Abstract
Reversible protein glutathionylation plays a key role in cellular regulation and cell signaling and protects protein thiols from hyperoxidation. Sulfiredoxin (Srx), an enzyme that catalyzes the reduction of Cys-sulfinic acid derivatives of 2-Cys peroxiredoxins (2-Cys Prxs), has been shown to catalyze the deglutathionylation of actin. We show that deglutathionylation of 2-Cys Prx, a family of peroxidases, is specifically catalyzed by Srx. Using the ubiquitously expressed member of 2-Cys Prx, Prx I, we revealed the following. (i) Among its four Cys residues, Cys52, Cys83, and Cys173 can be glutathionylated in vitro. Deglutathionylation with Cys mutants showed that Cys83 and Cys173 were preferentially catalyzed by Srx, with glutathionylated Srx as the reaction intermediate, whereas glutaredoxin I was more favorable for deglutathionylating Cys52. (ii) Studies using site-directed mutagenesis coupled with binding and deglutathionylation activities revealed that Pro174 and Pro179 of Prx I and Tyr92 of Srx are essential for both activities. Furthermore, relative to glutaredoxin I, Srx exhibited negligible deglutathionylation activity for glutathionylated cysteine and glutathionylated BSA. These results indicate that Srx is specific for deglutathionylating Prx I due to its favorable affinity for Prx I. To assess the biological relevance of these observations, we showed that Prx I is glutathionylated in A549 and HeLa cells under modest levels of H2O2. In addition, the level of glutathionylated Prx I was substantially elevated in small interfering RNA-mediated Srx-knocked down cells, whereas the reverse was observed in Srx-overexpressing cells. However, glutathionylation of Prx V, not known to bind to Srx, was not affected by the change in Srx expression levels.
Reversible covalent modifications of proteins play important roles in cellular regulation and in signal transduction due, in part, to their enormous capacity for integrating biological information and signal amplification (1, 2). Among them, protein glutathionylation/deglutathionylation is mediated by redox signals that are known to be generated in response to ligation of various cell surface receptors (3–6). Furthermore, protein glutathionylation can protect the protein thiol moiety from being hyperoxidized to its irreversible sulfinic acid derivative, due to the catalytic action of glutaredoxin in regenerating the deglutathionylated protein. Glutathionylation of various proteins was found to be involved in various physiological processes, such as growth, differentiation, cell cycle progression, transcriptional activation, cytoskeletal functions, and metabolism. Together with the fact that GSH is present in cells in the millimolar concentration range, protein glutathionylation constitutes a major cellular regulatory mechanism (for a review, see Refs. 7–9).
Sulfiredoxin (Srx),3 a small cysteine-containing protein, was first purified from Saccharomyces cerevisiae and characterized as an enzyme to catalyze the reduction of the cysteine sulfinic acid derivative in peroxiredoxin (Prx) (10). This enzyme is responsible for the in vivo reactivation of the inactive sulfinic acid derivative of 2-Cys Prx (11). Further investigation by Chang et al. (12) revealed that Srx is ubiquitously expressed in mammalian tissues. Recently, Findlay et al. (13) observed that HEK293 cells transfected with the pcDNA3.1/hismyc plasmid containing Srx were capable of lowering the levels of glutathionylated proteins, such as glutathionylated actin and PTP1B (protein-tyrosine phosphatase 1B), induced by the anticancer prodrug, PABA/NO. This observation was verified by showing that purified human Srx can catalyze the deglutathionylation of actin and PTP1B, although its catalytic efficiency was not examined.
Peroxiredoxins are thiol-specific antioxidant enzymes ubiquitously expressed in organisms (14, 15). These enzymes are known to catalyze the removal of H2O2 and lipid hydroperoxides. Mammalian cells express six Prxs, and they can be divided into three subclasses, namely typical 2-Cys Prxs that consist of Prx I–IV, atypical 2-Cys Prx (Prx V), and 1-Cys Prx (Prx VI). Among them, Prx I is highly abundant and ubiquitously expressed. The typical 2-Cys Prxs are the largest class of Prxs that contain a conserved N-terminal Cys residue and a conserved C-terminal Cys residue. During catalytic action, the conserved N-Cys-free thiol is selectively oxidized to Cys-SOH, which reacts with the C-Cys-free thiol of the other subunit to form an intermolecular disulfide bond, and the disulfide bond is reduced specifically by Trx (16, 17). The sulfenic intermediate can be further oxidized to the sulfinic acid derivative (Cys-SO2H), resulting in enzyme inactivation (18, 19). Srx was identified as the enzyme that catalyzes the reduction of the sulfinic acid derivative in the presence of Mg2+, ATP, and thiol as electron donors (10, 12).
Using diamide, a strong RSH oxidant, Fratelli et al. (20, 21) showed that Prx I and Prx V can be glutathionylated in human T lymphocytes and hepatoma cells, respectively. However, diamide is capable of extracting a hydride from thiols to form a reactive RS+ that can easily form a disulfide bond with another thiol, such as GSH (22). Therefore, the glutathionylated proteins so obtained may not be physiologically relevant.
In this study, we showed that in the absence of diamide and H2O2, Prx I can be glutathionylated at three (Cys52, Cys83, and Cys173) of the four cysteine residues by GSSG. Using site-directed mutagenesis, we revealed that Srx preferentially catalyzes the deglutathionylation of Cys83 and Cys173, whereas Grx is a preferred enzyme to catalyze the deglutathionylation of Cys52. The specificity of Srx as a deglutathionylating enzyme for Prx I derives from its favorable binding affinity to Prx I. The catalytic reaction involves the formation of a glutathionylated Srx, at its only and conserved cysteine residue, as a reaction intermediate. The biological relevance of this finding is supported by the observation that the level of glutathionylated Prx I in A549 cells was substantially elevated in Srx-siRNA-treated cells and substantially reduced in Srx-overexpressing cells.
EXPERIMENTAL PROCEDURES
Materials
NADPH, GSH, GSSG, dithiothreitol (DTT), iodoacetamide (IAM), acrylamide, ATP, glutathione ethyl ester, and MgCl2 were obtained from Sigma; glutathione reductase was from U.S. Biochemical Corp. (Cleveland, OH); sulfo-NHS-biotin was from Pierce; streptavidin-agarose was from Invitrogen; Lys-C and chymotrypsin were from Roche Applied Science; human Grx I, which has a specific activity of 97 units/mg (pure Grx I exhibits a specific activity of 100 units/mg using glutathionylated cysteine (CSSG) as substrate, and 1 unit is defined as 1 μmol of NADPH oxidized/GSSG formed/min), glutathionylated cysteine and glutathionylated BSA were prepared in the laboratory of Professor John J. Mieyal (Case Western Reserve University, Cleveland, OH); a monoclonal antibody to glutathione was from Virogen (Watertown, MA); and a polyclonal antibody to human Srx was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Preparation of Recombinant Proteins
Escherichia coli expression plasmids encoding wild type human Prx I and two mutants with Cys52 and Cys173 individually replaced by serine residues (C52S and C173S, respectively) were prepared as described (18). Three Prx I mutants with two cysteines replaced by serine residues (C52S/C83S, C83S/C173S, and C52S/C173S) were generated by standard PCR-mediated site-directed mutagenesis using Prx I (C52S) or Prx I (C173S) as templates. The procedure for constructing the bacterial expression vector for human wild type Srx was described previously (12). Mutant Srx proteins with His100, Arg101, and Tyr92 replaced by Asn, Met, and Arg, respectively, were generated by standard PCR-mediated site-directed mutagenesis with hSrx as a template. Escherichia coli BL21(DE3) cells harboring each plasmid were cultured at 37 °C in LB medium supplemented with ampicillin (100 μg/ml). After the addition of isopropyl-1-thio-β-d-galactopyranoside (0.4 mm), the cultures were incubated for 3 h at 25 °C, and then the cells were lysed. The recombinant proteins were purified as described (23).
