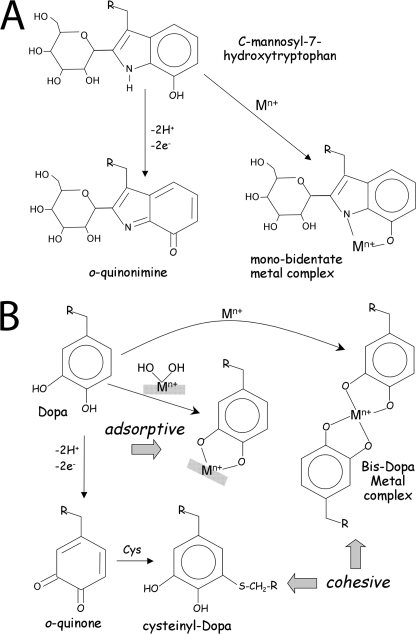
C-linked mannosylation of proteins is an unusual but not unprecedented modification of tryptophan. It was first reported in pancreatic ribonuclease and since then in over 47 other proteins including notably thrombospondin and complement proteins with the sequence motif WX1X2W (20, 22–26). Although the function of this modification remains elusive, mannosylation is known to render the tryptophan more polar and solvent-accessible (23). In complement proteins, C-ManTrp in the thrombospondin-like repeat domains has been proposed to have an adhesive function (33). The potential presence of up to 80 C-ManTrp residues in Pvfp-1, which functions as an adhesive and coating in byssus (9–11), begs the hypothesis that mannosylation makes tryptophan behave more like Dopa: sticky and prone to cross-link formation. This conjecture, however, is too simple because tryptophan needs to be hydroxylated before becoming redox active like Dopa. 5-Hydroxytryptophan in engineered proteins and as a free amino acid was reported to undergo oxidation resulting in the formation of p-quinonimine and di-Trp cross-links (27, 34). Indeed, based on these studies, we expected to find C2-mannosylated 5-HOTrp in peptide e and Pvfp-1. UV spectra, however, support the rarer 7-OH rather than the 5-OH isomer and represent the first report of naturally occurring 7-hydroxytryptophan in proteins (35). 7-OHTrp seems a better mimic of Dopa than the other isomers. Upon oxidation, it forms an o-quinonimine (Fig. 10A), and the indolic N-H and phenolic OH groups are close enough to chelate metal ions (Fig. 10A), a common capability of mussel byssus (9, 36).
FIGURE 10.
C2-hexosyl-7-OHTrp reactivity (A) has parallels with peptidyl-Dopa (B). In the redox pathway, C2-hexosyl-OHTrp loses two hydrogens to become an o-quinonimine (bottom); Dopa loses two hydrogens (2H+ + 2e−) to become an o-quinone that reacts with cysteine, histidine, lysine, and other Dopa residues to form cross-links (B). In the metal chelate pathway, both the indolic nitrogen and phenolic oxygen of C2-hexosyl-7-OHTrp should contribute electrons for a bidentate complexation (A, top right); Dopa forms stable bidentate complexes with metal ions and metal hydroxides (Mn+[OH]n) (B, center). The preferred metal (M) bound to Dopa in Mytilus byssus is FeIII; in Pvfp-1 of P. viridis byssus it is unknown. Covalent cross-links and bis-bidentate metal complexes contribute to adhesive cohesion, whereas mono-bidentate metal hydroxide complexes contribute to adhesive adsorption.
If 7-hydroxylation makes Trp more like Dopa, why bother with mannosylation? Trp mannosylation was observed to block cleavage in the vicinity of modified residues by exo- and endoproteases, hence rendering proteins more resistant to degradation (28). In addition, Trp-rich proteins are quite hydrophobic and thus prone to aggregation in solution (37). C-mannosylation of Trp residues appears to make them fully water accessible while preventing aggregation (38).
In summary, the green mussel P. viridis, employs an intriguing alternative to Dopa in Pvfp-1, one of its byssal adhesive proteins. Although much bulkier, C-Man-7-OHTrp resembles Dopa in having attributes that contribute to both cohesive and adsorptive interactions necessary for adhesion (39).
