Abstract
The metabolome of the model species Chlamydomonas reinhardtii has been analyzed during 120 h of sulfur depletion to induce anaerobic hydrogen (H2) production, using NMR spectroscopy, gas chromatography coupled to mass spectrometry, and TLC. The results indicate that these unicellular green algae consume freshly supplied acetate in the medium to accumulate energy reserves during the first 24 h of sulfur depletion. In addition to the previously reported accumulation of starch, large amounts of triacylglycerides were deposited in the cells. During the early 24- to 72-h time period fermentative energy metabolism lowered the pH, H2 was produced, and amino acid levels generally increased. In the final phase from 72 to 120 h, metabolism slowed down leading to a stabilization of pH, even though some starch and most triacylglycerides remained. We conclude that H2 production does not slow down due to depletion of energy reserves but rather due to loss of essential functions resulting from sulfur depletion or due to a build-up of the toxic fermentative products formate and ethanol.
A variety of unicellular eukaryotic green algae have the ability to produce H2 under anaerobic conditions (1, 2). This ability is greatly enhanced in the light (3). Studies using the model species Chlamydomonas reinhardtii have shown that H2 generation stems from the use of two oxygen (O2) sensitive Fe-hydrogenase enzymes, HydA1 and HydA2, that use reduced ferredoxin to catalyze the reduction of protons to yield H2 (4, 5). The electrons for the reduction of ferredoxin that is used in H2 production can come from endogenous substrates or from water oxidation (6, 7).
H2 production was originally studied by making algal cultures anaerobic by purging with inert gases or by incubation in the dark and then exposure to light (3, 8). H2 was produced during these experiments in small amounts and for a relatively short period of time. More recently, a method of sulfur depletion was devised in which cells are resuspended in sulfur-depleted medium, allowing H2 production to be observed for numerous days and in larger quantities (9). By depleting a sealed culture of sulfur, photosynthetic O2 evolution decreases and it becomes microxic, leading to anaerobic pathways becoming operative and to the onset of H2 production. It is proposed that sulfur depletion preferentially limits synthesis of the D1 protein of photosystem II (9). Sulfur depletion has to date proven to be the best procedure for inducing H2 production in terms of volume and purity.
Sulfur is transported into C. reinhardtii cells primarily as the sulfate anion, SO42−, and is required for a variety of lipids, proteins, and metabolites (10). Microarray studies have been performed on C. reinhardtii following transfer to sulfur-free medium (11) and in a sulfur-deplete anaerobic environment in which H2 production was observed (12). In both studies, extensive changes in transcript abundance were observed, with over 20% of analyzed transcripts showing a change of greater than 2-fold, many of them with putative or unknown functions. Both studies showed sulfur depletion to lead to a general increase in transcripts involved in sulfur assimilation, protein degradation and stress, a decrease in most transcripts encoding components of the photosynthetic apparatus, a decrease in transcripts for carbon metabolism through the Calvin-Benson cycle, an increase in those of the oxidative pentose phosphate cycle, and an increase in those of starch synthesis. Key additional observations made in work by Ngyuen et al. (12) showed that the added effect of anaerobicity led to repression of genes of the glyoxylate cycle, an up-regulation of the major light harvesting complex Lhcbm9 protein and transcript, hydrogenase-encoding transcripts HydA1, HydA2, and HydEF, and an up-regulation of genes of fermentative pathways.
The up-regulation of genes of fermentative pathways and hydrogenase-encoding transcripts are key processes that C. reinhardtii employs to respond to anaerobic conditions. However, these are not the sole processes, and it has been shown that there is a “whole cell” response such that hundreds to thousands of genes are regulated to deal with anoxic conditions. Analysis of the genome of C. reinhardtii has shown a large number of peptides to be involved in anaerobic metabolism (13), and microarray analysis has revealed over 500 transcripts involved in diverse processes such as transcription/translation regulators, prolylhydroxylases, hybrid cluster proteins, proteases, transhydrogenases, and catalases to be up-regulated by the onset of anoxia (14). It is clear that photosynthetic organisms respond to anoxia by adopting an integrated response rather than activating or inactivating only a few select pathways.
To better understand the response of C. reinhardtii to sulfur depletion and to anoxia as well as how such responses underpin H2 production, we undertook detailed metabolomic studies of C. reinhardtii. Metabolomics employs a non-targeted profiling approach and can potentially detect and quantify hundreds of metabolites, particularly the more abundant intermediates of primary metabolism. NMR, GC/MS,5 and TLC were used in parallel to obtain as much information as possible about key metabolites. The aim of this work was to provide first insight into the metabolic pathways employed to survive anoxia during illumination and to support H2 production.
EXPERIMENTAL PROCEDURES
Anaerobic Acclimation to Achieve H2 Production via Sulfur Depletion
Anaerobic H2 production conditions were achieved by the method of sulfur depletion as reported by Melis et al. (9). In brief, cultures of C. reinhardtii Stm6 (15) were harvested at late exponential phase by centrifugation (2500 × g, 3 min, 25 °C) and washed three times in sulfur-depleted TAP medium (16). Cells were resuspended to a density of 15 × 106 cells/ml in sulfur-depleted TAP medium (pH 7.3) and sealed in 600-ml conical flasks fitted with gas collection tubes. Cultures were exposed to 500 μE·m−2·s−1 continuous white light and stirred with magnetic stirrers at 150 rpm. Dissolved O2 concentration, H2 gas evolution, and pH of the cultures were monitored for 120 h post sulfur depletion. Samples were taken for metabolomic analysis at 0, 24, 48, 72, 96, and 120 h post sulfur depletion. For GC/MS analyses, 6 samples were taken at each time point (3 biological × 2 technical replicates). For NMR analyses, 16 samples were taken at each time point (4 biological × 4 technical replicates). In addition, analysis by NMR required the removal of Tris from the growth medium, so TAP medium was modified so that Tris was replaced with high concentrations of phosphate (NaH2PO4·H2O, 2.33 g l−1 and Na2HPO4, 3.27 g l−1) and acetate concentration was set at 15 mm; all other conditions were kept identical.
Dissolved O2, pH, and H2 Evolution Measurements
Dissolved O2 concentrations and pH of algal cultures were measured every 5 min using a D130 data logger system with SZ10T DO and SP11X pH electrodes (Consort, Belgium). Gas composition was measured by extraction with a Hamilton gas-tight syringe and injection into an Agilent Micro GC3000 gas chromatograph fitted with a PlotU pre-column (3 m × 0.32 mm) and MolSieve 5APlot column (10 m × 0.32 mm). Argon (32.5 p.s.i.) was used as the carrier gas.