Preparation of Glutathionylated Proteins
Glutathionylation was carried out by disulfide exchange with GSSG. Typically, 40 μm recombinant protein was incubated overnight with 10 mm GSSG at 4 °C in 50 mm Tris (pH 7.4) buffer containing 100 mm NaCl. After overnight incubation, excess GSSG was removed using either size exclusion chromatography (TSK-GEL G3000SW; TOSOH Corp., Tokyo, Japan) or strong anion exchange chromatography (TSK Super Q-5PW; TOSOH). The glutathionylated products were identified using reverse phase high performance liquid chromatography (HPLC) mass spectrometric methods.
LC-MS Analysis
Proteins and peptides were analyzed using reverse phase HPLC with a Vydac C18 column coupled with an electrospray mass spectrometer (Agilent 1100 Series LC/MSD). The column was pre-equilibrated with 0.05% trifluoroacetic acid in water. The elution was performed over 35 min with a linear gradient of 0–70% acetonitrile in 0.05% trifluoroacetic acid. The effluent from the spectrophotometric detector was mixed in a tee with 100 ml/min acetic acid pumped by another model 1100 pump, and the mixture was introduced into the mass spectrometer (24). Mass spectra were deconvoluted using the software provided by the instrument manufacturer (Chemstation Version 9, Agilent Technologies, Palo Alto, CA). Theoretical average m/z values of singly charged ions, [M + H]+, were calculated using GPMAW software (Lighthouse Data, Odense, Denmark).
Double Alkylation and Digestion of Prx I
Dried protein (20 μg) was redissolved in 100 mm Tris buffer (pH 8.5), containing 6 m guanidine HCl, 1 mm EDTA, and 10 mm iodoacetamide, followed by a 30-min incubation at 37 °C. After precipitation in 20% trichloroacetic acid and washing with ethanol/ethyl acetate, the protein was incubated with 5 mm DTT for 30 min and subsequently reacted with 25 mm acrylamide at 37 °C for 30 min. The alkylated proteins were digested with Lys-C overnight at room temperature, and half of the sample was further digested with chymotrypsin to smaller peptides.
Standard Coupled Assay for Measuring Deglutathionylation
The time course of the deglutathionylation catalyzed by either Srx or Grx I was monitored by the production of GSSG, which was measured by the GSSG-mediated oxidation of NADPH catalyzed by GSSG reductase, modified from a procedure described previously (25). The reaction mixtures for the coupled assay contained 50 mm Tris (pH 7.4), 100 mm NaCl, 150 μm NADPH, 0.5 mm GSH, 2 units/ml GSSG reductase, and the indicated concentration of glutathionylated substrate and Srx or Grx I. The deglutathionylation reactions were carried out at 37 °C. The rate of NADPH oxidation was measured from the slope of the linear portion of the time course of decreasing A340 nm. Each experiment was repeated at least five times, and a typical one is shown.
Identification of Reaction Intermediate
To identify the reaction intermediate for the deglutathionylation of Prx I catalyzed by Srx, 5 μm glutathionylated Prx I (C52S/C83S) or Prx I (C83S/C173S) was incubated with 5 μm Srx at 37 °C for 1 h. The reaction was terminated by the addition of trifluoroacetic acid until the pH reached 2–3. The resulting products were analyzed using the LC-MS method described above.
Cell Culture
A549 (human lung carcinoma) cells were obtained from the American Type Culture Collection (Manassas, VA) (ATCC number CCL-185) and maintained in RPMI 1640 medium (Quality Biological, Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin (Invitrogen). HeLa (human cervical adenocarcinoma) cells were maintained in Dulbecco's minimum essential medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (CCL-2TM; ATCC).
Biotinylation of Glutathione Ethyl Ester
BioGEE was prepared as described previously (26). Briefly, sulfo-NHS-biotin was reacted with glutathione ethyl ester at a 1:1 molar ratio in 50 mm NaHCO3, pH 8.5. After 1 h of incubation at room temperature, the reaction was terminated, and the remaining biotinylation reagent was quenched by the addition of NH4HCO3 to a 5-fold molar excess with respect to the starting sulfo-NHS-biotin concentration.
Purification of Glutathionylated Proteins Using BioGEE
BioGEE was added to the medium 1 h prior to the addition of H2O2. Soluble proteins covalently bound to biotin were extracted in batches using streptavidin-agarose. The agarose beads were washed three times with radioimmune precipitation buffer (1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 150 mm NaCl, and 50 mm Tris, pH 8.0) and twice with phosphate-buffered saline containing 2 mm EDTA and 0.1% SDS. Proteins bound to streptavidin via a disulfide bond were then eluted from the beads by incubation for 30 min with phosphate-buffered saline/EDTA/SDS containing 10 mm DTT. Proteins in the eluent were resolved by SDS-PAGE and detected by Western blotting.
Depletion and Overexpression of hSrx in A549 Cells
A small interfering RNA (siRNA) duplex targeting the 5′-GGAGGUGACUACUUCUACU-3′ sequence in the open reading frame of hSrx mRNA as well as a control RNA duplex of random sequence were obtained from Dharmacon Research (Chicago, IL). The A549 cells (2 × 106) were suspended in 100 μl of solution T (Amaxa Biosystems, Walkersville, MD) with 1 μm siRNA, and this mixture was subjected to electroporation using the U-17 program of the Nucleofector instrument (Amaxa Biosystems, Walkersville, MD). For overexpression of Srx, A549 cells were transfected with 2 μg of pCDNA/Srx or an empty vector using the same procedure as described for siRNA.
RESULTS AND DISCUSSION
In Vitro Glutathionylation of Prx I
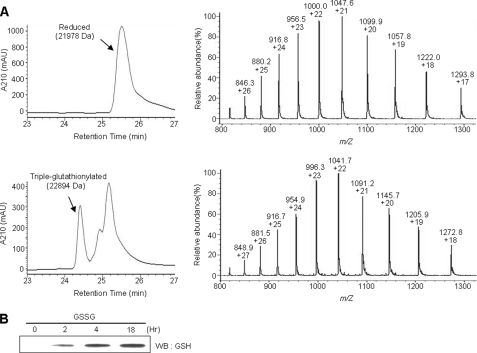
Human Prx I contains four cysteine residues: Cys52, Cys71, Cys83, and Cys173. To investigate whether these cysteine residues can be glutathionylated, recombinant Prx I protein was incubated with GSSG at 4 °C for 18 h in 50 mm Tris (pH 7.4) buffer containing 100 mm NaCl. The oxidized end products were analyzed using reverse phase HPLC coupled with electrospray mass spectrometry. The theoretical mass for the reduced form of recombinant Prx I is 21,979.3 Da, and we observed a value of 21,977.7 Da (Fig. 1A). After 18 h of incubation with 10 mm GSSG, two new peaks were observed in the chromatogram. Mass analysis revealed that the major new peak had a mass of 22,894, which is 916 Da higher than that of the reduced form. Since the theoretical mass for three covalently bound glutathiones is 915.9 Da, this peak corresponds to a triple-glutathionylated Prx I. The second peak contained several species, including mono- and diglutathionylated Prx I. Glutathionylation of Prx I was also observed in a time-dependent manner by Western blot analysis using antibodies specific for glutathione (Fig. 1B). Considering the fact that Prx I has two active site cysteines, N-Cys (Cys52) and C-Cys (Cys173), it was expected that Cys52 and Cys173 would be easily glutathionylated. However, except for those two cysteines, it was not clear which ones of the remaining two might be glutathionylated in Prx I. Therefore, additional experiments were carried out to identify the glutathionylated cysteine residues.