With respect to cohesion, the two-electron oxidation of C-Man-7-OHTrp to an o-quinonimine (Fig. 10A) mimics Dopa-o-quinone, which is known to form covalent cross-links with other Dopa, cysteine, lysine, and histidine residues (Fig. 10B) (7, 40). The coordination of metal ions via the o-hydroxyl groups in Dopa also contributes to cohesion, particularly in the byssal cuticle of Mytilus galloprovincialis (9). The ortho-placement of indolic-N and phenolic OH groups in C-Man-7-OHTrp seems well suited to metal ion chelation, but this has yet to be investigated.
The stickiness of adhesive molecules is determined by their adsorptive tendencies. Dopa complexation of metal hydroxides on surfaces provides strong and reversible interactions (Fig. 10B) (41). Again, given the ortho-placement of electronegative elements, such interactions also seem likely with C-Man-7-OHTrp. Future studies need to closely examine what adaptive advantages C-Man-7-OHTrp offers over Dopa.
Acknowledgments
We thank S. Manabe and Y. Ito (RIKEN) for synthetic C2-(α-d-mannopyranosyl)-tryptophan and K. Ohkawa (Shinshu University) for numerous discussions. C. Ross and M. Gilg (University of North Florida) and A. Benson and M. Blouin (U.S. Geological Survey) generously provided P. viridis from Tampa, FL.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DE018468. This work was also supported by National Science Foundation Grant MRSEC DMR05-20415.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S9.
- Dopa
- 3,4-dihydroxyphenyl-l-alanine
- C-ManTrp
- C2-[α-d-mannopyranosyl]-tryptophan
- MALDI
- matrix-assisted laser desorption ionization
- TOF
- time-of-flight
- OHTrp
- hydroytryptophan
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1.Waite J. H. (2002) Integr. Comp. Biol. 42,1172–1180 [DOI] [PubMed] [Google Scholar]
- 2.Waite J. H. (1992) Results Probl. Cell Differ. 19,27–54 [DOI] [PubMed] [Google Scholar]
- 3.Dalsin J. L., Messersmith P. B. (2005) Materials Today 8,38–46 [Google Scholar]
- 4.Yu M., Deming T. J. (1998) Macromolecules 31,4739–4745 [DOI] [PubMed] [Google Scholar]
- 5.Westwood G., Horton T. N., Wilker J. J. (2007) Macromolecules 40,3960–3964 [Google Scholar]
- 6.Lee H., Lee B. P., Messersmith P. B. (2007) Nature 448,338–341 [DOI] [PubMed] [Google Scholar]
- 7.Liu B., Burdine L., Kodadek T. (2006) J. Am. Chem. Soc. 128,15228–15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holten-Andersen N., Fantner G. E., Hohlbauch S., Waite J. H., Zok F. W. (2007) Nat. Materials 6,669–672 [DOI] [PubMed] [Google Scholar]
- 9.Holten-Andersen N., Mates T. E., Toprak M. S., Stucky G. D., Zok F. W., Waite J. H. (2009) Langmuir 25,3323–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holten-Andersen N., Waite J. H. (2008) J. Dent. Res. 87,701–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Q., Gourdon D., Sun C., Holten-Andersen N., Anderson T. H., Waite J. H., Israelachvili J. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104,3782–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohkawa K., Nishida A., Yamamoto H., Waite J. H. (2004) Biofouling 20,101–115 [DOI] [PubMed] [Google Scholar]
- 13.Benson A. J., Marelli D. C., Frischer M. E., Danforth J. M., Williams J. D. (2001) J. Shellfish Res. 20,21–29 [Google Scholar]
- 14.Waite J. H. (1991) Anal. Biochem. 192,429–433 [DOI] [PubMed] [Google Scholar]