Hot Medium Metabolite Extraction from C. reinhardtii for NMR Analysis
Algal culture (1 ml) was removed from the bioreactor side-port with a gas-tight lockable syringe and immediately immersed in a beaker full of boiling water for 3 min to inactivate enzyme activity. Cell culture was transferred to a microcentrifuge tube, and cellular debris was centrifuged at 16,000 × g for 3 min. For NMR analysis, 500 μl of the supernatant solution was transferred to an NMR tube, and 50 μl of D2O was added.
Hot Methanol Metabolite Extraction from C. reinhardtii for GC/MS Analysis
Algal culture (2 ml) was transferred to a pre-chilled microcentrifuge tube and centrifuged at 3,000 × g for 1 min at 4 °C. The supernatant was discarded, and the pellet was washed once with 32.5% (v/v) methanol kept at −20 °C. 500 μl of 100% (v/v) methanol supplemented with 2 μg of ribitol for sample normalization was added to the pellet. Metabolite extraction was performed by 15 min of shaking in a thermomixer (1,200 rpm) at 70 °C. Cell debris was centrifuged at 16,000 × g for 5 min, and 100 μl of the supernatant solution was dried in a vacuum evaporator for 3 h.
GC/MS Sample Preparation for Total Metabolomic Analysis
Dried samples were derivatized by the addition of 20 μl of a 20 mg/ml solution of methoxylamine hydrochloride (Sigma-Aldrich) in pyridine (30 °C for 90 min). 30 μl of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) was then added and shaken for a further 30 min at 37 °C. 10 μl of an alkane standard mix containing 50 ng each of C12, C15, C19, C22, C28, C32, and C36 in chloroform was added for retention index determination.
GC/MS Sample Preparation for Amino Acid Analysis and Quantifier Ion Selection
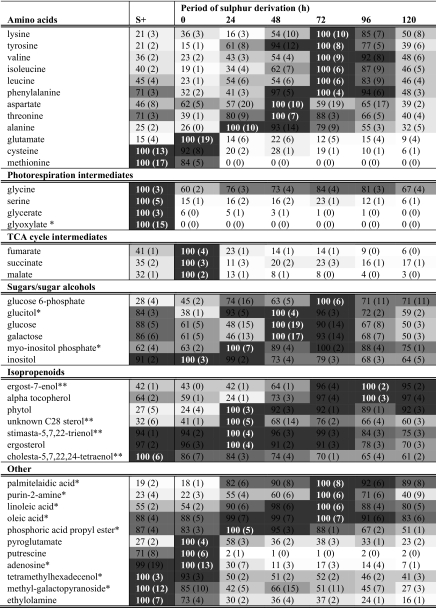
To enhance the signal of amino acids for GC/MS analysis, cell extracts were derivatized with the agent N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide (MBTSTFA), which generates higher signals than MSTFA for the amino acids. Its use permits molecular formula determination of the quantifier ion from spectral analysis and allows for calculation of isotopic signal contribution of unlabeled metabolites toward the labeled pool. For amino acid analysis, dried samples were derivatized with 20 μl of a 20 mg/ml solution of methoxylamine hydrochloride at 30 °C for 90 min. 20 μl of MBTSTFA was then added, and samples were shaken (750 rpm) at 85 °C for 60 min. No other compounds were added. Table 1 lists the amino acids that could be identified that generated both labeled (15N) and unlabeled (14N) signals and shows the molecular formula of the quantifier ion (Q-ion) as well as the theoretical isotopic contribution of the unlabeled component toward the labeled component.
TABLE 1.
Molecular formula of ions used for quantification of labeled and unlabeled amino acids
Isotopic contribution of unlabeled metabolite signal was subtracted from the signal response of 15N-labeled metabolites in all cases.
| Metabolite | Q-ion molecular formula | 14N Q-ion | 15N Q-ion | Isotope % contribution |
|---|---|---|---|---|
| Alanine | Si2C11H26NO2 | 260 | 261 | 23 |
| Aspartate | Si3C18H40NO4 | 418 | 419 | 36 |
| Glutamate | Si3C19H43N2O3 | 432 | 433 | 38 |
| Glycine | Si2C10H24NO2 | 246 | 247 | 22 |
| Isoleucine | Si2C14H32NO2 | 302 | 303 | 27 |
| Leucine | Si2C14H32NO2 | 302 | 303 | 26 |
| Lysine | Si3C20H42N3O2 | 431 | 433 | 18 |
| Phenylalanine | Si2C17H30NO2 | 336 | 337 | 30 |
| Proline | SiC8H14NO2 | 184 | 185 | 15 |
| Serine | Si3C17H40NO3 | 390 | 391 | 35 |
| Threonine | Si3C18H42NO3 | 404 | 405 | 36 |
| Tyrosine | Si3C23H44NO3 | 466 | 467 | 42 |
| Valine | Si2C11H26NO2 | 260 | 261 | 23 |
GC/MS Instrument Settings
Samples were randomized, and 1 μl of derivatized sample was injected splitless into an Agilent 6890 GC fitted with an Agilent 5975 MSD. Helium was used as the carrier gas at a constant flow of 1 ml/min. Inlet temperature was set at 300 °C. Oven temperature was initially set at 70 °C for 1 min, ramped at 1 °C/min until 76 °C, then ramped at 6 °C/min until 325 °C, with a final hold of 10 min. A Varian Factor 4 capillary column (VF-5ms, 30 m × 0.25 mm, 0.25 μm plus 10 m EZ-Guard) was used. The MSD transfer line heater was kept at 300 °C. MS quadrupole temperature was kept at 150 °C and source temperature at 230 °C. Mass detection range was set from 40 to 600 atomic mass units.
GC/MS Metabolite Peak Identification and Quantification
Spectral data files were processed with AMDIS (version 2.65) for metabolite identification. Metabolites were identified by retention index and spectral comparison to pre-run standards or by searching the NIST library (indicated with asterisks). All identified metabolites were entered into MSD ChemStation (version E.02.00.493), and a quantitation data base was created using specific target ions and qualifier ions unique to each metabolite. All spectra were manually reviewed. Normalization was performed to the internal standard ribitol and to the cell number.