FIGURE 1.
Glutathionylation of Prx I. A, LC-MS analysis of Prx I glutathionylation. Glutathionylation was carried out by disulfide exchange with GSSG. The reaction mixtures containing 40 μm Prx I and 10 mm GSSG in Tris buffer, pH 7.4, were incubated at 4 °C for 18 h. The protein products were then subjected to reverse phase HPLC-MS analysis. B, Western blot analysis of Prx I glutathionylation. The time course of Prx I glutathionylation described in A was monitored by Western blot (WB) using an antibody specific for glutathione.
Identification of Glutathionylated Cysteine Residues in Prx I
To determine the glutathionylated cysteine residues in Prx I, a double alkylation method was employed. In view of the fact that thiol groups on cysteines can be modified easily by alkylating reagents, such as iodoacetamide and acrylamide, the redox state of individual thiols can be determined by using a stepwise alkylation technique in combination with mass spectrometric analysis.
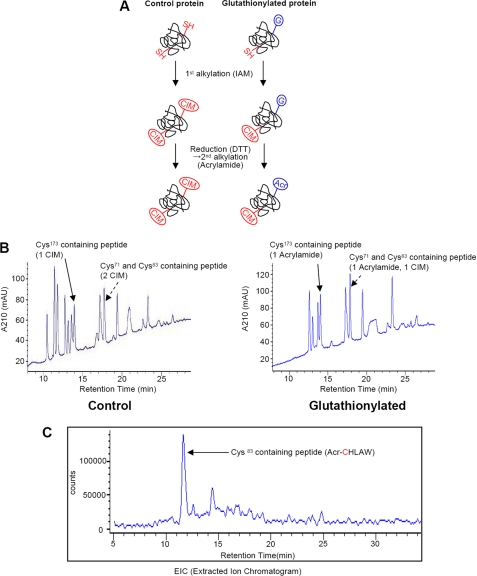
The scheme for this double alkylation procedure is illustrated in Fig. 2A and shows how glutathionylated cysteine was identified. Prx I (C52S) mutant protein, in which Cys52 was replaced by serine, was subjected to glutathionylation and alkylation. The use of this Prx I mutant instead of Prx I (WT) was to prevent disulfide bond formation during the reaction. The protein was confirmed to be glutathionylated on two cysteine residues by LC-MS analysis (data not shown). The reduced (control) and glutathionylated Prx I (C52S) were redissolved in 0.1 m Tris (pH 8.5) buffer containing 6 m guanidine HCl, 1 mm EDTA, and 10 mm iodoacetamide to alkylate non-glutathionylated cysteine residues. After 30 min of incubation at 37 °C, the protein was precipitated by the addition of 10% trichloroacetic acid, followed by washing to remove the unreacted reagents. The protein was dried and then redissolved in 6 m guanidine HCl in Tris buffer as it was in the first alkylation step but without iodoacetamide. The glutathionylated residue was then reduced with 5 mm DTT for 30 min at 37 °C. Subsequently, the reduced cysteine was alkylated with 25 mm acrylamide. Thus, the cysteine residues modified by iodoacetamide were initially reduced, and the glutathionylated cysteine was the one modified by acrylamide.
FIGURE 2.
Identification of glutathionylated cysteine residues in Prx I using Prx I (C52S) mutant. A, a scheme for the double alkylation procedure. SH, G, IAM, CIM, and Acr, free thiol, glutathione, iodoacetamide, carbamidomethyl group, and acrylamide, respectively. B, LC-MS analysis of Lys-C digest from control and glutathionylated Prx I (C52S). Reduced (control) and glutathionylated Prx I (C52S) proteins were alkylated, as shown in A, and the resulting proteins were subjected to LC-MS analysis after Lys-C digestion. Peaks corresponded to two peptides, one containing both Cys71 and Cys83 and the other containing Cys173, which are highlighted with arrows. C, extracted ion chromatogram of Cys83-containing peptide from Lys-C/chymotrypsin digest of glutathionylated Prx I (C52S). A portion of the Lys-C digest from glutathionylated Prx I (C52S) was further incubated with chymotrypsin to hydrolyze a peptide bond between Cys71 and Cys83. The resulting digests were subjected to LC-MS analysis. The peak corresponding to the peptide containing Cys83 modified by acrylamide was extracted from the mass window.
The alkylated proteins were incubated with Lys-C at room temperature overnight, and then one-half was used for LC-MS analysis (Fig. 2B), and the other half was further incubated with chymotrypsin because Lys-C could not hydrolyze the peptide bonds between Cys71 and Cys83. Fig. 2B shows the elution patterns obtained after the alkylated control and glutathionylated Prx I (C52S) were digested with Lys-C. The peaks identified with an arrow contained the cysteine residue(s). Results from the LC-MS analysis of the cysteine- and the Ser52-containing peptides obtained from the glutathionylated Prx I (C52S) are summarized in Table 1. The theoretical masses for peptides 69–93 and 169–190 with the sequence shown in Table 1 should be 2767.4 and 2349.16 Da, respectively. However, the observed values were 2896.0 Da for peptide 69–93 and 2420.5 Da for peptide 169–190. These results indicate that Cys71 and Cys83 in peptide 69–93 were modified by one acrylamide and one iodoacetamide, since alkylation by one iodoacetamide and one acrylamide would add 57.02 and 71.04 Da to the unmodified peptide, respectively. Together, they correspond to a mass increase of 128.06 Da. In addition, based on the observed mass of 2420.5 Da in peptide 169–190, it indicates that Cys173 was alkylated with acrylamide, since the observed mass is 71.34 Da higher than that expected for the unmodified peptide 169–190. These results indicate that there are two glutathionylation sites in Prx I (C52S); one is Cys173, and the other could either be Cys71 or Cys83. To identify which of these two cysteines is the glutathionylation site, the Lys-C digest was subjected to chymotrypsin digestion, since it cleaves the peptide at the carboxyl site of Tyr, Trp, and Phe. Thus, from peptide 69–93, one would obtain peptides 69–82, 83–87, and 88–93. The mass analysis showed that peptide 83–87 was alkylated with an acrylamide, since its observed mass was 699.32 Da, which is 71 Da higher than the theoretical value of 628.32 Da calculated for the peptide CHLAW. These results indicate that the glutathionylation sites in Prx I are Cys83, Cys52, and Cys173, since Cys71 is not glutathionylated. Interestingly, Cys83 in Prx I is located at the putative dimer-dimer interface (27) and is susceptible to glutathionylation. As a consequence, glutathionylation may exert an effect on Prx I oligomerization.
