Abstract
Mouse embryonic stem cells (mESCs) maintain pluripotency and indefinite self-renewal through yet to be defined molecular mechanisms. Leukemia inhibitory factor has been utilized to maintain the symmetrical self-renewal and pluripotency of mESCs in culture. It has been suggested that molecules with significant cellular effects on retinoblastoma protein (pRb) or its related pathways should have functional impact on mESC proliferation and differentiation. However, the involvement of pRb in pluripotent differentiation of mESCs has not been extensively elaborated. In this paper, we present novel experimental data indicating that Cdk2ap1 (cyclin-dependent kinase 2-associating protein 1), an inhibitor of G1/S transition through down-regulation of CDK2 and an essential gene for early embryonic development, confers competency for mESC differentiation. Targeted disruption of Cdk2ap1 in mESCs resulted in abrogation of leukemia inhibitory factor withdrawal-induced differentiation, along with altered pRb phosphorylation. The differentiation competency of the Cdk2ap1−/− mESCs was restored upon the ectopic expression of Cdk2ap1 or a nonphosphorylatable pRb mutant (mouse Ser788 → Ala), suggesting that the CDK2AP1-mediated differentiation of mESCs was elicited through the regulation of pRb. Further analysis on mESC maintenance or differentiation-related gene expression supports the phosphorylation at serine 788 in pRb plays a significant role for the CDK2AP1-mediated differentiation of mESCs. These data clearly demonstrate that CDK2AP1 is a competency factor in the proper differentiation of mESCs by modulating the phosphorylation level of pRb. This sheds light on the role of the establishment of the proper somatic cell type cell cycle regulation for mESCs to enter into the differentiation process.
Maintenance of pluripotency is essential to guarantee proper differentiation of cells and embryo development. ES3 cells can be maintained pluripotent in culture, and leukemia inhibitory factor (LIF) has been utilized to maintain the symmetrical self-renewal of mESCs (1, 2). Binding of LIF to its receptor, LIF-R, causes heterodimerization of LIF-R and gp130 and triggers downstream activation of Jak (3). Several lines of evidence show that Stat3 (signal transducer and activator of transcription 3) is the key downstream mediator of the LIF/gp130 signaling pathway, leading to the maintenance of self-renewal and pluripotency (4–6). However, other lines of evidence show that LIF/gp130/Stat3 are not essential for pluripotency, and the existence of unidentified novel pathways that maintain pluripotency has been suggested (7). In undifferentiated embryonic stem cells, pRb is known to remain as an inactive form, and CDK2 and E2F1 remain constitutively active, allowing rapid self-regeneration of cells. When the cells differentiate, the cell cycle regulatory machinery becomes active and initiates pRb-dependent cell cycle control (8, 9).
p12CDK2AP1 (CDK2AP1) is a highly conserved, ubiquitously expressed gene that is down-regulated in ∼70% of oral cancers (10, 11). Murine Cdk2ap1 with only three amino acid deviations from the human CDK2AP1 is located at chromosome 5 (12, 13). CDK2AP1 has been shown to be an S-phase regulator through two important cellular partners: CDK2 and DNA polymerase-α/primase (14, 15). Murine embryonic stem cells with disrupted expression of Cdk2ap1 showed an increased proliferation and an altered cell cycle profile with a reduced G2/M phase along with an increased CDK2 activity (13). Recently, Cdk2ap1 has been identified as one of the stem cell-specific genes that are enriched in both embryonic and adult stem cells (16). Cdk2ap1 has been categorized as one of genes that are expressed in early stage preimplantation embryos (17). In addition, Cdk2ap1 mRNA has been found to be elevated upon estrogen treatment during early implantation process, suggesting its role in uterine decidualization (18).
In this paper, we analyzed phenotypic characteristics of Cdk2ap1−/− mESCs. Upon withdrawal of LIF, Cdk2ap1−/− mESCs showed significantly abrogated differentiation phenotype and hyperphosphorylation of pRb. The differentiation competency of the Cdk2ap1−/− mESCs was restored upon the ectopic expression of Cdk2ap1 or a pRb phosphorylation mutant, S788A (equivalent to human pRb S795A (19, 20)). It links pRb phosphorylation to the regulation of differentiation competency in mESCs. Further analysis on mESC maintenance or differentiation-related gene expression suggests that the phosphorylation at serine 788 in pRb plays a significant role for the CDK2AP1-mediated differentiation of mESCs. Taken together, we conclude that CDK2AP1 is a competency factor in mESC differentiation through the regulation of pRb phosphorylation. Our current data support the unique role of CDK2AP1 in the proper regulation of cellular differentiation of mESCs during early embryonic development.
EXPERIMENTAL PROCEDURES
Cell Culture
Murine embryonic stem cells were grown on a gelatin-coated plate and maintained in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, 1.2 ml/500 ml β-mercaptoethanol, 1% l-glutamine, 0.2% (v/v) LIF, 1% (v/v) penicillin/streptomycin, and 0.1% (v/v) gentamycin sulfate. Generation of Cdk2ap1 knock-out ES cells was described by Kim et al. (13).
AP Activity Assay and Staining
ES cells were cultured in medium with or without LIF and gently washed with 1× PBST (1× PBS with 0.05% Tween 20) before staining with the StemTAG AP staining kit (CHEMICON International, Inc., Temecula, CA). The AP activity assay was performed according to the manufacturer's instructions. Briefly, cells were washed twice with cold PBS and lysed in Cell Lysis Buffer in a StemTAG AP activity assay kit (Cell Biolabs, San Diego, CA). After a 10-min incubation at 4 °C, the lysate was transferred to a fresh tube and spun down at 12,000 × g for 10 min. Protein concentration was determined. Each 50 μl of cell lysate was transferred to a 96-well plate in triplicate and mixed with 50 μl of StemTAG AP activity assay substrate. After incubating for 20 min at 37 °C, the reaction was stopped by adding 50 μl of 1× stop solution and mixed by placing the plate on an orbital plate shaker for 30 s. The activity was determined by measuring A405 and normalized against the amount of protein in the reaction.