- 15.Hugli T. E., Moore S. (1972) J. Biol. Chem. 247,2828–2834 [PubMed] [Google Scholar]
- 16.Simpson R. J., Neuberger M. R., Liu T. Y. (1976) J. Biol. Chem. 251,1936–1940 [PubMed] [Google Scholar]
- 17.Manabe S., Marui Y., Ito Y. (2004) Chem. Eur. J. 9,1435–1447 [DOI] [PubMed] [Google Scholar]
- 18.Knight J. A., Robertson G., Wu J. T. (1983) Clin. Chem. 29,1969–1971 [PubMed] [Google Scholar]
- 19.Waite J. H., Tanzer M. L. (1981) Anal. Biochem. 111,131–136 [DOI] [PubMed] [Google Scholar]
- 20.Li J. S., Cui L., Rock D. L., Li J. (2005) J. Biol. Chem. 280,38513–38521 [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-de Peredo A., Klein D., Macek B., Hess D., Peter-Katalinic J., Hofsteenge J. (2002) Mol. Cell. Proteomics 1,11–18 [DOI] [PubMed] [Google Scholar]
- 22.Gutsche B., Grun C., Scheutzow D., Herderich M. (1999) Biochem. J. 343,11–19 [PMC free article] [PubMed] [Google Scholar]
- 23.Hofsteenge J., Müller D. R., de Beer T., Löffler A., Richter W. J., Vliegenhart J. F. (1994) Biochemistry 33,13524–13530 [DOI] [PubMed] [Google Scholar]
- 24.Julenius K. (2007) Glycobiology 17,868–876 [DOI] [PubMed] [Google Scholar]
- 25.Furmanek A., Hofsteenge J. (2000) Acta Biochim. Pol. 20,781–789 [PubMed] [Google Scholar]
- 26.Hofsteenge J., Blommers M., Hess D., Furmanek A., Miroshnichenko O. (1999) J. Biol. Chem. 274,32786–33794 [DOI] [PubMed] [Google Scholar]
- 27.Wu Z., Dryhurst G. (1996) Bio-org. Chem. 24,127–149 [Google Scholar]
- 28.Hofsteenge J., Löffler A., Müller D. R., Richter W. J., de Beer T., Vliegenthart J. F. (1996) Tech. Protein Chem. 8,163–171 [Google Scholar]
- 29.Ek A., Witkop B. (1953) J. Am. Chem. Soc. 75,500–501 [Google Scholar]
- 30.Kodama T., Freeman M., Rohrer L., Zabrecky J., Matsudaira P., Krieger M. (1990) Nature 343,531–535 [DOI] [PubMed] [Google Scholar]
- 31.Kishore U., Reid K. B. (2000) Immunopharmacology 49,159–170 [DOI] [PubMed] [Google Scholar]
- 32.Mays C., Rosenberry T. L. (1981) Biochemistry 20,2810–2817 [DOI] [PubMed] [Google Scholar]
- 33.Maves K. K., Weiler J. M. (1993) Immunol. Res. 12,233–243 [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z., Alfonta L., Tian F., Bursulaya B., Uryu S., King D. S., Schultz P. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101,8882–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuda M., Takahashi Y., Fromont J., Mikami Y., Kobayashi J. (2005) J. Nat. Prod. 68,1277–1278 [DOI] [PubMed] [Google Scholar]
- 36.Nicholson S., Szefer P. (2003) Marine Pollution Bulletin 46,1040–1043 [DOI] [PubMed] [Google Scholar]
- 37.Liu J., Yong W., Deng Y., Kallenbach N. R., Lu M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101,16156–16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munte C. E., Gäde G., Domogalla B., Kremer W., Kellner R., Kalbitzer H. R. (2008) FEBS J. 275,1163–1173 [DOI] [PubMed] [Google Scholar]
- 39.Waite J. H., Holten-Andersen N., Jewhurst S., Sun C. (2005) J. Adhesion 81,297–317 [Google Scholar]
- 40.Sagert J., Sun C. J., Waite J. H. (2006) in Biological Adhesives ( Smith A. M., Callow J. A. eds) pp. 125–143, Springer Verlag, Heidelberg [Google Scholar]
- 41.Lee H., Scherer N. F., Messersmith P. B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103,12999–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]