TABLE 1.
Cysteine-containing peptides from control (upper lines) or glutathionylated (lower lines) Prx I (C52S) analyzed by LC-MS
Theoretical average m/z values of singly charged ions ([M + H]+) were calculated using GPMAW software (Lighthouse Data, Odense, Denmark).
| Residues | Retention time | Modification | Mass |
Sequence | |
|---|---|---|---|---|---|
| Theoretical | Observed | ||||
| min | m/z | ||||
| 37–66 | 26.7 | None | 3626.1 | 3625.3 | YVVFFFYPLDFTFVSPTEIIAFSDRAEEFK |
| 69–93 | 17.7 | 2 CIM | 2881.4 | 2881.9 | LNCQVIGASVDSHFCHLAWVNTPKK |
| 17.8 | 1 ACR/1 CIM | 2895.5 | 2896.0 | ||
| 169–190 | 13.9 | 1 CIM | 2406.2 | 2406.4 | HGEVCPAGWKPGSDTIKPDVQK |
| 14.0 | 1 ACR | 2420.2 | 2420.5 | ||
Prx I Deglutathionylation Catalyzed by Srx and Grx I
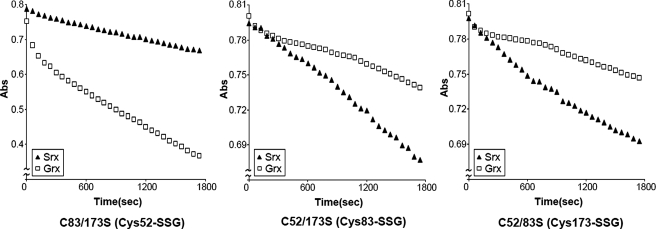
Although Srx was discovered as an enzyme that catalyzes the reduction of the cysteine sulfinic acid derivative in Prx (10, 11), it has also been shown to possess glutathionyl mixed disulfide oxidoreductase activity (13). We investigated whether Srx can function as an enzyme to catalyze the deglutathionylation of glutathionylated Prx I and compared its catalytic efficiency with that of Grx I (7, 28). To avoid complexity, characterization of the deglutathionylation process for each of the three cysteine residues in Prx I was carried out with three double mutants in which two of the glutathionylation sites were replaced with serine residues. With these mutants, the rate of deglutathionylation can be determined individually for each glutathionylated cysteine in Prx I. Fig. 3 shows the time courses for the deglutathionylation of Cys52, Cys173, and Cys83 catalyzed by Srx or Grx I. The deglutathionylation reaction was monitored by a coupled enzyme system in which GSSG formed during the deglutathionylation reaction was used as substrate to oxidize NADPH catalyzed by GSSG reductase. The data reveal that at 37 °C, the initial rate for the deglutathionylation of the glutathionylated Cys52, Cys83, and Cys173 catalyzed by Srx ranged from 0.58 to 0.63 μm/min, whereas that catalyzed by fully active human Grx I for Cys52 was 1.51 μm/min, and that for Cys83 and Cys173 was 0.21 and 0.20 μm/min, respectively. In other words, relative to Grx I, Srx is ∼3-folds more efficient in catalyzing the deglutathionylation of Cys83 and Cys173. However, Grx I was found to be about 2.5-folds more efficient than Srx in catalyzing the deglutathionylation of Cys52. It should be pointed out that the initial rapid decrease in A340 nm shown in some of the time courses in Fig. 3 was caused by the reduction of the contaminated GSSG by NADPH. This GSSG was generated by the oxidation of GSH prior to the addition of GSSG reductase, followed by substrate addition to initiate the deglutathionylation reaction.
FIGURE 3.
Deglutathionylation of Prx I mutants catalyzed by Srx and Grx I. The time course of the deglutathionylation reaction of Prx I mutants was measured at 37 °C with a coupled assay enzyme, GSSG reductase (2 units/ml), which catalyzed the reduction of GSSG generated during the deglutathionylation reaction in the presence of NADPH (0.15 mm). In addition, the reaction mixtures also contained 50 mm Tris (pH 7.4), 100 mm NaCl, 0.5 mm GSH, 1 μm human Grx I (□) or Srx (▴), and 20 μm glutathionylated Prx I mutant. Using CSSG as substrate, the specific activity of pure human Grx I is 100 ± 10 units/mg, and the enzyme used in this assay has a specific activity of 97 units/mg.
Identification of Glutathionylated Srx as Reaction Intermediate
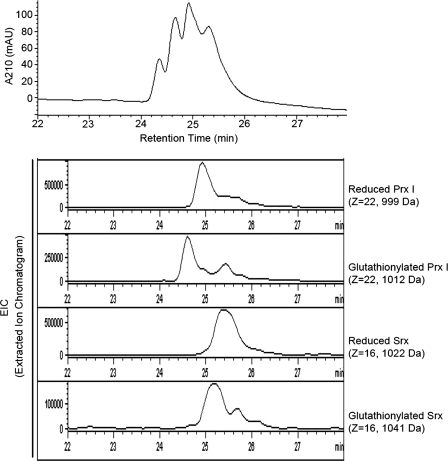
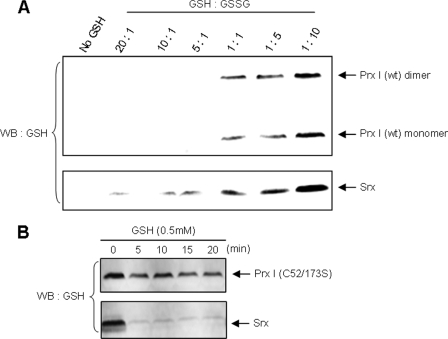
The mechanism of Grx-mediated deglutathionylation of proteins is well established (29). The catalytic process includes the formation of glutathionylated Grx as intermediate. However, Findlay et al. (13) reported that, although the conserved Cys99 residue in Srx is required for Srx-mediated deglutathionylation, Srx is not an acceptor molecule for glutathione. In an attempt to identify the reaction intermediate, we incubated a mixture containing 5 μm glutathionylated Prx I (C83/C173S) mutant and 5 μm Srx in 50 mm Tris, 100 mm NaCl, pH 7.4, buffer, at 37 °C for 1 h. The reaction was terminated by the addition of trifluoroacetic acid to a final pH of 2–3. The resulting mixture was subjected to LC-MS analysis. The chromatogram in Fig. 4 indicates the presence of four protein species in the reaction mixture. They correspond to the reduced and glutathionylated Prx I mutant and Srx. Based on the corresponding mass of each protein peak, we extracted each protein from the mass spectrum. The extracted ion chromatogram for the four expected proteins is shown at the bottom of Fig. 4. Together, the four protein species constituted the observed LC profile. In other words, the reaction products clearly showed the presence of glutathionylated Srx generated from the reaction of Srx with the glutathionylated Prx I mutant. It should be pointed out that a separate glutathionylation experiment, carried out at pH 7.4 without added trifluoroacetic acid but where the products were immediately analyzed by LC-MS methods after incubation for 1 h, yielded similar results. In addition, Srx is readily glutathionylated by GSSG, as shown by LC-MS analysis (data not shown). Fig. 5A shows that in the physiological concentration range of glutathione (i.e. 5 mm), both wild type Prx I and Srx were found to be increasingly glutathionylated with a decreasing [GSH]/[GSSG] ratio. Interestingly, Srx appeared to be more susceptible than Prx I to glutathionylation, as indicated by the Western blot showing that only glutathionylated Srx was observed when the [GSH]/[GSSG] ratios were present in the range from 20 to 10. At a 1:1 ratio, both Prx I and Srx were found to be significantly glutathionylated.