Gene Expression Profiling of Embryoid Bodies
Cdk2ap1+/+ or Cdk2ap1−/− mESCs were subjected to embryo body formation by the aggregation method, as described (21). Embryo bodies were maintained for up to 10 days in culture and harvested for total RNA preparation by using the RNeasy kit as instructed by manufacturer (Qiagen, Valencia, CA). An equal amount of total RNA was used to quantitatively compare the mRNA expression levels between Cdk2ap1+/+ and Cdk2ap1−/− embryoid bodies by using Power SYBR Green 2× PCR mix (Applied Biosystems, Foster City, CA). The experiment was done in biological duplicate, and the level of expression was normalized against glyceraldehyde-3-phosphate dehydrogenase. The -fold change was determined by comparing the normalized ΔCt values from differentiating embryoid bodies against undifferentiating cells grown in monolayer.
For quantitative real time PCR analysis on cells grown in monolayer culture, cells were seeded on 24-well plate and transduced with lentivirus. After 3–5-day transduction, total cellular RNA was prepared by using the RNeasy minikit following the manufacturer's instructions, including on-column DNA digestion procedure (Qiagen, Valencia, CA). The concentration of RNA was determined by using NanoDrop (NanoDrop Technologies, Wilmington, DE), and reverse transcription was performed with 2–3 μg of RNA/20-μl reaction by using oligo(dT) and murine leukemia virus reverse transcriptase (PerkinElmer Life Sciences). Real time PCR was done in duplicate by using Power SYBR Green 2× PCR mix (Applied Biosystems, Foster City, CA) with 1 μl of cDNA/10-μl reaction, and the level of mRNA was determined after normalizing against ΔCt from glyceraldehyde-3-phosphate dehydrogenase internal control. -Fold change was empirically calculated by employing 2−ΔΔCt, and the relative ratio was obtained against the control set. The amplification of specific product was confirmed from the melting curve analysis and also by examining the final product on agarose gel.
Generation of Inducible mESC Clones Expressing pRb Phosphorylation Mutant
To generate Tet-off-inducible pRb mutant clones, Cdk2ap1+/+ or Cdk2ap1−/− mESCs were stably electroporated with tTA-Advanced plasmid (Clontech, Palo Alto, CA) along with a puromycin selection marker. The selected clones were further tested for the presence of functional tTA expression by transiently transfecting a pTRE-GFP construct in the presence or absence of doxycyclin (100 ng/ml). Clones that showed the expression of GFP only in the absence of doxycyclin were selected and maintained for the second electroporation with pTRE-HA-pRb constructs. pTRE-HA-pRb mutant constructs were made by PCR amplification of mouse pRb with HA tag from the parental wild type pECE-ΔB/X-HA (from Brenda Gallie, University of Toronto) and subcloning into pTRE-Advanced vector (Clontech, Palo Alto, CA). The appropriate mutation at T246A, S773A, S788A, S800A/S804A, and T814A was introduced by using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Purified plasmid DNA was electroporated into tTA-expressing WT or Cdk2ap1−/− mESCs along with the hygromycin resistance marker and screened with medium containing hygromycin (100 μg/ml) and doxycyclin (100 ng/ml). The inducibility of mutant pRb expression was determined by Western analysis with anti-HA antibody (Sigma). For differentiation analysis, cells were seeded on a gelatin-coated 24-well plate in quadruplicate (100 cells/well) in the presence or absence of doxycyclin. After 2 days, cells were fed with complete ES culture medium with or without LIF and cultured for up to 10 days before microscopic examination.
RESULTS
Altered Differentiation Competency in Cdk2ap1−/− mESCs
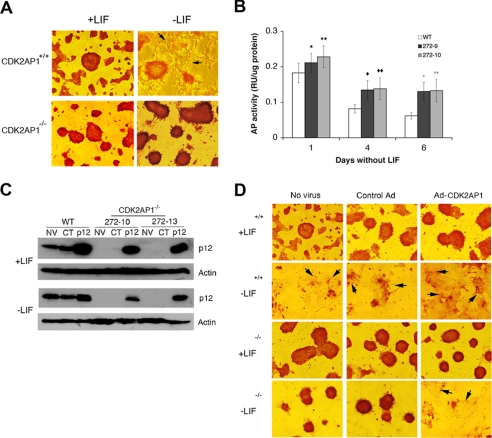
Heterozygous and homozygous Cdk2ap1 knock-out mESCs were generated as reported previously (13). We found the homozygous knock-out of Cdk2ap1 in mice resulted in the embryonic lethality at embryonic days 3.5–5.5 (22). To gain insight into the molecular and biochemical alterations resulting through Cdk2ap1 knock-out, we examined phenotypic and biochemical changes in vitro by using Cdk2ap1−/− mESC clones. Based on the known effect of CDK2AP1 on CDK2 activity and pRb phosphorylation and previous findings on CDK2 activity and pRb phosphorylation in mESCs, we hypothesized that CDK2AP1 has a functional role in the differentiation competency of mESCs (15, 23). As shown in Fig. 1A, both Cdk2ap1+/+ and Cdk2ap1−/− cells showed similar morphologies in the presence of LIF. Upon withdrawal of LIF, wild type cells showed differentiated morphologies largely devoid of alkaline phosphatase staining. On the contrary, Cdk2ap1−/− mESCs showed no evidence of differentiation, and cells retained alkaline phosphatase staining, indicative of an undifferentiated state and sustained self-renewal (Fig. 1A). We were able to maintain the phenotype up to 20 days without any noticeable changes (data not shown). In accordance with the sustained AP positivity, a quantitative biochemical AP activity assay analysis revealed that Cdk2ap1−/− mESCs showed higher AP activity compared with wild type mESCs (Fig. 1B). We have also demonstrated that the level of Oct3/4 was sustained in Cdk2ap1−/− mESCs even under differentiation conditions (24). This result showed that the knock-out of the Cdk2ap1 gene in mESCs altered the differentiation program and maintains an undifferentiated phenotype even under differentiation stimulus. This suggests that CDK2AP1 is involved in differentiation of mESCs.
FIGURE 1.