NMR Settings and Data Analysis
High resolution proton NMR spectroscopy was performed on a 500-MHz spectrometer (Avance 500, Bruker, Karlsruhe, Germany). Standard one-dimensional 1H Carr-Purcell-Meiboom-Gill spectra with 0.5-ms T2 spin echo time and water presaturation during the relaxation delay were recorded with 512 scans. Fourier transformation was performed, and all 1H NMR spectra were manually phased and baseline-corrected using TOPSPIN (Bruker Biospin, Karlsruhe, Germany). Chemical shift referencing was performed on the formate peak at 8.44 ppm. Data analysis was carried out using the program AMIX (version 3.6.8, Bruker Biospin). The chemical shift region from 0.2 to 9.0 ppm of each NMR spectrum was divided into buckets (spectral integral regions) of 0.04-ppm width. Integral intensities were normalized to the total intensity of the regions examined for each spectrum. The resulting bucket tables were used without further scaling for principal component analysis with the multivariate data analysis program Pirouette (version 4, Infometrix). Metabolites were identified based on chemical shift peak similarities to standards in public databases. Relative abundances were calculated from spectral integrals.
Starch Analysis
Algal cultures (2 ml) were centrifuged at 16,000 × g for 3 min, the supernatant solution was discarded, and the pellets were washed with 1 ml of 100% methanol (v/v) and 1 ml of sodium acetate buffer (100 mm, pH 4.8) consecutively. The pellets were re-suspended in 500 μl of sodium acetate buffer and autoclaved (15 min 120 °C) for starch solubilization. Starch was hydrolyzed by incubation with 50 units of amyloglucosidase (Sigma-Aldrich) at 60 °C overnight. Cellular debris was removed by centrifugation (16,000 × g, 3 min), and 500 μl of the supernatant solution was taken for glucose analysis via NMR. Glucose concentration was calculated based on integral comparisons to standards and converted to starch mass.
TLC
Algal culture (2 ml) was centrifuged at 16,000 × g for 3 min. Non-polar lipids were extracted from the pellet by the separate addition of 400 μl of methanol, 400 μl of chloroform, and 400 μl of water. Samples were sonicated in a water bath for 20 min and vortexed between methanol and chloroform additions. Phase separation was performed by centrifugation at 16,000 × g for 5 min, and 200 μl of the chloroform layer was taken and dried in a vacuum evaporator. Samples were resuspended to a volume of 20 μl in chloroform, and 3 μl was loaded onto a 10-cm × 10-cm HPTLC-HL normal phase silica gel plate (Analtech); the plate was developed in an 80:20:2 hexane:ether:acetic acid running buffer. The plate was dried thoroughly (using a hair dryer) and sprayed with 80% (v/v) acetone supplemented with 0.5 mg liter−1 primuline (Sigma-Aldrich). Once again the plate was dried, and stained lipids were visualized under UV illumination.
Fatty Acid Analysis
Algal cultures (2 ml) were centrifuged at 16,000 × g for 3 min. The supernatant solution was discarded, and the pellets were hydrolyzed and methyl-esterified by shaking (1,200 rpm) with 300 μl of a 49:1 100% methanol (v/v):18 m H2SO4 solution for 2 h at 80 °C; 100 μg of nonadecanoic acid was added as internal standard. 300 μl of 0.9% (w/v) NaCl and 300 μl of hexane was then added, and the mixture was sonicated for 20 min and vortexed. Phase separation was performed by centrifugation at 16,000 × g for 3 min. 1 μl of the hexane layer was injected into an Agilent 6890 GC fitted with a 5975 MSD. A DB-Wax column (Agilent, 122-7032) was used with running conditions as described for Agilent's RTL DB-Wax method. Fatty acid methyl esters were quantified using the area of the total ion current compared with that of the internal standard. Identification of fatty acid methyl esters was based on mass spectral profiles, comparison to standards, and expected retention time from Agilent's RTL method and verified by comparison to those previously described in C. reinhardtii by Poerschmann et al. (17).
RESULTS
H2 Production, pH, and O2 Levels during Sulfur Depletion
For metabolic profiling, photobiological H2 production was induced in C. reinhardtii by sulfur depletion. H2 production, pH, and O2 levels were continuously measured through 120 h of sulfur depletion (Fig. 1). Samples for metabolic profiling were taken at 0, 24, 48, 72, 96, and 120 h post re-suspension of C. reinhardtii culture in sulfur-depleted medium. TAP medium was used for culture cultivation, but for NMR analysis it was necessary to replace Tris with phosphate buffer (see “Experimental Procedures”) resulting in a 50% reduction of H2 production; H2 initiation, peak production, and termination times matched to within 10% of those in TAP medium, as did pH and dissolved O2 levels (data not shown).
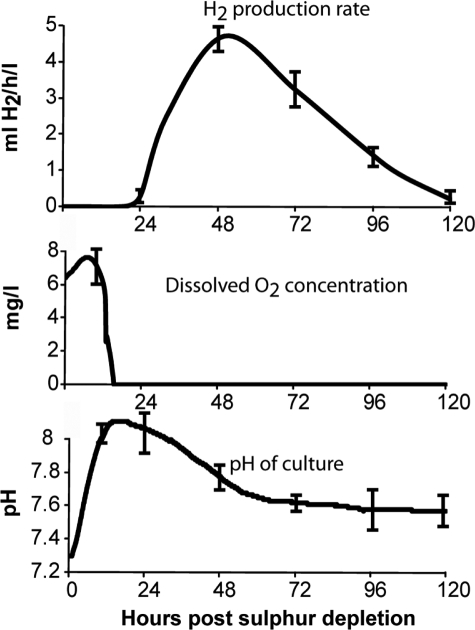
FIGURE 1.
H2 production, change in dissolved O2 concentration, and pH of C. reinhardtii cultures following sulfur depletion. The figures represent averages from six biological replicates. Standard error bars are shown.
Once C. reinhardtii cells were subjected to sulfur depletion there was an initial increase in the dissolved O2 concentration and pH of the growth medium. After ∼12 h, the dissolved O2 concentration began to drop rapidly and the culture became anoxic at ∼20 h. The pH began to decline once anoxic conditions were reached. At ∼24 h, H2 could be measured in the gas collection tube above the bioreactors. H2 production rates peaked at ∼4.5 ml of H2/h/liter of culture at 48 h after sulfur depletion and then declined steadily until 120 h, at which point H2 production rates were below 0.3 ml H2/h/liter. Peak H2 production rates coincided with a drop of pH from 7.9 to 7.6. In vitro studies have shown hydrogenase activity to be optimal in the pH range of 7.0–7.4 but still maintain high activity up to pH 8.0 (18). The decline in pH could potentially increase hydrogenase activity but may also affect other facets of metabolism that control rates of H2 production.