FIGURE 4.
Identifying glutathionylated Srx as the reaction intermediate. Glutathionylated Prx I (C83S/C173S) mutant was incubated with Srx at 37 °C for 1 h in 50 mm Tris (pH 7.4) buffer, and the resulting mixture was subjected to LC-MS analysis. The extracted ion chromatogram shows each peak corresponding to each of the four different proteins: reduced Prx I (C83S/C173S), glutathionylated Prx I (C83S/C173S), reduced Srx, and glutathionylated Srx.
FIGURE 5.
Comparative study of the glutathionylation and deglutathionylation of Prx I and Srx. A, glutathionylation of Prx I and Srx. Prx I or Srx (10 μm each) was incubated with the GSH/GSSG mixture at the indicated ratios with the total glutathione concentration fixed at 5 mm. The reaction was carried out in 50 mm Tris buffer (pH 7.4) for 1 h. The proteins were separated by SDS-PAGE and monitored with anti-glutathione antibody. B, deglutathionylation of Prx I and Srx. To analyze the deglutathionylation reaction, glutathionylated Prx I (C52S/C173S) or Srx was incubated with 0.5 mm GSH for the indicated times, and the proteins were subjected to SDS-PAGE followed by Western blot analysis (WB) using the anti-GSH antibody.
We have shown that glutathionylated Srx is an intermediate for the Srx-catalyzed deglutathionylation reaction and that Srx is readily glutathionylated. However, for Srx to function efficiently as an enzyme for the deglutathionylation, its glutathionylated intermediate must readily undergo deglutathionylation. Fig. 5B shows that relative to the Prx I (C52S/C173S) mutant, Srx was rapidly deglutathionylated by 0.5 mm GSH. Together, we revealed that Srx has a potential to serve as an efficient catalyst for protein deglutathionylation with glutathionylated Srx as the reaction intermediate, since Srx is readily glutathionylated and deglutathionylated.
Specificity of Srx as a Deglutathionylating Enzyme for Prx I
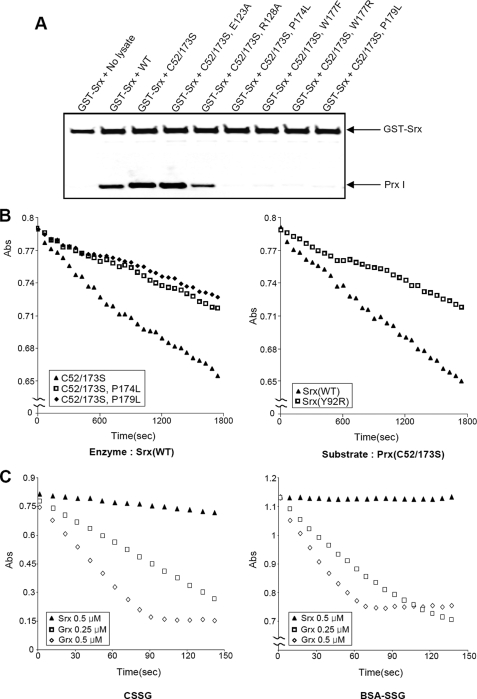
Grx is the most extensively investigated deglutathionylating enzyme, and it is a more efficient catalyst than other thiol disulfide oxidoreductases for reducing protein-SSG (30). However, the efficiency of Grx I-mediated deglutathionylation varies about 100-fold among various protein substrates (29). The variability is dependent upon the different apparent Km values for the substrate and its turnover rate constant. Among the three glutathionylated cysteine residues in Prx I, relative to Srx, Grx I works well only for catalyzing the deglutathionylation of Cys52. This suggests that Srx is a better enzyme for catalyzing the deglutathionylation of Prx I. This selectivity can be attributed to the fact that Srx interacts specifically with 2-Cys Prx. To verify the argument that specific interaction between Srx and Prx I plays a crucial role in determining the specificity of Srx over Grx I as a deglutathionylating enzyme for Prx I, site-directed mutagenesis was carried out on amino acid residues known to interact with Srx and Prx I. Lee et al. (31) showed that mutation at the putative Srx-binding interface of Prx I drastically reduced their interaction. Furthermore, a crystal structural study of the Prx I·Srx complex revealed that the C-terminal tail of Prx I (residues 172–186) was completely unfolded and packed onto the backside of Srx, away from the Srx active site (32). In this study, Jönsson et al. (32) also showed that Srx mutants, such as Y92R and L117R, exhibited a significant reduction in their binding affinity to Prx I. Based on the observation of four point mutations in the C-terminal tail (P174L, W177F, W177R, and P179L) and two in another binding interface (E123A and R128A) of Prx I (C52/C173S), mutants were prepared for the binding and deglutathionylation activity studies. Srx (Y92R) was also obtained to determine its effect on deglutathionylation activity. Fig. 6A shows that double mutation in C52S and C173S of Prx I does not reduce (and, if anything, slightly enhances) its interaction with GST-Srx. However, additional mutation in the C-terminal residues, such as Pro174, Trp177, or Pro179, totally abolished its binding to GST-Srx. In contrast, mutations on Glu123 and Arg128 did not significantly alter their interaction with GST-Srx.
FIGURE 6.
A, the effect of Prx I mutations on interactions with Srx. Wild-type and mutant Prx I-expressing cell lysates (500 μg) were incubated for 2 h at 4 °C with GST-hSrx fusion protein (5 μg) in 1 ml of binding buffer (50 mm HEPES buffer (pH 7.0), 150 mm NaCl, l% Triton X-100, 1 mm EDTA, and 1 mm DTT). Samples precipitated with GSH-Sepharose resins were subjected to Western blot analysis with antibodies to Prx I or GST. B, deglutathionylation of Prx I mutants catalyzed by Srx or Srx (Y92R). The reaction rates were monitored by a coupled assay system as described in Fig. 3. In these reactions, 1 μm Srx or Srx(Y92R) was used to catalyze the deglutathionylation of 20 μm glutathionylated Prx I mutants. The data show the decrease in A340 nm due to NADPH oxidation by GSSG generated during the glutathionylation reaction. C, deglutathionylation assay with CSSG and BSA-SSG as substrates. The deglutathionylation of CSSG and BSA-SSG (100 μm each) was catalyzed by 0.5 μm Srx (▴), 0.25 μm human Grx I (□), or 0.5 μm human Grx I (◇). The reactions were monitored by the coupled assay system described in B. The reaction time courses were obtained at 37 °C.