Altered differentiation of Cdk2ap1−/− mESCs upon LIF withdrawal. A, in the absence of LIF, Cdk2ap1+/+ cells are differentiated and became negative to alkaline phosphatase staining, which is an indication of undifferentiation. However, Cdk2ap1−/− cells showed resistance to differentiation stimuli and maintained undifferentiated phenotype. B, biochemical assessment of alkaline phosphatase activity showed an induced level in Cdk2ap1−/− cells (272-9 and 272-10). (*, p = 0.08; **, p = 0.002; ♦, p = 0.03; ♦♦, p = 0.004; +, p = 0.05; ++, p = 0.03). The error bar represents S.E. C, Western analysis confirmed the restored level of CDK2AP1 after adenoviral delivery of Cdk2ap1 into mESCs. D, the specificity of the role of CDK2AP1 in mESC differentiation was evaluated by restoring the expression of CDK2AP1 in Cdk2ap1−/− cells using adenovirus. Cells (WT or Cdk2ap1−/− (272-10 and 272-13)) were transduced with no virus (NV), control virus (CT), or Ad-Cdk2ap1 and grown in the presence or absence of LIF for 10 days before staining for AP activity. The restoration of CDK2AP1 in Cdk2ap1−/− mESCs resulted in differentiation of cells, whereas the control virus did not induce differentiation. Differentiated unstained cells are shown with arrows.
The role of CDK2AP1 in mESC differentiation has been confirmed by restoring the expression of CDK2AP1 in Cdk2ap1−/− mESCs (Fig. 1C). As shown in Fig. 1D, when the Cdk2ap1−/− mESCs transduced with AdPL-Cdk2ap1 were forced to differentiate upon LIF withdrawal, there was evidence of noticeable differentiation of cells and reduction of AP activity staining, implying that the restoration of CDK2AP1 in Cdk2ap1−/− mESCs returned the cells back to the proper differentiation competency. These results link CDK2AP1 to mESC differentiation. They also show that the cells devoid of CDK2AP1 become independent of LIF in self-renewal and differentiation. We further confirmed that the compromised differentiation phenotype observed with Cdk2ap1−/− cells was not due to the hyperproduction of LIF in differentiating cells in culture (supplemental Fig. S1). In addition, we found that the level of LIF-R was not altered in Cdk2ap1−/− mESCs (data not shown). We also found that the function of LIF-R in mediating the phosphorylation of Stat3 was not altered in Cdk2ap1−/− mESCs (supplemental Fig. S2). We also found that Cdk2ap1−/− mESCs normally differentiate in vitro upon the induction of differentiation with chemical inducers, such as DMSO or retinoic acid (data not shown). These results suggest that CDK2AP1 plays a role in the decision-making between self-renewal versus differentiation under nonrenewing conditions, which should have cellular effect on further downstream lineage commitment process.
Gene Expression Profiling of Differentiating Cdk2ap1−/− Embryoid Bodies
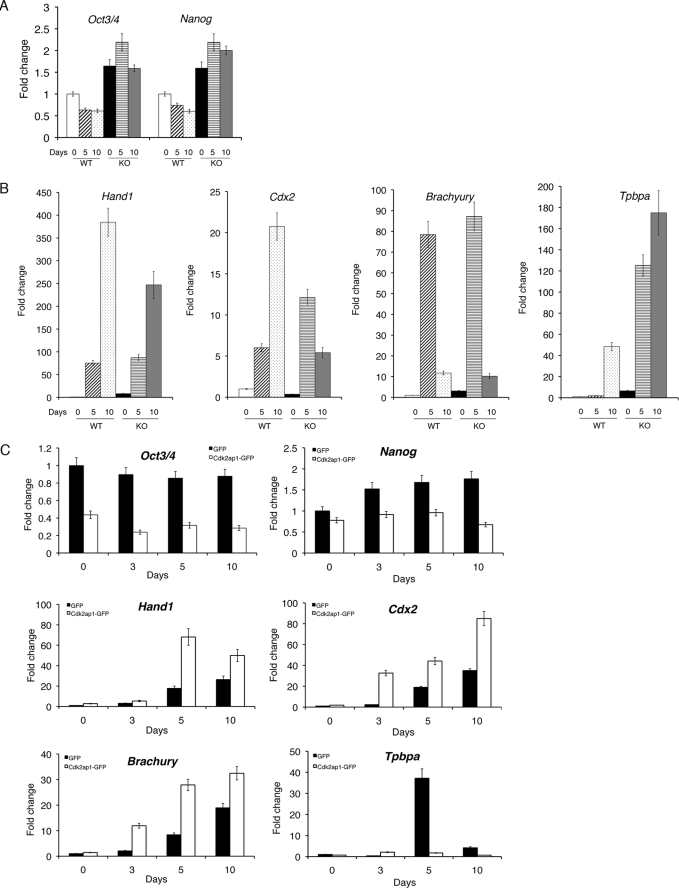
To assess molecular changes stemming from the loss of Cdk2ap1, we have examined the mRNA expression of most of the stem cell-related genes by quantitative reverse transcription-PCR and compared Cdk2ap1+/+ and Cdk2ap1−/− embryoid bodies differentiated for different time points (0, 5, and 10 days) (Fig. 2 and supplemental Fig. S3). The level of Oct3/4 and Nanog was decreased in Cdk2ap1+/+ embryoid bodies upon differentiation (∼2-fold decrease in both cases compared with undifferentiated Cdk2ap1+/+ mESCs) (Fig. 2A). However, we noticed that Cdk2ap1−/− embryoid bodies maintained a significantly higher level of both Oct3/4 and Nanog (∼3-fold higher than Cdkap1+/+ embryoid bodies with p < 0.01 and 0.04, respectively) after differentiation. As shown in Fig. 2B, among several differentiation-related genes we examined, we found that the induction of Hand1 and Cdx2 upon differentiation of EB was significantly reduced in Cdk2ap1−/− mESCs compared with Cdk2ap1+/+ mESCs (Hand1, p < 0.03; Cdx2, p < 0.005), whereas the level of Tpbpa (p < 0.01) was significantly increased in Cdk2ap1−/− mESC embryoid bodies. The expression levels of other stem cell marker genes and differentiation-related genes are presented in supplemental Fig. S3, A and B.
FIGURE 2.