NMR Analysis of the Metabolome of C. reinhardtii during Sulfur-depleted H2 Production
For NMR analysis a hot culture medium extraction was performed. This procedure extracted metabolites from the cells into the medium, so both the extracellular and internal metabolomes were analyzed as a whole. This extraction procedure resulted predominantly in analysis of the extracellular metabolome. However, preliminary experiments indicated that, for the compounds detected by NMR, the extracellular and intracellular metabolomes were very similar, but the cells contributed only 5–10% to the total (data not shown). Because it proved difficult to rapidly isolate cells in sufficient quantity under anaerobic conditions, we analyzed the unfractionated culture. Samples were taken for NMR analysis at 24-h intervals over 120 h of sulfur-depleted H2 production. Generated spectra were integrated over 0.04-ppm regions, and principal component analysis was performed to reduce the number of variables in the data set and to isolate those that contributed most to metabolic variance (Fig. 2). Data base searching of chemical shifts that contributed to variance allowed the positive identification of seven metabolites; their changes in abundance are shown in Fig. 3. Starch concentration was determined separately following digestion to glucose.
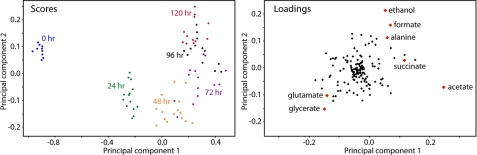
FIGURE 2.
Scores and loadings plots from the first two principal components after outliers were removed. Samples are color-coded on the scores plot to indicate sample points. Buckets belonging to key components that contribute to spectral variance and that could be identified are shown on the loadings plot.
FIGURE 3.
Changes in relative abundance of metabolites during 120 h of sulfur depletion. Standard error bars from 16 replicates (4 biological × 4 technical) are shown. Starch concentrations were calculated from four biological replicates in independent experiments.
Analysis of the scores plot from the first two principal components revealed that the largest changes occurred during the first 24 h. This is largely attributable to the consumption of acetate during this period, which is the key contributor to metabolic variance throughout the time course. The consumption of acetate was fastest during the first 24 h then progressively slowed until 72 h; after that its concentration increased marginally. The grouping of samples in the scores plot reflects this trend, and it is interesting to note the reversal of movement along principal component 1 between 72 and 120 h, likely a result of an overall production of acetate in this period, notably when H2 is still being produced. The fermentation products ethanol and formate were the next largest contributors to spectral variance. These products increased steadily once anoxic conditions were reached. Succinate and alanine also responded in a manner similar to ethanol and formate. Their abundances steadily increased throughout the time course. Glutamate decreased to approximately half its abundance in the first 24 h and continued to decline slowly until 96 h, at which point it remained at a steady level. Glycerate and starch were seen to increase in the first 24 h and then declined steadily until the end of the time course.
Metabolome of C. reinhardtii Analyzed by GC/MS during Sulfur-depleted H2 Production
Using GC/MS, the metabolome of C. reinhardtii was analyzed at 24-h intervals over 120 h of sulfur depletion. In addition, samples were taken immediately prior to washing and re-suspending cells in sulfur-depleted medium. Using a hot methanol extraction, ∼200 putative metabolites could be detected, of which 43 were identified. Relative abundances of these metabolites are shown in Table 2.
TABLE 2.
Relative abundances of metabolites from C. reinhardtii following 120 h of sulphur depletion
Metabolite abundances have been normalized with the highest amount of each metabolite detected at any time point set to 100; other values represent a percent of this amount. Darker shades of grey indicate higher abundance. The column labeled S+ shows metabolite abundances just prior to sulfur depletion. Standard errors from six replicates (3 independent biological × 2 technical replicates) are shown in parentheses. Metabolites have been sorted into classes and ordered from highest increase to greatest decrease. Metabolites annotated with single asterisks have been identified from database searches; double asterisks indicate identification from spectral analysis; all other metabolites have been identified from retention time and spectral profiles of pre-run standards.
Following re-suspension of cells in sulfur-depleted medium (see columns S+ and 0), a number of metabolites changed in abundance, particularly polar metabolites. The process of re-suspension takes ∼2 h and has two likely consequences. First, there were three wash steps employed so that leakage of metabolites during this step is expected; this is likely responsible for the decrease in abundance noted for many metabolites. Second, stress to the cells from the wash and centrifugation steps is likely to result in changes in metabolism. These factors are probably key to the observed increase in abundance for particular metabolites such as tricarboxylic acid intermediates and glutamate.
GC/MS analysis performed during this work was semiquantitative, and as such no molar amounts have been deduced. However, from analysis of total ion current chromatograms, it should be noted that amino acids were low in abundance compared with most other metabolites. Of the amino acids, serine, glycine, alanine, lysine, and glutamate gave the highest signal, followed by isoleucine, valine, tyrosine, and threonine. Leucine and phenylalanine gave very low signals and proline, aspartate, cysteine, and methionine were only just detectable.
The results establish that the sulfur-containing amino acids methionine and cysteine decline in amount following transfer to sulfur-depleted medium. With the exception of glutamate, other amino acids generally increased in amount following the onset of anaerobic conditions. The general trend was for abundance to increase between 0 and 72 h and then decline in the last 48 h of sulfur depletion. Alanine was the exception to this trend, which peaked at 24 h. This result is contrary to that seen via NMR analysis, in which alanine continually increased until the end of the time course. Because NMR analysis was performed on aliquots of whole culture, whereas GC/MS analysis was performed on cells isolated from culture medium, it implies that alanine is released into the medium during the time course. This same effect was noted for succinate, and it appears that the release of these metabolites into the media is a strategy C. reinhardtii employs to cope with anaerobic stress.
Similar to the response observed for most amino acids, sugars and sugar alcohols increased in abundance upon anaerobiosis. Levels typically peaked at 24 or 48 h of anaerobiosis and then declined toward the end of the experiment. Tricarboxylic acid cycle intermediates all decreased from 24 to 120 h of sulfur depletion, but with an exception for succinate, which is believed to show an intracellular decrease and an extracellular increase.