For the deglutathionylation studies, we selected the Prx I (C52S/C173S and P174L) and Prx I (C52S/C173S and P179L) mutants with the Prx I (C52S/C173S) mutant as a control to investigate the effects of substrate mutation and selected Srx (Y92R), which has been shown to exhibit very low affinity for Prx I (32), with wild type Srx as the control for the effects of enzyme mutation. In all cases, the substrates were first purified and glutathionylated at Cys83, and their extent of glutathionylation was verified using LC-MS analysis. Fig. 6B shows that mutation on Pro174 or Pro179 of Prx I and mutation on Tyr92 of Srx drastically inhibited the deglutathionylation activity of Srx. In addition, to ensure that Srx (Y92R) can readily form a glutathionylated intermediate, Srx (Y92R) was glutathionylated by GSSG, and the formation of glutathionylated Srx (Y92R) was verified using LC-MC analysis (data not shown). Together, these results reveal that mutation in either Prx I or Srx, which leads to the reduction of their binding affinity, also causes a great decrease in deglutathionylation activity.
To further demonstrate that Srx, relative to Grx I, is a specific deglutathionylation enzyme for Prx I, which can be attributed to its favorable binding affinity for 2-Cys Prx, we carried out comparative studies for the deglutathionylation of CSSG and glutathionylated BSA (BSA-SSG) catalyzed by Srx or Grx I. The reaction was initiated by the addition of 100 μm CSSG to a buffer containing either Grx I or Srx or the indicated concentration of the enzyme in the case of BSA-SSG. Fig. 6C shows that Grx I deglutathionylated both CSSG and BSA-SSG in an enzyme dose-dependent manner. In addition, Grx I is clearly a much more efficient enzyme than Srx. At 0.5 μm, Srx exhibited negligible activity with CSSG as substrate, whereas no activity was observed with BSA-SSG. Considering that both CSSG and BSA-SSG do not possess any binding motifs for Srx, these results support the notion that Srx is most likely the enzyme for catalyzing Prx I under physiological conditions.
Prx I Is Glutathionylated in Cells
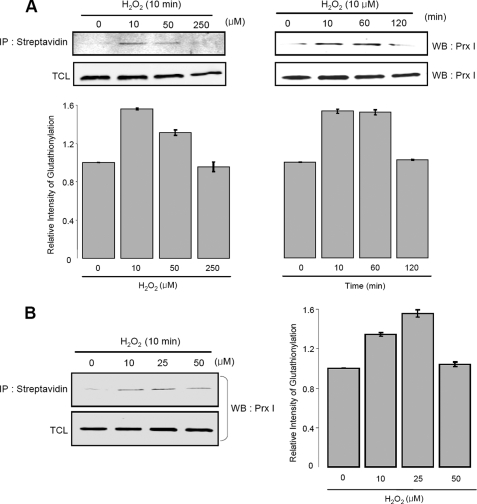
Glutathionylated peroxiredoxins I and V have been observed in human lymphocytes and in rat hepatocytes, respectively, after the cells were treated with a 1 mm diamide (20, 21). However, because diamide can rapidly extract a hydride from thiols to yield a reactive RS+ that can easily form a disulfide bond with another thiol, such as GSH, the glutathionylated proteins so obtained may not be physiologically relevant. Since Prx I can easily be hyperoxidized by H2O2 to form its inactive sulfinic acid derivative and lead to mobilization of Srx to catalyze the reactivation of this 2-Cys Prx sulfinic acid derivative, we determined the range of H2O2 at which hyperoxidation of Prx I was minimized under our experimental conditions. With A549 cells, we found that Prx I started to undergo hyperoxidation after 1 min of incubation with 50 μm H2O2 (data not shown). Therefore, the glutathionylation of Prx I was investigated with 10–50 μm H2O2 to avoid complications from hyperoxidized Prx I. In this study, the glutathionylated proteins were monitored with biotinylated glutathione. The A549 cells were treated with BioGEE, biotinylated glutathione ethyl ester, a membrane-permeable analogue of glutathione (26), for 1 h prior to H2O2 treatment. After the reaction, the biotinylated proteins were precipitated using streptavidin-agarose beads, followed by washing and elution with DTT. The glutathionylated Prx I was identified and quantitated by Western blot analysis with Prx I antibody. Fig. 7A shows that 10 μm H2O2 can induce a significant increase in glutathionylated Prx I after 10 min of incubation in A549 cells. At higher H2O2 concentrations and 2-h incubation times, the level of glutathionylated Prx I decreases, probably due to hyperoxidation of Prx I. Consistently, Prx I in HeLa cells was also glutathionylated but at a slightly higher concentration of H2O2 (Fig. 7B). This observation is in accordance with the fact that the level of thioredoxin found in HeLa cells was about 40% higher than that in A549 cells (33).
FIGURE 7.
Glutathionylation of Prx I in A549 or HeLa cells. A, glutathionylation of Prx I in A549 cells. To induce glutathionylation with biotinylated glutathione, cells were preincubated with 250 μm BioGEE for 1 h and subsequently exposed to H2O2 in a concentration- or time-dependent manner. Proteins that bound covalently to biotin were extracted using streptavidin-agarose and eluted with DTT. The eluted proteins were subjected to SDS-PAGE followed by Western blot analysis using the Prx I antibody. The histogram represents the means ± S.E. (n = 3) of the glutathionylation content relative to the untreated control. B, glutathionylation of Prx I in HeLa cells. Glutathionylation in HeLa cells was analyzed using the same technique as in Fig. 7A. The histogram represents means ± S.E. (n = 3) of the glutathionylation content relative to the untreated control. TCL, total cell lysates. IP, immunoprecipitation.
Effects of the Changes in Srx Levels on Prx I Glutathionylation in A549 Cells
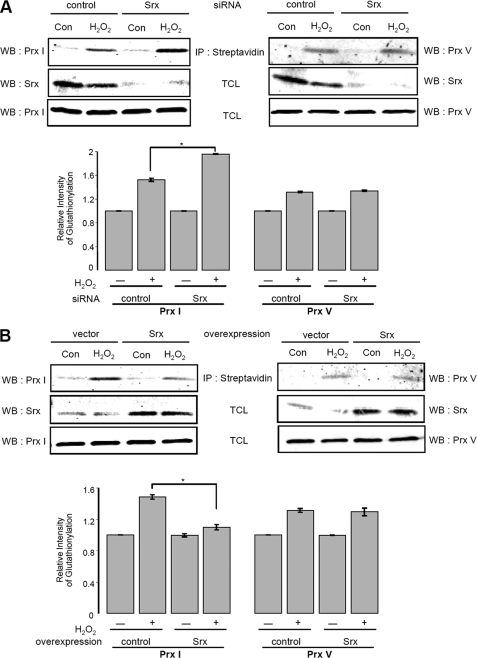
We have shown in vitro that Srx is a more efficient catalyst for deglutathionylating Prx I, relative to Grx I, and Prx I was found to be glutathionylated in both A549 and HeLa cells when subjected to oxidative stress with about 10 μm H2O2. To assess the biological relevance of Srx-mediated deglutathionylation of Prx I, we investigated the effects on the levels of glutathionylated Prx I by overexpressing and knocking down cellular Srx. When A549 cells were transfected with an siRNA specific for hSrx mRNA for 49 h, the level of Srx was drastically reduced (Fig. 8A). After the cells were transfected with siRNA for 48 h, they were then treated with BioGEE for 1 h and subsequently exposed to 10 μm H2O2 in the case of Prx I and 250 μm H2O2 for Prx V for 10 min. A higher concentration of H2O2 was used in the case of Prx V, because Prx V is more resistant to hyperoxidation than Prx I. The BioGEE-labeled proteins were isolated as described above and subjected to Western blot analysis with Prx I or Prx V antibodies. The results (Fig. 8A) revealed that the level of glutathionylated Prx I was substantially increased in Srx-depleted cells after H2O2 treatment. In contrast, the level of glutathionylated Prx V was hardly affected by Srx knockdown, consistent with the fact that Srx does not bind to Prx V. These observations indicate that, in the cells, Srx can regulate the extent of glutathionylated Prx I via its deglutathionylation activity or by forming a complex with Prx I to prevent its glutathionylation. However, the latter argument requires that Srx form a stable Prx I·Srx complex and that the concentration of cellular Prx I and Srx be present at nearly stoichiometric levels. The concentration of Prx isoforms in cultured cells has been shown to be high, particularly in transformed cells (e.g. in HeLa cells, the level of Prx I and Prx II represents 0.4 and 0.33% of the total cytosolic proteins, respectively) (34). On the other hand, the quantity of Srx, an enzyme using modified Prx I and II as its substrates, is present at a level close to 5% of the total Prx I (data not shown). Considering that Srx can bind to both Prx I and Prx II, one would expect the fraction of Srx·Prx I in the pool of Prx I plus Prx II to be about 2.5%. This is a negligibly low level to account for the hypothesis that the increase observed in glutathionylated Prx I in Srx knocked down cells is due to the decrease in the formation of Srx·Prx I complex to prevent glutathionylation of Prx I. Together these results support the notion that the deglutathionylation activity of Srx is responsible for the observed decrease in glutathionylated Prx I levels in Srx-knocked down A549 cells.