Gene expression profiles of Cdk2ap1−/− mESC embryoid bodies. The molecular effects of the deletion of Cdk2ap1 on the regulation of stem cell genes or differentiation-related genes were examined by quantitative reverse transcription-PCR analysis in differentiating embryoid bodies. A, WT (Cdk2ap1+/+) or Cdk2ap1−/− mESCs were subjected to embryoid body formation and differentiation for 0, 5, and 10 days in culture. Total RNA was isolated after the indicated differentiation time period, and the level of Oct3/4 (p < 0.01) and Nanog (p < 0.04) was determined in biological duplicates. The statistical p value was determined by Student's t test by comparing values from two different genotypes for each differentiation time point. B, the level of differentiation-related genes, Hand1 (p < 0.03), Cdx2 (p < 0.005), Brachyury (p < 0.02), and Tpbpa (p < 0.01) was determined. Significant down-regulation of Hand1 and Cdx2 was observed in Cdk2ap1−/− mESC embryoid bodies, whereas Tpbpa was up-regulated in Cdk2ap1−/− mESC embryoid bodies. C, the effect of restoring Cdk2ap1 in Cdk2ap1−/− mESC embryoid bodies was examined at the molecular level. Selected clones stably expressing Cdk2ap1 were generated from Cdk2ap1−/− mESCs and subjected to differentiation as described above. The expression of Hand1 (p < 0.007), Cdx2 (p < 0.03), and Brachyury (p < 0.08) was up-regulated, whereas the expression of Tpbpa (p < 0.001) was down-regulated by restoring Cdk2ap1. The error bar represents S.E. The statistical p value was determined by comparing values against day 0. KO, knock-out.
We further examined the effect of restoring Cdk2ap1 expression by generating stable GFP-expressing or CDK2AP1-GFP-expressing clones from two Cdk2ap1−/− mESCs (272-9 and 272-10) (supplemental Fig. S4). The clones were then subjected to pluripotent differentiation by embryoid body formation for different time periods (0, 3, 5, and 10 days). The restoration of Cdk2ap1 expression resulted in down-regulation of stem cell marker genes, such as Oct3/4 and Nanog (Oct3/4, p < 0.09; Nanog, p < 0.01) (Fig. 2C). This finding is very consistent with the sustained expression of Oct3/4 and Nanog in differentiating Cdk2ap1−/− mESC embryoid bodies (Fig. 2A) and supports a role of CDK2AP1 in the regulation of stem cell marker gene expressions in differentiating embryoid bodies.
We also examined the effect of restoring Cdk2ap1 in Cdk2ap1−/− mESCs on the expression of differentiation-related genes. Most profound changes were observed with Hand1 (p < 0.007), Cdx2 (p < 0.03), and Brachyury (p < 0.08) (Fig. 2C). Especially, the ectopic expression of Cdk2ap1 resulted in the restoration of differentiation-dependent induction of Hand1 and Cdx2 in Cdk2ap1−/− mESC embryoid bodies. The expression of Brachyury was not significantly altered in Cdk2ap1−/− mESC embryoid bodies (Fig. 2A), but the expression was significantly induced with the ectopic expression of Cdk2ap1. This implies that the expression of some of differentiation-related genes can be regulated not just by the presence of CDK2AP1, but also by the level of CDK2AP1 in the cells. This hypothesis was supported by the comparison of other differentiation-related genes (supplemental Fig. S5, 1, 2, and 3). Collectively, Cdk2ap1−/− embryoid bodies showed a sustained level of Oct3/4 and Nanog mRNA after differentiation, whereas there were significantly reduced levels of changes in some of the differentiation-related genes. In this experiment, we measured the consequences of the targeted gene mutation. From comparing the expression profiles, we have demonstrated that some, if not all, stem cell marker gene expressions are sustained, and several differentiation marker gene expressions are down-regulated in Cdk2ap1−/− mESCs even under differentiation conditions. More interestingly, some of these molecular events are reversibly regulated in the restoration experiment, which demonstrated the specificity of molecular effect elicited through CDK2AP1. This result demonstrated a compromised differentiation competency in Cdk2ap1−/− embryoid bodies at the molecular level, and it suggested a role of Cdk2ap1 in the mESC differentiation process.
Ectopic Expression of Nonphosphorylatable pRb S788A Resulted in Spontaneous Differentiation of Cdk2ap1−/− mESCs in Adherent Culture
Based on our previous findings, we hypothesized that there should be a sustained phosphorylation of pRb in Cdk2ap1−/− mESCs with elevated CDK2-associated kinase activities (15). We further hypothesized that the sustained hyperphosphorylation of pRb in the Cdk2ap1−/− mESCs is responsible for the differentiation blockade, since pRb has been implicated in the differentiation control of somatic cells as well as a potential role in stem cell biology (9, 25–31). Interestingly, we observed spontaneous differentiation of both WT and Cdk2ap1−/− mESCs after transducing with miCDK2 even in the presence of LIF (supplemental Fig. S6). This result demonstrates that CDK2 activity is essential in the maintenance of LIF-mediated pluripotent self-renewal and also suggests that the compromised differentiation in Cdk2ap1−/− cells is functionally related to the elevated CDK2 activity.
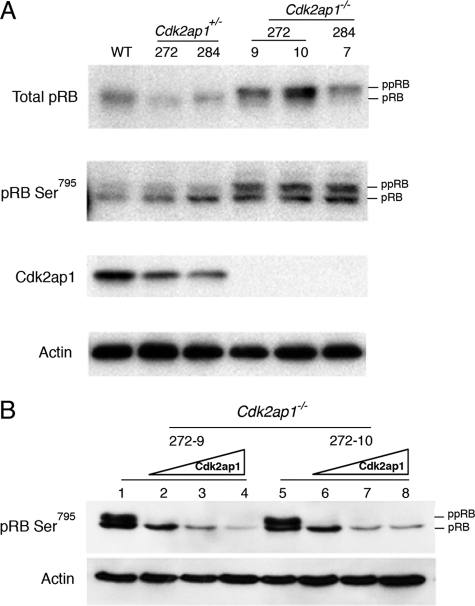
The level of pRb and its phosphorylation status were examined in Cdk2ap1+/+, Cdk2ap1+/−, and Cdk2ap1−/− mESCs, as shown in Fig. 3. Cdk2ap1−/− cells showed a dramatic increase in the phosphorylated form of pRb. To see if the phosphorylation of pRb is related to the increased activity of CDK2, the anti-pRb Ser795 antibody (Cell Signaling Technology, Inc., Danvers, MA), which is known to be one of the targets of CDK2 kinase upon pRb phosphorylation, was used to stain the immunoblots. As shown in Fig. 3A, there was a clear hyperphosphorylated form of pRb in the Cdk2ap1−/− cells compared with Cdk2ap1+/+ WT and Cdk2ap1+/− cells. Although the pRb is detected with anti-pRb Ser795, the hyperphosphorylation in Cdk2ap1−/− mESCs might not be solely due to the phosphorylation of Ser795 (equivalent to mouse Ser788). Potential phosphorylation on other sites can be also contributing. In addition, the deletion of CDK2AP1 in mESCs seems to induce the total level of pRb expression, as indicated in Fig. 3. We further confirmed the specificity of the effect of CDK2AP1 on pRb phosphorylation. As shown in Fig. 3B, the ectopic expression of CDK2AP1 in Cdk2ap1−/− mESCs resulted in hypophosphorylation of pRb detected with anti-Ser795 antibody. This demonstrates that there is a direct linkage between CDK2AP1 and pRb phosphorylation at Ser795.