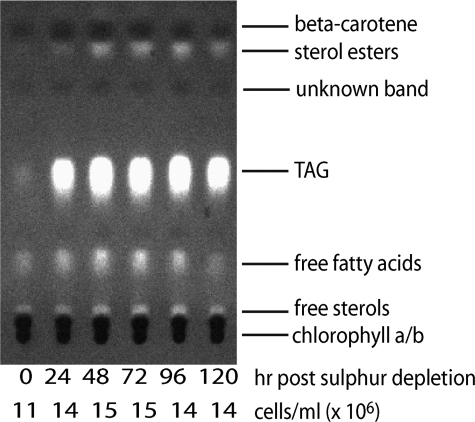
As observed from GC/MS total ion current signal generation, the most abundant sterols in C. reinhardtii were ergosterol, ergost-7-enol, and stigmasta-5,7,22-trienol. Ergosterol and stigmasta-5,7,22-trienol showed little change during the first 72 h of anaerobiosis but declined toward the end of the time course. These two sterols are known to be relatively stable metabolic end products, but the appearance of sterol esters during later stages of the experiment (Fig. 5) suggests that they may have been modified by esterification. The other major sterol in C. reinhardtii, ergost-7-enol, increased in abundance throughout the assay with a peak at 96 h. Ergost-7-enol can theoretically be converted to stigmasta-5,7,22-trienol via a methyltransferase reaction that would require S-adenosyl methionine. Methionine is a sulfur-containing amino acid, and its abundance was reduced during sulfur depletion. The lack of sulfur may cause the increase in ergost-7-enol due to an inability for it to be further processed. Maintenance of membrane selectivity and integrity has been postulated to be a key priority for cells suffering anoxia (19). Loss of these functions can result in leakage of molecules and ions and movement of destructive proteins such as proteases and lipases through membranes and sub-cellular compartments. The modes of action of these sterols toward increased fitness under anaerobic conditions are not understood but are hypothesized to be important in maintenance of membrane integrity.
FIGURE 5.
Analysis of the non-polar fraction of C. reinhardtii by TLC following sulfur depletion.
Amino Acid Metabolism of C. reinhardtii during Sulfur-depleted H2 Production
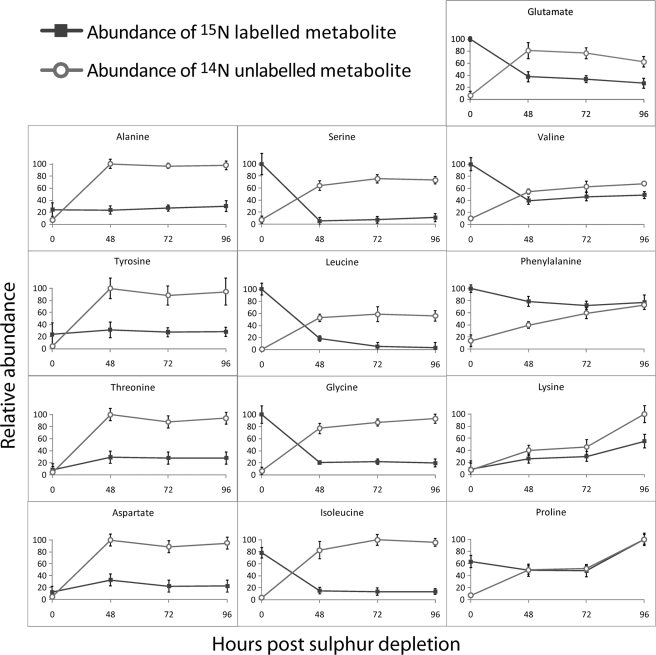
Analysis of the metabolome of C. reinhardtii during sulfur-depleted H2 production revealed that amino acids play a dominant role in adaptive responses. The roles of amino acids are not clear, and their rates of change could be a result of a changing balance between protein degradation and synthesis, or they may play a more direct role in balancing energy needs, nitrogen utilization, and redox states. Further detailed analysis of amino acid metabolism was performed by growing cells in TAP medium with isotopically labeled nitrogen salts (15N) followed by re-suspension in unlabeled (14N) medium upon sulfur depletion. Fig. 4 shows the changes in abundance of 13 labeled and unlabeled amino acids through 96 h of sulfur depletion.
FIGURE 4.
Relative abundances of 15N-labeled and unlabeled amino acids during 96 h of sulfur depletion. Cells were grown aerobically in medium containing 15N salts and resuspended in 14N salts upon sulfur depletion. Standard error bars are shown from four independent biological replicates.
Upon sulfur depletion, the medium was replenished with fresh nutrients. If amino acids were naturally being turned over, an increase in 14N amino acids and a decrease in 15N amino acids would be expected. This pattern was measured for serine, leucine, glycine, glutamate, isoleucine, valine, and phenylalanine. Two amino acids, lysine and proline, showed an increase in both 15N and 14N isotopes. The increase in 15N-labeled amino acids may come from protein breakdown or synthesis from pre-existing amino acids, whereas the increase in 14N amino acids indicates that they were also synthesized de novo. Alanine, tyrosine, threonine, and aspartate showed little change in 15N levels but had elevated levels of the 14N isotope. Thus, these amino acids increased in amount through de novo synthesis. The increase in abundance of these amino acids during anaerobic H2 production compared with their normal levels is suggestive of a particular role under anoxic conditions or H2 production.
Lipid Analysis of C. reinhardtii during Sulfur Depletion
The lipid-soluble fraction of C. reinhardtii was analyzed at 24-h intervals following sulfur depletion by TLC (Fig. 5). Triacylglyceride (TAG) abundance increased substantially between 0 and 24 h of sulfur depletion and remained high throughout the remainder of the time course. Sterol esters showed an increase during the H2 production phase.
The cell pellet from each sample point was hydrolyzed and derivatized to form the methyl-esters for analysis by GC/MS. Quantification of detectable fatty acids is shown in Table 3. Similar to analysis by TLC, an increase in fatty acid concentration was evident. Total abundance of fatty acids peaked at 24 h post re-suspension in sulfur-free medium and declined toward the end of the time course. No dramatic changes could be observed in ratios of saturated, monounsaturated, and polyunsaturated fatty acids. Because most of the fatty acids quantified are apparently derived from TAG (Fig. 5), interestingly the amount of energy accumulated in TAG was greater than the amount accumulated in starch (Fig. 3) during the first 24 h of sulfur depletion; although it should be noted that starch measurements were made from cells grown in a medium optimized for applicability to NMR analysis and not H2 production.
TABLE 3.