FIGURE 8.
Effect of Srx expression level on glutathionylated Prxs in A549 cells. A, effect of Srx depletion by siRNA on Prxs glutathionylation. Forty-eight hours after transfection with a control siRNA or human Srx siRNA, A549 cells were incubated with 250 μm BioGEE for 1 h and subsequently exposed to 10 μm H2O2 for Prx I or 250 μm H2O2 for Prx V for 10 min. Biotinylated proteins were precipitated (IP) with streptavidin-agarose, eluted with DTT, and then separated by SDS-PAGE, followed by Western blot analysis (WB) with Srx, Prx I, or Prx V antibodies. The histogram represents means ± S.E. (n = 3) of the glutathionylation content relative to the untreated control. *, two groups are significantly different from each other at p < 0.005. B, effect of Srx overexpression on Prxs glutathionylation. A549 cells were subjected to transient transfection with an expression vector for human Srx or with the corresponding empty vector (pCDNA) for 36 h. Cells were treated with 250 μm BioGEE prior to the addition of H2O2 (10 μm for Prx I, 250 μm for Prx V). Samples were prepared as described in A and then subjected to Western blot analysis with Srx, Prx I, or Prx V antibodies. The histogram represents means ± S.E. (n = 3) of the glutathionylation content relative to the untreated control. *, two groups are significantly different from each other at p < 0.005. TCL, total cell lysates.
To further demonstrate that cellular levels of Srx play an important role in regulating the state of glutathionylation of Prxs that exhibit selective binding affinities for Srx, we investigated the effects of overexpressing Srx on the glutathionylation levels of Prx I and Prx V in A549 cells. Here A549 cells were transfected with an expression vector for human Srx or the corresponding empty vector. Fig. 8B shows that the level of Srx was significantly elevated in Srx-overexpressing cells. In addition, consistent with the results obtained with siRNA-mediated depletion of Srx, the amount of glutathionylated Prx I in Srx-overexpressing cells was reduced compared with that in control cells after H2O2 treatment. However, the amount of glutathionylated Prx V did not show any change in Srx-overexpressing cells compared with vector only-transfected cells. Together, these results reveal that cellular levels of Srx regulate the levels of glutathionylated Prx I via its deglutathionylation activity. However, glutathionylation of Prx V, not known to bind to Srx, was not affected by the changes in Srx expression levels.
Conclusions
We demonstrated here that human Prx I can undergo glutathionylation in vitro and in living cells, such as A549 and HeLa cells. Among the four cysteines of Prx I, Cys52, Cys83, and Cys173 were identified as glutathionylated residues by stepwise double alkylation methods and combined mass spectrometry. Although little is known about the specificity of protein glutathionylation, it is generally agreed that most glutathionylated cysteine residues are reactive and/or accessible cysteine residues (8, 35). Both Cys52 and Cys173 are reactive cysteines in Prx I. Glutathionylation of these two cysteines will inhibit the peroxidase activity of Prx I. In addition, structural analysis of Prx from various species (36, 37) indicates that the Cys83 in human Prx I is probably located at the dimer-dimer interface and accessible. It has been shown that the Cys83 at the dimer-dimer interface can form a disulfide bond with the Cys83 of another subunit and shift the dimer-decamer equilibrium in favor of decamer formation. Blockage of the Cys83-Cys83 disulfide formation by substituting Cys83 with Ser reversed the equilibrium to favor the dimer. Thus, glutathionylation of Cys83 is expected to shift the equilibrium to favor the dimer. It should be pointed out that Noguera-Mazon et al. (38) have reported that glutathionylation of plant 1-Cys Prx from Populus tremula, which exists in a very different structure with a subunit interface different from that reported for mammalian 2-Cys Prx (39), could induce the dissociation of this dimeric 1-Cys Prx.
The mechanism of deglutathionylation catalyzed by Srx is not known. Our findings reveal that it proceeds via a mechanism similar to that of Grx by forming a glutathionylated intermediate with its conserved Cys99 residue. Kinetic and binding analyses reveal that Srx is a preferred enzyme for deglutathionylating Prx I due to its favorable binding affinity with Prx I. The physiological relevance of these findings is supported by the observation that Prx I is glutathionylated in A549 and HeLa cells when they were subjected to mild oxidative stress by a few tenths of a micromolar concentration of H2O2. In addition, the level of Prx I glutathionylation in A549 cells was drastically elevated when Srx was knocked down by the siRNA technique and reduced when Srx was overexpressed in A549 cells. On the other hand, the amount of Prx V glutathionylation was not affected by the change in Srx expression level. Together, these results suggest that glutathionylation of Prx I can be regulated specifically by Srx. In view of our findings, it would be interesting to learn whether glutathionylation can alter the biological function of Prx I.
Acknowledgments
We thank Dr. Rodney L. Levine for invaluable help during the course of identifying the glutathionylated cysteine residues, and we thank Dr. Ho Zoon Chae for valuable discussions.
This work was supported, in whole or in part, by the National Institutes of Health, NHLBI, Intramural Research Program and by National Institutes of Health Research Grant 2 PO1 AG 15885 (to J. J. M.). This study was also supported in part by a Brain Korea21 grant (to J. W. P.), by BioR and D Program grants of Korean Science and Engineering Foundation (to S. G. R.), and by a Merit Review research grant from the Department of Veterans Affairs (to J. J. M.).
- Srx
- sulfiredoxin
- Prx
- peroxiredoxin
- Grx
- glutaredoxin
- DTT
- dithiothreitol
- IAM
- iodoacetamide
- BioGEE
- biotinylated glutathione ethyl ester
- HPLC
- high performance liquid chromatography
- LC
- liquid chromatography
- MS
- mass spectrometry
- siRNA
- small interfering RNA
- CSSG
- glutathionylated cysteine
- BSA-SSG
- glutathionylated BSA
- hSrx
- human Srx.