FIGURE 3.
Hyperphosphorylation of pRb in Cdk2ap1−/− mESCs. A, the level of pRb and phosphorylation status of serine 795 of pRb was examined by Western analysis by using specific antibodies (anti-pRb G3–245 from BD Pharmingen and anti-pRb Ser795 antibody from Cell Signaling Technology, Inc. (Danvers, MA)). Cdk2ap1−/− cells (clones 272-9, 272-10, and 284-7) showed induced hyperphosphorylation of pRb compared with WT or their parental Cdk2ap1+/− clones (272 and 284). B, the effect of restoring Cdk2ap1 on pRb phosphorylation was examined in Cdk2ap1−/− mESCs. Cells were transduced with control virus (lanes 1 and 5) or Ad-CDK2AP1 virus (lanes 2 and 6, MOI 10; lanes 3 and 7, MOI 20; lanes 4 and 8, MOI 30) as shown in Fig. 1. The result showed hypophosphorylation of pRb upon the expression of CDK2AP1, demonstrating the specificity of the role of CDK2AP1 in pRb phosphorylation.
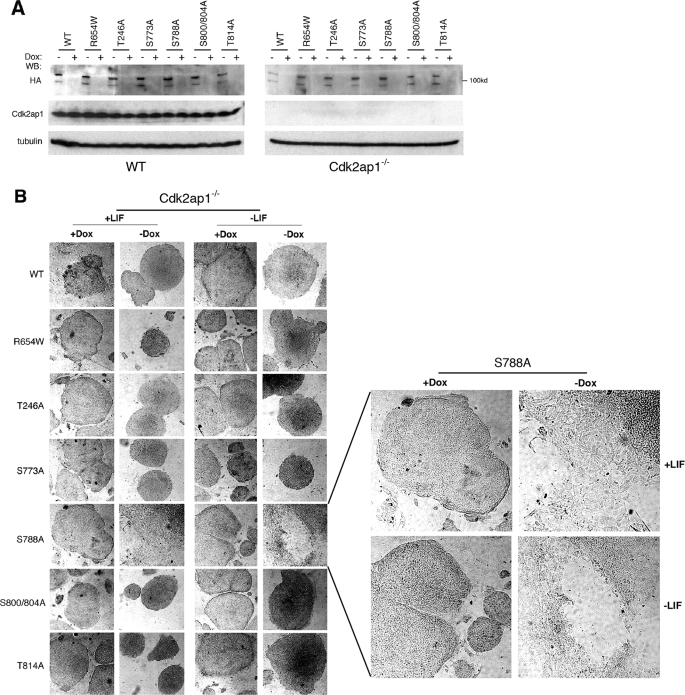
We next determined if the differentiation blockade in Cdk2ap1−/− mESCs was specifically due to the hyperphosphorylation of pRb or the induced expression of pRb by examining the effect of the ectopic expression of nonphosphorylatable mutants of pRb along with WT pRb. We have generated the Tet-off-inducible pRb phosphorylation mutant-expressing cell lines from WT and Cdk2ap1−/− mESCs (Fig. 4A). These cell lines were then used for a differentiation assay to examine the specific effect of pRb mutant expression on mESC differentiation. Of all of the pRb mutants (WT, R654W T214A, S773A, S788A, S800/804A, and T814A), only the inducible expression of S788A mutant in the absence of doxycyclin specifically induced spontaneous differentiation in Cdk2ap1−/− mESCs, whereas there were no noticeable changes when the expression was shut off with the addition of doxycyclin (Fig. 4B). In addition, there was no differentiation observed with other pRb mutants and also in Cdk2ap1+/+ mESCs. This finding clearly demonstrated that the phosphorylation of pRb at serine 788 in Cdk2ap1−/− mESCs was in part responsible for the compromised differentiation competency elicited upon the knock-out of Cdk2ap1.
FIGURE 4.
Inducible expression of pRb S788A mutant rescued Cdk2ap1−/− cells from differentiation blockade. The specific effect of pRb phosphorylation on Ser788 was demonstrated by using the inducible expression system. A, Tet-off inducible cell lines were generated from both Cdk2ap1+/+ and Cdk2ap1−/− cells by stably expressing tTA transactivator. The inducible expression of various pRb mutants (WT, T214A, R654W, S773A, S788A, S800A/S804A, and T814A) was determined by Western analysis. Treatment of cells with doxycyclin resulted in turning off the HA-tagged pRb mutant expression. B, cellular effect of the inducible pRb mutant expression on mESC differentiation was examined in Cdk2ap1−/− mESCs. Cells were grown on chamber slide and cultured in the presence or absence of LIF up to 10 days. Morphological changes of cells were monitored after staining cells for alkaline phosphatase activity. Undifferentiated cells maintain tightly packed morphology, whereas differentiated cells showed flattened and spread morphology. The right panel shows an enlarged image of S788A mutant morphology. Dox, doxycyclin.