Fatty acid composition of C. reinhardtii cells during 120 h of sulfur depletion
Average values (micrograms/ml culture) from three biological replicates are shown. Double bond indicates distance from the terminal methyl group.
| Fatty acid | Period of sulfur depletion (h) |
|||||
|---|---|---|---|---|---|---|
| 0 | 24 | 48 | 72 | 96 | 120 | |
| μg/ml culture | ||||||
| 14:0 | 0.3 | 0.3 | 0.4 | 0.3 | 0.3 | 0.5 |
| 16:0 | 13.6 | 27.6 | 26.5 | 27.1 | 25.1 | 23.7 |
| 16:1ω9c | 2.3 | 6.4 | 6.5 | 7 | 6.4 | 6 |
| 16:1ω7c | 0.7 | 1.9 | 1.9 | 2 | 1.8 | 1.7 |
| 16:2ω6,9cc | 1.1 | 3.6 | 3.5 | 4 | 3.5 | 3.8 |
| 16:3ω6,9,13 | 0.9 | 1.1 | 1 | 1.1 | 1 | 1.2 |
| 16:3ω3,6,9,1ccc | 1.3 | 2.9 | 2.6 | 2.6 | 1.9 | 1.9 |
| 16:4ω3,6,9,13 | 7.8 | 8.3 | 6.7 | 7 | 5.3 | 5.5 |
| 18:0 | 2.4 | 4 | 3.7 | 3.6 | 3.5 | 3.4 |
| 18:1ω9c | 4 | 14.6 | 11.6 | 11.7 | 11.2 | 11 |
| 18:1ω7c | 4.8 | 9.9 | 9.5 | 9.6 | 9.2 | 8.9 |
| 18:2ω6,9cc | 4.7 | 19.4 | 19.6 | 20.6 | 18 | 19.1 |
| 18:3ω3,6,9cc | 13.6 | 16.5 | 13.6 | 13.3 | 10 | 10.1 |
| Total fatty acid (μg/ml) | 57.6 | 116.7 | 106.9 | 110 | 97.2 | 96.8 |
Visualization of Changes to the Metabolic Network upon Induction of H2 Production
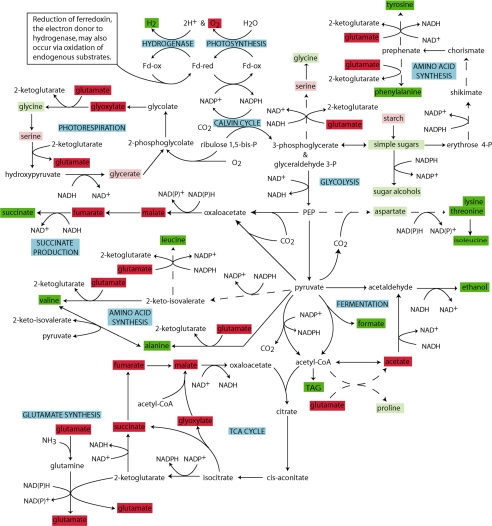
To gain a visual representation of the changes in the metabolome during transition to anaerobic H2 production, trends of abundances of individual metabolites were mapped onto a network manually created from selected reactions present in the Kyoto Encyclopedia of Genes and Genomes and MetaCyc databases, and in the literature (Fig. 6). Data sets generated from both NMR and GC/MS analysis were collated, and abundances of metabolites at 72 h post sulfur depletion, which was approximately halfway through the H2 production phase, were compared with abundances at 0 h. Metabolite abundances are illustrated on the network map in green if their values increased during anaerobic H2 production and in red if they declined. Darker colors are reflective of a stronger response.
FIGURE 6.
Diagrammatic representation of changes in levels of metabolites from C. reinhardtii during transition to anaerobic H2 production. Metabolites that showed an increase in abundance of >2-fold are shown in dark green; an increase of between 1.2- and 2-fold is shown in light green. Metabolites exhibiting a decline in abundance of >2-fold are shown in red, and those that decreased by between 1.2- and 2-fold are shown in pink. Metabolites not colored could not be detected with our measurement systems. When multiple reaction steps have been omitted the reaction pathway is shown with a broken line.
Visual representation aims to show the status of the metabolome during H2 production as compared with its status prior to anaerobic induction. Observations indicate a general down-regulation of the tricarboxylic acid cycle and an increase in abundance of amino acids, sugars, sugar alcohols, and fermentation products. Starch is broken down potentially to supply energy, but there is no indication that TAG is used for this process. Although a small increase in glycine was observed, other photorespiratory intermediates decreased, suggesting that the increase did not result from higher rates of photorespiration but from synthesis via alternate pathways.
The observation of an increase of succinate in the media but not in the cell pellet indicates that cells excrete this metabolite during anaerobic metabolism. This is represented in Fig. 6 by a coloring of succinate in red (decrease) when depicted in the tricarboxylic acid cycle and a coloring of succinate in green (increase) in a separate succinate production pathway.
DISCUSSION
Metabolic Flux during Sulfur-depleted Photobiological H2 Production Involves Three Different Stages
The metabolome of C. reinhardtii during sulfur depletion may be interpreted as undergoing changes in three distinct phases. The first is an aerobic energy conversion stage during the first 24 h; the second an anaerobic H2 production phase from 24 to 72 h; and the third phase a stabilization of pH and decline in H2 production up to 120 h.
During the first 24 h of sulfur depletion, cells accumulate energy stores in the form of starch and TAG. This is probably due to lack of sulfur for synthesis of amino acids and other cell components required for growth, in which circumstances carbon is stored in a form that is readily available when sulfur becomes available again. This is a well characterized adaptive response and is also seen upon nitrogen depletion (20).
The second phase is the anaerobic H2 production phase; during this time cells appear to respond to the lack of O2 by breaking down endogenous substrates (starch and some amino acids) and entering fermentative metabolism. The latter is characterized by a decrease in pH of the medium and increases in amounts of fermentation products (e.g. ethanol and formate). The amounts of metabolites “upstream” of pyruvate (sugars, sugar alcohols, and some amino acids) were high while tricarboxylic acid cycle intermediates were low, reflecting a block in mitochondrial oxidative phosphorylation.
The third and final stage results in a slower rate of starch breakdown, stabilization of pH, and a reduction in rate of H2 production. This third phase may be interpreted as a response to depleted energy supply, although the medium still contained acetate and the cells still contained starch and TAG. It could also reflect toxicity of some products of fermentation. Alternatively, the third phase may reflect the long term consequences of sulfur depletion, which will prevent growth and replenishment of key enzymes or other cell components.
Metabolic Changes during Anaerobic H2 Production
By reducing O2 levels, the ability to oxidize NADH in the mitochondria is diminished, and the conversion of pyruvate to acetyl-CoA by pyruvate dehydrogenase is inhibited. Glycolysis becomes the major source of ATP generation, and for this to continue there is a need to oxidize NADH formed during this process. Fermentation to ethanol and formate can achieve this and is a necessary survival mechanism under such anaerobic conditions (Fig. 3). As pyruvate is inhibited in its conversion to acetyl-CoA, alternate pathways of carbon flow must become operative to ensure glycolysis continues. In addition to fermentative pathways, carbon appears to be directed toward amino acid, succinate, and sugar alcohol synthesis. These reactions allow for the oxidation of NAD(P)H, a potentially important process in balancing redox states under anaerobic conditions.