REFERENCES
- 1.Hunter T. (1995) Cell 80,225–236 [DOI] [PubMed] [Google Scholar]
- 2.Chock P. B., Stadtman E. R. (1996) Principles of Medical Biology ( Bittar E. E., Bittar N. eds) pp. 201–220, JAI Press, Greenwich, CT [Google Scholar]
- 3.Sundaresan M., Yu Z. X., Ferrans V. J., Irani K., Finkel T. (1995) Science 270,296–299 [DOI] [PubMed] [Google Scholar]
- 4.Bae Y. S., Kang S. W., Seo M. S., Baines I. C., Tekle E., Chock P. B., Rhee S. G. (1997) J. Biol. Chem. 272,217–221 [PubMed] [Google Scholar]
- 5.Barrett W. C., DeGnore J. P., Keng Y. F., Zhang Z. Y., Yim M. B., Chock P. B. (1999) J. Biol. Chem. 274,34543–34546 [DOI] [PubMed] [Google Scholar]
- 6.Rhee S. G. (2006) Science 312,1882–1883 [DOI] [PubMed] [Google Scholar]
- 7.Shelton M. D., Chock P. B., Mieyal J. J. (2005) Antioxid. Redox Signal. 7,348–366 [DOI] [PubMed] [Google Scholar]
- 8.Ghezzi P. (2005) Free Radic. Res. 39,573–580 [DOI] [PubMed] [Google Scholar]
- 9.Gallogly M. M., Mieyal J. J. (2007) Curr. Opin. Pharmacol. 7,381–391 [DOI] [PubMed] [Google Scholar]
- 10.Biteau B., Labarre J., Toledano M. B. (2003) Nature 425,980–984 [DOI] [PubMed] [Google Scholar]
- 11.Woo H. A., Chae H. Z., Hwang S. C., Yang K. S., Kang S. W., Kim K., Rhee S. G. (2003) Science 300,653–656 [DOI] [PubMed] [Google Scholar]
- 12.Chang T. S., Jeong W., Woo H. A., Lee S. M., Park S., Rhee S. G. (2004) J. Biol. Chem. 279,50994–51001 [DOI] [PubMed] [Google Scholar]
- 13.Findlay V. J., Townsend D. M., Morris T. E., Fraser J. P., He L., Tew K. D. (2006) Cancer Res. 66,6800–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee S. G., Kang S. W., Chang T. S., Jeong W., Kim K. (2001) IUBMB Life 52,35–41 [DOI] [PubMed] [Google Scholar]
- 15.Hofmann B., Hecht H. J., Flohé L. (2002) Biol. Chem. 383,347–364 [DOI] [PubMed] [Google Scholar]
- 16.Chae H. Z., Chung S. J., Rhee S. G. (1994) J. Biol. Chem. 269,27670–27678 [PubMed] [Google Scholar]
- 17.Chae H. Z., Uhm T. B., Rhee S. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91,7022–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang K. S., Kang S. W., Woo H. A., Hwang S. C., Chae H. Z., Kim K., Rhee S. G. (2002) J. Biol. Chem. 277,38029–38036 [DOI] [PubMed] [Google Scholar]
- 19.Rabilloud T., Heller M., Gasnier F., Luche S., Rey C., Aebersold R., Benahmed M., Louisot P., Lunardi J. (2002) J. Biol. Chem. 277,19396–19401 [DOI] [PubMed] [Google Scholar]
- 20.Fratelli M., Demol H., Puype M., Casagrande S., Eberini I., Salmona M., Bonetto V., Mengozzi M., Duffieux F., Miclet E., Bachi A., Vandekerckhove J., Gianazza E., Ghezzi P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,3505–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fratelli M., Demol H., Puype M., Casagrande S., Villa P., Eberini I., Vandekerckhove J., Gianazza E., Ghezzi P. (2003) Proteomics 3,1154–1161 [DOI] [PubMed] [Google Scholar]
- 22.Kosower N. S., Kosower E. M., Wertheim B., Correa W. S. (1969) Biochem. Biophys. Res. Commun. 37,593–596 [DOI] [PubMed] [Google Scholar]
- 23.Chae H. Z., Kang S. W., Rhee S. G. (1999) Methods Enzymol. 300,219–226 [DOI] [PubMed] [Google Scholar]
- 24.Apffel A., Fischer S., Goldberg G., Goodley P. C., Kuhlmann F. E. (1995) J. Chromatogr. A 712,177–190 [DOI] [PubMed] [Google Scholar]
- 25.Gravina S. A., Mieyal J. J. (1993) Biochemistry 32,3368–3376 [DOI] [PubMed] [Google Scholar]
- 26.Sullivan D. M., Wehr N. B., Fergusson M. M., Levine R. L., Finkel T. (2000) Biochemistry 39,11121–11128 [DOI] [PubMed] [Google Scholar]
- 27.Lee W., Choi K. S., Riddell J., Ip C., Ghosh D., Park J. H., Park Y. M. (2007) J. Biol. Chem. 282,22011–22022 [DOI] [PubMed] [Google Scholar]
- 28.Holmgren A. (2000) Antioxid. Redox Signal. 2,811–820 [DOI] [PubMed] [Google Scholar]
- 29.Mieyal J. J., Srinivaan U., Starke D. W., Gravina S. A., Mieyal P. A. (1995) in Biothiols in Health and Disease ( Packer L., Cadenas E. eds) pp. 305–372, Marcel Dekker, Inc., New York [Google Scholar]
- 30.Chrestensen C. A., Starke D. W., Mieyal J. J. (2000) J. Biol. Chem. 275,26556–26565 [DOI] [PubMed] [Google Scholar]
- 31.Lee D. Y., Park S. J., Jeong W., Sung H. J., Oho T., Wu X., Rhee S. G., Gruschus J. M. (2006) Biochemistry 45,15301–15309 [DOI] [PubMed] [Google Scholar]
- 32.Jönsson T. J., Johnson L. C., Lowther W. T. (2008) Nature 451,98–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andoh T., Chock P. B., Chiueh C. C. (2002) J. Biol. Chem. 277,9655–9660 [DOI] [PubMed] [Google Scholar]
- 34.Chae H. Z., Kim H. J., Kang S. W., Rhee S. G. (1999) Diabetes Res. Clin. Pract. 45,101–112 [DOI] [PubMed] [Google Scholar]
- 35.Casagrande S., Bonetto V., Fratelli M., Gianazza E., Eberini I., Massignan T., Salmona M., Chang G., Holmgren A., Ghezzi P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,9745–9749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsonage D., Youngblood D. S., Sarma G. N., Wood Z. A., Karplus P. A., Poole L. B. (2005) Biochemistry 44,10583–10592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumura T., Okamoto K., Iwahara S., Hori H., Takahashi Y., Nishino T., Abe Y. (2008) J. Biol. Chem. 283,284–293 [DOI] [PubMed] [Google Scholar]
- 38.Noguera-Mazon V., Lemoine J., Walker O., Rouhier N., Salvador A., Jacquot J. P., Lancelin J. M., Krimm I. (2006) J. Biol. Chem. 281,31736–31742 [DOI] [PubMed] [Google Scholar]
- 39.Hirotsu S., Abe Y., Okada K., Nagahara N., Hori H., Nishino T., Hakoshima T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96,12333–12338 [DOI] [PMC free article] [PubMed] [Google Scholar]