Differential Gene Regulation by pRb Ser788 Mutant in Cdk2ap1 Knock-out mESCs
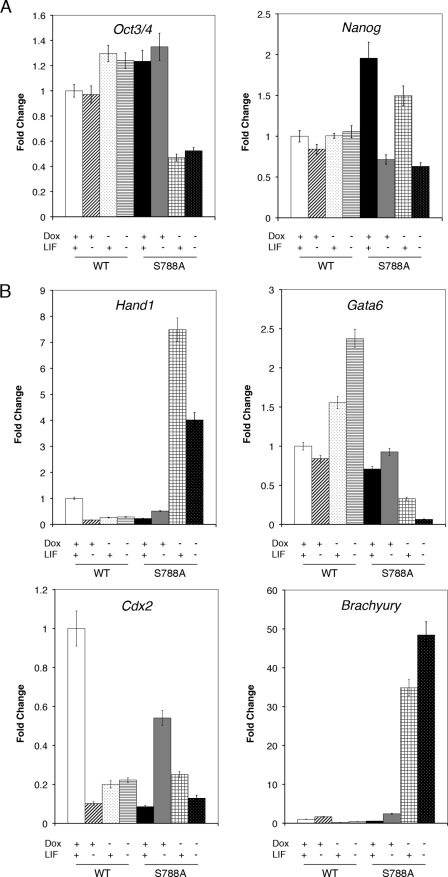
To gain insight into the potential molecular mechanism leading to the differentiation alteration in the Cdk2ap1 knock-out mESCs, the effect of nonphosphorylatable pRb on the expression of these stem cell genes was examined (Fig. 5 and supplemental Fig. S7). As presented in Fig. 5A, the expression of pRb WT into Cdk2ap1−/− mESCs did not induce significant changes or elicit marginal changes if any, at the level of genes involved in stem cell pluripotency and self-renewal (Oct3/4, p < 0.04; Nanog, p < 0.03) compared with the uninduced control. In addition, the level of other stem cell marker genes, such as Sox2, Socs3, and Rex1, was not noticeably altered. Overall, this implies that pRb WT overexpression either did not induce cellular differentiation or induced a marginal change in Cdk2ap1−/− mESCs as morphologically evidenced in Fig. 4B. However, as shown in Fig. 5A, an ectopic expression of pRb S788A mutant in Cdk2ap1−/− mESCs specifically resulted in a decreased level of Oct4 (p < 0.04) while showing no noticeable changes of Nanog (p < 0.01), Sox2 (p < 0.01), Socs3 (p < 0.05), and Rex1 (p < 0.4) (supplemental Fig. S7). It was noticed that the expression of Nanog (p < 0.01) in the pRb S788A mutant showed more noticeable LIF dependence than pRb WT clones, but it was not specific to the induction of pRb S788A mutant. In addition, the expression of Socs3 (p < 0.05) and Rex1 (p < 0.4) showed a higher degree of LIF dependence in both pRb WT and S788A mutant (supplemental Fig. S7). This finding suggests that phosphorylation of pRb at Ser788 plays a role only in the LIF-independent regulation of Oct3/4. As shown in Fig. 5B, although there exist considerable variations in gene expression levels, we found that the expression of Hand1 (p < 0.02) and Brachyury (p < 0.05) was most noticeably affected by the induction of pRb S788A expression, independent of LIF treatment. The expression of other differentiation-related genes was quite variably affected by the expression of either pRb WT or pRb S788A mutant (Fig. 5B). Therefore, although it was not possible to clearly differentiate the potential cellular effect of site-specific pRb phosphorylation on these genes, it seems that each site-specific pRb phosphorylation has some overlapping yet unique effect on the expression of mESC differentiation-related genes, which implies a complex nature of the signaling network exerted by pRb on differentiation.
FIGURE 5.
The effect of the inducible pRb S788A on the level of stem cell marker genes and differentiation-related genes. The inducible pRb WT or pRb S788A mutant clones generated from Cdk2ap1−/− mESCs were grown in the presence or absence of LIF and with or without the induction of gene activation. The effect of the induction of pRb WT or S788A mutant expression on stem cell marker gene or differentiation-related genes was determined by real time quantitative reverse transcription-PCR analysis. A, the level of Oct3/4 and Nanog was determined in biological duplicates. The expression of pRb WT did not show any changes at the level of Oct3/4 in Cdk2ap1−/− mESCs during differentiation, but the expression of pRb S788A showed significant down-regulation of Oct3/4 regardless of LIF treatment (Oct3/4; p < 0.04). The analysis on the level of Nanog showed LIF withdrawal-dependent down-regulation only in the pRb S788A mutant (Nanog; p < 0.01). B, the levels of differentiation-related gene (Hand1, Gata6, Cdx2, and Brachyury) were compared in the inducible pRb WT and pRb S788A mESCs. The expression of Hand1 (p < 0.02) and Brachyury (p < 0.05) was most noticeably affected by the induction of pRb S788A expression, independent of LIF treatment. Statistical p values were determined by Student's t test by comparing values against the samples treated with doxycyclin and leukemia inhibitory factor (+Dox+LIF). Error bar, S.E.
Overall, this finding implicates that the differentiation blockade in Cdk2ap1 knock-out mESCs is through the disrupted regulation of pRb activity via phosphorylation at serine 788. It supports our hypothesis that CDK2AP1 plays a critical role in mESC differentiation by modulating pRb activity and suggests that pRb regulation is an effector in mESC differentiation.
DISCUSSION
Embryonic stem (ES) cells undergo unique self-renewal process and maintain pluripotent competency for specification into different cell lineages (32, 33). Recent findings suggest potential molecular mechanisms for how ES cells maintain pluripotency and control the self-renewal process, but their details are still largely unknown (6, 7, 33–36). One of the prominent features of cell cycle regulation in ESCs is the lack of the pRb pathway. Embryonic stem cells have a short G1 phase in which hypophosphorylated pRb is virtually undetectable (8). It is likely that Rb is rephosphorylated immediately after mitosis in ES cells. ESCs express Rb and p107, but not p130 (37, 38). ESCs fail to arrest in G1 after DNA damage but arrest at the Rb-independent G2/M checkpoint (39, 40). Evidence suggests that ESCs are not controlled by pRb in G1 (41, 42). The cellular mechanism underlying functional inactivation of pRb is being elucidated. ES cells show a low level of cyclin D, and Cdk4-associated kinase activity is virtually undetectable. In contrast, ESCs show constitutive cyclin E/CDK2 kinase activity. During differentiation, there is a robust expression of D-type cyclins and Cdk4-associated kinase activity along with a reduction of cyclin E/CDK2 kinase activity, resulting in G1/S control by pRb pathway (28, 43). From these findings, it is evident that molecules with significant cellular effects on pRb or its related pathways should have functional impact on mESC proliferation and differentiation. However, the involvement of pRb in pluripotent differentiation of mESCs has not been extensively elucidated.