The conversion of pyruvate to alanine can occur via an aminotransferase reaction, probably also involving the conversion of glutamate to 2-ketoglutarate. 2-Ketoglutarate may be converted back to glutamate during reductive assimilation of NH3. In this manner the conversion of pyruvate to alanine indirectly allows for the oxidation of one molecule of NAD(P)H. In addition to alanine, the amino acids tyrosine, threonine, proline, lysine, and aspartate were formed de novo during sulfur-depleted H2 production and accumulated to higher levels than were present during aerobic growth, as indicated by the elevated 14N-labeled forms following transfer from 15N to 14N salts. Pre-existing reserves of these amino acids did not decline during sulfur depletion, indicating that they were not broken down or incorporated into protein. Although glutamate abundance decreased during anaerobiosis, there was continual flow of ammonium from the medium into various amino acids. Assimilation of ammonium takes place by the glutamine synthetase/glutamate synthase cycle (21), and therefore it must be reasoned that glutamate synthesis is occurring to a degree equal to that of total new amino acid synthesis. The overall decrease in abundance of glutamate may occur as a result of a decrease in the level of its precursor 2-ketoglutarate. We could not measure 2-ketoglutarate but speculate that its amount declined similarly to other tricarboxylic acid cycle intermediates. Amino acids are in a more reduced state than their organic acid counterparts, and their increased production under anaerobic conditions may have one of the following three functions: 1) to serve as a carbon and nitrogen store until aerobic conditions are restored; 2) to remove pyruvate or other glycolytic intermediates; or 3) to provide a means of oxidizing NAD(P)H.
An increase of succinate under dark anaerobic H2-producing conditions was observed by Dubini et al. (22). They suggested fermentative succinate production to be an alternative mechanism of pyruvate metabolism; allowing reoxidation of NAD(P)H and a continuation of glycolysis. This idea is supported in this work, and the observation of an increase of succinate in the media by NMR, which is not a relatively sensitive analytical technique, suggests that an alternate succinate production pathway is important. In particular, the work of Dubini et al. showed that the formation of succinate increased when hydrogenase activity was depleted. The ability to switch between fermentative pathways should offer C. reinhardtii a clear advantage in survival under anaerobic conditions.
Sugar alcohols increased during sulfur depletion, except inositol, which decreased. Sugar alcohols have been implicated in responses to drought and salinity stresses and also may serve as antioxidants (23). Their increased amounts observed during anoxia may form part of a general stress response. Alternatively, the conversion of sugars to their alcoholic reduced forms may function to oxidize NAD(P)H and serve as a stored energy source. It will be important in future work to obtain fully quantitative results from metabolomic studies so as to determine whether such changes can indeed make a significant contribution to these processes.
The reduction of the plastoquinone pool through NAD(P)H derived from oxidation of various endogenous substrates has been suggested (8, 24, 25). If NAD(P)H can reduce the plastoquinone pool, then presumably any reaction that oxidizes these molecules reduces the amount of H2 that can be generated. Knocking out reactions that involve NAD(P)H oxidation have the potential to result in increased H2. Knocking out fermentation steps, sugar alcohol synthesis, and amino acid synthesis appear to be capable of increasing the amount of reductant that may be channeled to the hydrogenase. However, as glycolysis is a key energy-generating pathway during anoxic conditions, cells may suffer from an energy crisis if the ability to oxidize NAD(P)H by hydrogenase is not equal to the rates achievable with fermentative or other steps. In the current work we noted starch consumption to decrease simultaneously with H2 production. This correlation, as well as other studies (26, 27), shows a clear involvement of starch in H2 production. Whether starch degradation leads to reduction of the plastoquinone pool or whether it is important for maintaining anoxic conditions through reduction of O2 generated during photosynthesis is yet to be determined. In either scenario, its importance toward H2 production is fundamental.
Advantages of H2 Production
This work has been discussed with the view that H2 production is a result of balancing ATP and NAD(P)H levels. An alternative reason for H2 production is that it may be a consequence of cytosolic acidification. The imposition of anaerobic conditions stimulates a number of responses, one of which is the acidification of the cytosol (19). Anoxia tolerance has been correlated with an ability to limit drops in cytosolic pH and initiate cytosolic re-alkalinization (28). C. reinhardtii may use the hydrogenase system to combat acidosis. The reduction of excess H+ ions would allow for a return to a neutral cytosolic pH.
Photorespiratory and tricarboxylic acid/glyoxylate cycle metabolites were largely seen to be reduced in amount under anoxic conditions. Photorespiration requires O2 and understandably is down-regulated under anoxia. Production of glycine during H2 production is assumed to come from other synthetic pathways. Lack of O2 causes a buildup of NADH and limits the oxidation of tricarboxylic acid cycle intermediates. This subsequently inhibits the conversion of pyruvate to tricarboxylic acid cycle intermediates. A reduction of flow through the tricarboxylic acid cycle results in decreased production of CO2, and this can have implications for H2 production. Low levels of CO2 would limit flow through the Calvin cycle and this in turn would lead to a buildup of NADPH and reduction of the photosynthetic electron transport chain. With the photosynthetic chain fully reduced and no avenue of escape through NADP+ reduction, hydrogenase can serve as an electron valve. Use of hydrogenase in this manner reduces oxidative damage to the photosynthetic apparatus and allows for ATP generation through creation of a proton gradient that can be utilized by ATP synthase.
The Role of Acetate for H2 Production
The contribution of acetate to H2 production appears to be important. Because H2 production can only be observed in trace amounts without acetate or light (data not shown) it is probable that both sources act together in a concerted fashion. Results here suggest that if acetate does contribute directly toward H2 production, it does so only during the period 24–72 h of sulfur depletion. The increase in acetate after 72 h decreases the likelihood of it being used for H2 production after this point; its increase is likely due to its formation on route to the fermentative products formate and ethanol. Consumption of acetate during the first 24 h may occur through oxidative phosphorylation after entry via the glyoxylate cycle; this may be crucial toward H2 production by reducing O2 levels. It is also possible that cells funnel acetate toward TAG production, because a large increase in TAG was noticed between 0 h and 24 h of sulfur depletion. The abundance of TAG is of interest due to the potential for more energy reserves to be funneled toward H2 production and also the potential of the algal culture to be used for oil production following H2 production.
Optimization of H2 Production
Hydrogenase essentially acts as an H+/e− release valve, offering cells an ability to enhance ATP production via ATP synthase during anaerobic times. Photosystem I activity is the most likely explanation that can accommodate both light dependence and anaerobicity. Photosystem II may be important for basal O2 generation and to ensure that the photosystems and electron carriers remain reduced, but ultimately any O2 generated must be consumed to prevent inhibition of HydA. As consumption of O2 requires endogenous substrate, the maximal amount of H2 that can be produced cannot exceed that permitted by the endogenous substrate stores if the enzymes remain O2-sensitive, and no additional CO2 can enter the system. Therefore, for increased amounts of H2 it will be important to maximize the amount of substrate that can be funneled toward hydrogenase, limit any competition diverting electrons away from hydrogenase, and pursue novel methods of breaking down metabolites to H2.