The biological role of the specific phosphorylation site for pRb regulation still remains elusive. Especially, its contribution to the differentiation of mESCs is largely unknown. The differential expression of stem cell maintenance or differentiation-related genes mediated by the preferential phosphorylation of pRb at serine 788 implicates that the knock-out of Cdk2ap1 in mESCs leads to the collective alteration of differentiation potential, leading to the observed differentiation blockade in the Cdk2ap1−/− mESCs (Fig. 5). The most significant and interesting molecular changes observed upon the inducible expression of pRb S788A in Cdk2ap1−/− mESCs were the down-regulation of Oct3/4 and up-regulation of Hand1 (Fig. 5). The regulation of these two genes showed consistent changes before and after the restoration of CDK2AP1 and the pRb S788A mutant. Although we convincingly show that CDK2AP1 plays a role in the regulation of CDK2 activity and further in pRb phosphorylation, it must be one of many mechanisms regulating pRb phosphorylation. It is also speculated that the activation of CDK2 itself in mESCs may have different cellular effects from expressing S788A, one of its downstream targets. The restoration of Cdk2ap1 in Cdk2ap1−/− mESCs should be more specific to the pathway involving CDK2AP1-mediated pRb phosphorylation. Since CDK2 itself has many downstream targets and also Ser788 is a target of multiple effectors, it is expected that the manipulation of these two will have somewhat different downstream molecular effects. One way to appreciate their concurrent effects would be by identifying any overlapping events elicited by the two.
Although ectopic expression of Cdk2ap1 did not elicit spontaneous differentiation in WT and Cdk2ap1−/− mESCs in the presence of LIF, it induced differentiation in Cdk2ap1−/− mESCs only in the absence of LIF. This demonstrates that CDK2AP1 is a downstream regulator of LIF-dependent self-renewal/differentiation of mESCs. In contrast, the ectopic expression of pRb S788A mutant resulted in the spontaneous differentiation of Cdk2ap1−/−, but not WT mESCs, regardless of LIF treatment. This suggests that the deletion of Cdk2ap1 in mESCs drives the cells toward an LIF-independent cascade. We speculate that expression of the pRb S788A mutant in WT mESC has no phenotypic effect, because the cells maintain normal LIF-dependent signaling in the presence of WT Cdk2ap1. Deletion of Cdk2ap1 results in the mESCs becoming capable of self-renewing even in the absence of LIF signaling, with the phosphorylation of pRb at Ser788 as one of the downstream events. This is in addition to other possible molecular alterations in the absence of Cdk2ap1 that could mediate the function of Cdk2ap1 in LIF-dependent mESC self-renewal/differentiation, such as the epigenetic control of Oct3/4 promoter, as demonstrated by our group (24).
Further study is required to unveil how Cdk2ap1 responds to a differentiation signal and transmits it further downstream. It is possible that Cdk2ap1 itself is under the regulation of mESC differentiation through either transcriptional or post-translational regulation, such as molecular dimerization that has been shown to influence the activity of CDK2AP1 (44). Also the signaling and molecular mechanism needs to be delineated to understand how pRb mediates or triggers differentiation of mESCs through modulation of downstream genes. It is speculated that the activation of pRb should elicit its effect on gene regulation through either the regulation of a certain transcription factor or via epigenetic regulation, such as DNA methylation and histone acetylation. Our preliminary data suggest that the hyperphosphorylation of pRb in Cdk2ap1−/− mESCs is accompanied by the activation of E2F1 promoter (data not shown). Further detailed examination of E2F1 activation should reveal the consequence and significance of this event in stem cell maintenance and differentiation. As an extension of this study, it will be meritorious and highly translational to examine if CDK2AP1 functions as a competency factor in human embryonic stem cell differentiation.
Supplementary Material
Acknowledgments
We thank Dr. Brenda Gallie (University of Toronto) and Dr. David Goodrich (Rosewell Park Cancer Institute) for kindly providing murine pRb plasmids. We thank Dr. J. Wade Harper (Harvard Medical School) and Dr. Philip Hinds (Tufts University) for human pRb WT and mutant plasmids. We also thank Dr. Philip Hinds and Dr. Cun-Yu Wang (UCLA) for valuable comments and suggestions regarding our work.
This work was supported, in whole or in part, by National Institutes of Health Public Service Grants T32 DE 007296-08 (to Y. K.) and R01 DE 14857 (to D. T. W.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- ES
- embryonic stem
- ESC
- embryonic stem cell
- LIF
- leukemia inhibitory factor
- mESC
- mouse embryonic stem cell
- LIF-R
- LIF receptor
- pRb
- retinoblastoma protein
- AP
- alkaline phosphatase
- GFP
- green fluorescent protein
- WT
- wild type
- HA
- hemagglutinin
- tTA
- tetracycline-controlled transactivator.
REFERENCES
- 1.Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. (1988) Nature 336,688–690 [DOI] [PubMed] [Google Scholar]
- 2.Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988) Nature 336,684–687 [DOI] [PubMed] [Google Scholar]
- 3.Ernst M., Oates A., Dunn A. R. (1996) J. Biol. Chem. 271,30136–30143 [DOI] [PubMed] [Google Scholar]
- 4.Niwa H., Burdon T., Chambers I., Smith A. (1998) Genes Dev. 12,2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst M., Novak U., Nicholson S. E., Layton J. E., Dunn A. R. (1999) J. Biol. Chem. 274,9729–9737 [DOI] [PubMed] [Google Scholar]
- 6.Raz R., Lee C. K., Cannizzaro L. A., d'Eustachio P., Levy D. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96,2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003) Cell 113,631–642 [DOI] [PubMed] [Google Scholar]
- 8.Savatier P., Huang S., Szekely L., Wiman K. G., Samarut J. (1994) Oncogene 9,809–818 [PubMed] [Google Scholar]
- 9.White J., Stead E., Faast R., Conn S., Cartwright P., Dalton S. (2005) Mol. Biol. Cell 16,2018–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shintani S., Mihara M., Terakado N., Nakahara Y., Matsumura T., Kohno Y., Ohyama H., McBride J., Kent R., Todd R., Tsuji T., Wong D. T. (2001) Clin. Cancer Res. 7,2776–2782 [PubMed] [Google Scholar]
- 11.Tsuji T., Duh F. M., Latif F., Popescu N. C., Zimonjic D. B., McBride J., Matsuo K., Ohyama H., Todd R., Nagata E., Terakado N., Sasaki A., Matsumura T., Lerman M. I., Wong D. T. (1998) J. Biol. Chem. 273,6704–6709 [DOI] [PubMed] [Google Scholar]
- 12.Kim Y., Tsuji T., Elovic A., Shintani S., Mihara M., Salih E., Kohno Y., Chin B. R., Patel V., Wong D. T. W., Todd R. (2001) Int. J. Oral Biol. December 2001, 87–91 [Google Scholar]
- 13.Kim Y., McBride J., Zhang R., Zhou X., Wong D. T. (2005) Oncogene 24,407–418 [DOI] [PubMed] [Google Scholar]
- 14.Matsuo K., Shintani S., Tsuji T., Nagata E., Lerman M., McBride J., Nakahara Y., Ohyama H., Todd R., Wong D. T. (2000) FASEB J. 14,1318–1324 [DOI] [PubMed] [Google Scholar]
- 15.Shintani S., Ohyama H., Zhang X., McBride J., Matsuo K., Tsuji T., Hu M. G., Hu G., Kohno Y., Lerman M., Todd R., Wong D. T. (2000) Mol. Cell. Biol. 20,6300–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R. C., Melton D. A. (2002) Science 298,597–600 [DOI] [PubMed] [Google Scholar]
- 17.Sharov A. A., Piao Y., Matoba R., Dudekula D. B., Qian Y., VanBuren V., Falco G., Martin P. R., Stagg C. A., Bassey U. C., Wang Y., Carter M. G., Hamatani T., Aiba K., Akutsu H., Sharova L., Tanaka T. S., Kimber W. L., Yoshikawa T., Jaradat S. A., Pantano S., Nagaraja R., Boheler K. R., Taub D., Hodes R. J., Longo D. L., Schlessinger D., Keller J., Klotz E., Kelsoe G., Umezawa A., Vescovi A. L., Rossant J., Kunath T., Hogan B. L., Curci A., D'Urso M., Kelso J., Hide W., Ko M. S. (2003) PLoS Biol. 1,E74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S., Lee S. A., Shim C., Khang I., Lee K. A., Park Y. M., Kang B. M., Kim K. (2003) Mol. Reprod. Dev. 64,405–413 [DOI] [PubMed] [Google Scholar]
- 19.Connell-Crowley L., Harper J. W., Goodrich D. W. (1997) Mol. Biol. Cell 8,287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams P. D., Li X., Sellers W. R., Baker K. B., Leng X., Harper J. W., Taya Y., Kaelin W. G., Jr. (1999) Mol. Cell. Biol. 19,1068–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller G. M. (1995) Curr. Opin. Cell Biol. 7,862–869 [DOI] [PubMed] [Google Scholar]
- 22.Kim Y., McBride J., Kimlin L., Pae E. K., Deshpande A., Wong D. T. (2009) PLoS ONE 4,e4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu M. G., Hu G. F., Kim Y., Tsuji T., McBride J., Hinds P., Wong D. T. (2004) Cancer Res. 64,490–499 [DOI] [PubMed] [Google Scholar]
- 24.Deshpande A. M., Dai Y. S., Kim Y., Kim J., Kimlin L., Gao K., Wong D. T. (2009) J. Biol. Chem. 284,6043–6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benevolenskaya E. V., Murray H. L., Branton P., Young R. A., Kaelin W. G., Jr. (2005) Mol. Cell 18,623–635 [DOI] [PubMed] [Google Scholar]
- 26.Sidle A., Palaty C., Dirks P., Wiggan O., Kiess M., Gill R. M., Wong A. K., Hamel P. A. (1996) Crit. Rev. Biochem. Mol. Biol. 31,237–271 [DOI] [PubMed] [Google Scholar]
- 27.Slack R. S., El-Bizri H., Wong J., Belliveau D. J., Miller F. D. (1998) J. Cell Biol. 140,1497–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stead E., White J., Faast R., Conn S., Goldstone S., Rathjen J., Dhingra U., Rathjen P., Walker D., Dalton S. (2002) Oncogene 21,8320–8333 [DOI] [PubMed] [Google Scholar]
- 29.McLear J. A., Garcia-Fresco G., Bhat M. A., Van Dyke T. A. (2006) Mol. Cell Neurosci. 33,260–273 [DOI] [PubMed] [Google Scholar]
- 30.Jori F. P., Galderisi U., Napolitano M. A., Cipollaro M., Cascino A., Giordano A., Melone M. A. (2007) Mol. Cell Neurosci. 34,299–309 [DOI] [PubMed] [Google Scholar]
- 31.Kotake Y., Cao R., Viatour P., Sage J., Zhang Y., Xiong Y. (2007) Genes Dev. 21,49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Shea K. S. (1999) Anat. Rec. 257,32–41 [DOI] [PubMed] [Google Scholar]
- 33.Chambers I. (2004) Cloning Stem Cells 6,386–391 [DOI] [PubMed] [Google Scholar]
- 34.Hanna L. A., Foreman R. K., Tarasenko I. A., Kessler D. S., Labosky P. A. (2002) Genes Dev. 16,2650–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Genes Dev. 17,126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A. H. (2004) Nat. Med. 10,55–63 [DOI] [PubMed] [Google Scholar]
- 37.Robanus-Maandag E., Dekker M., van der Valk M., Carrozza M. L., Jeanny J. C., Dannenberg J. H., Berns A., te Riele H. (1998) Genes Dev. 12,1599–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeCouter J. E., Whyte P. F., Rudnicki M. A. (1996) Oncogene 12,1433–1440 [PubMed] [Google Scholar]
- 39.Aladjem M. I., Spike B. T., Rodewald L. W., Hope T. J., Klemm M., Jaenisch R., Wahl G. M. (1998) Curr. Biol. 8,145–155 [DOI] [PubMed] [Google Scholar]
- 40.Prost S., Bellamy C. O., Clarke A. R., Wyllie A. H., Harrison D. J. (1998) FEBS Lett. 425,499–504 [DOI] [PubMed] [Google Scholar]
- 41.Dannenberg J. H., van Rossum A., Schuijff L., te Riele H. (2000) Genes Dev. 14,3051–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sage J., Mulligan G. J., Attardi L. D., Miller A., Chen S., Williams B., Theodorou E., Jacks T. (2000) Genes Dev. 14,3037–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savatier P., Lapillonne H., van Grunsven L. A., Rudkin B. B., Samarut J. (1996) Oncogene 12,309–322 [PubMed] [Google Scholar]
- 44.Kim Y., Ohyama H., Patel V., Figueiredo M., Wong D. T. (2005) J. Biol. Chem. 280,23273–23279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.