At the end of 120 h of sulfur depletion there remain substantial energy reserves in the form of starch, organic acids, formate, ethanol, TAG, and amino acids. To increase H2 production it is necessary to break these components down and funnel the reducing equivalents formed during these processes through the photosynthetic electron transport chain and ultimately to hydrogenase. Optimization of H2 production certainly requires a firm understanding of the process of directing reducing equivalents derived from substrate oxidation to hydrogenase. Identification of the NAD(P)H:plastoquinone oxidoreductase is key to this process. In addition, converting substrate to H2 likely requires NAD(P)H produced in the cytosol to be converted to NAD(P)H in the plastid. Because these molecules are too large to cross the plastid membrane one should consider overexpression of enzymes responsible for shuttling reducing equivalents into the plastid.
Blocking pathways that compete with hydrogenase for reductant has the potential to increase H2 production. The multiple fermentative pathways that Chlamydomonas possesses all offer potential sites of attack. Key fermentative pathways that C. reinhardtii employs appear to be directed toward production of ethanol, formate, succinate, alanine, and H2. Knocking out or partially silencing enzymes responsible for catalyzing key steps in the production of these fermentative products may result in higher H2 levels provided the cells do not suffer from an energy crisis once a pathway is disrupted. Finally, expression of foreign genes not present in Chlamydomonas may be utilized to break down some metabolites directly to H2. For example, formate can be broken down to H2 and CO2 in some bacterial species using formate lyase; prolonged survival and overall H2 production may be increased if formate lyase is expressed in C. reinhardtii.
CONCLUSION
This work used metabolomic data to map the operative pathways during anaerobic H2 production. The developed model aims to provide a framework that can assist researchers in the process of identifying key components for metabolic engineering to improve H2 production rates.
Acknowledgment
We are grateful to work performed by personnel employed through Metabolomics Australia at the University of Western Australia.
This work was supported in part by the Australian Research Council (ARC Grants FF0457721 and CE0561495). This work was also supported by the Solar Biofuels Consortium, the Centres of Excellence Program of the Government of Western Australia, the German Federal Ministry of Education and Science (BMBF-ForSys Partner 0315165A), and the Deutsche Bundesstiftung Umwelt (Grant DBU 20006/828).
- GC/MS
- gas chromatography coupled to mass spectrometry
- CPMG
- Carr-Purcell-Meiboom-Gill
- MBTSTFA
- N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide
- MSD
- mass selective detector
- MSTFA
- N-methyl-N-(trimethylsilyl)trifluoroacetamide
- TAG
- triacylglycerides
- Tris
- tris(hydroxymethyl)aminomethane.
REFERENCES
- 1.Boichenko V. A., Hoffmann P. (1994) Photosynthetica 30,527–552 [Google Scholar]
- 2.Timmins M., Thomas-Hall S. R., Darling A., Zhang E., Hankamer B., Marx U. C., Schenk P. M. (2009) J. Exp. Bot. 60,1691–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaffron H., Rubin J. (1942) J. Gen. Physiol. 219–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Happe T., Naber J. D. (1993) Eur. J. Biochem. 214,475–481 [DOI] [PubMed] [Google Scholar]
- 5.Forestier M., King P., Zhang L., Posewitz M., Schwarzer S., Happe T., Ghirardi M. L., Seibert M. (2003) Eur. J. Biochem. 270,2750–2758 [DOI] [PubMed] [Google Scholar]
- 6.Stuart T. S., Gaffron H. (1972) Planta 106,101–112 [DOI] [PubMed] [Google Scholar]
- 7.Melis A. (2007) Planta 226,1075–1086 [DOI] [PubMed] [Google Scholar]
- 8.Gfeller R. P., Gibbs M. (1984) Plant Physiol. 75,212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melis A., Zhang L., Forestier M., Ghirardi M. L., Seibert M. (2000) Plant Physiol. 122,127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollock S. V., Pootakham W., Shibagaki N., Moseley J. L., Grossman A. R. (2005) Photosynth. Res. 86,475–489 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z., Shrager J., Jain M., Chang C. W., Vallon O., Grossman A. R. (2004) Eukaryot. Cell 3,1331–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen A. V., Thomas-Hall S. R., Malnoë A., Timmins M., Mussgnug J. H., Rupprecht J., Kruse O., Hankamer B., Schenk P. M. (2008) Eukaryot. Cell 7,1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman A. R., Croft M., Gladyshev V. N., Merchant S. S., Posewitz M. C., Prochnik S., Spalding M. H. (2007) Curr. Opin. Plant Biol. 10,190–198 [DOI] [PubMed] [Google Scholar]
- 14.Mus F., Dubini A., Seibert M., Posewitz M. C., Grossman A. R. (2007) J. Biol. Chem. 282,25475–25486 [DOI] [PubMed] [Google Scholar]
- 15.Schönfeld C., Wobbe L., Borgstädt R., Kienast A., Nixon P. J., Kruse O. (2004) J. Biol. Chem. 279,50366–50374 [DOI] [PubMed] [Google Scholar]
- 16.Harris E. (1989) The Chlamydomonas Sourcebook, p. 780, Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 17.Poerschmann J., Spijkerman E., Langer U. (2004) Microb. Ecol. 48,78–89 [DOI] [PubMed] [Google Scholar]
- 18.Kosourov S., Seibert M., Ghirardi M. L. (2003) Plant Cell Physiol. 44,146–155 [DOI] [PubMed] [Google Scholar]
- 19.Greenway H., Gibbs J. (2003) Funct. Plant Biol. 30,999–1036 [DOI] [PubMed] [Google Scholar]
- 20.Ball S. G., Dirick L., Decq A., Martiat J. C., Matagne R. F. (1990) Plant Sci. 66,1–9 [Google Scholar]
- 21.Fernandez E., Galvan A. (2008) Eukaryot. Cell 7,555–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubini A., Mus F., Seibert M., Grossman A. R., Posewitz M. C. (2009) J. Biol. Chem. 284,7201–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoop J. M., Williamson J. D., Pharr D. M. (1996) Trends Plant Sci. 1,139–144 [Google Scholar]
- 24.Bernard L., Desplats C., Mus F., Cuiné S., Cournac L., Peltier G. (2006) FEBS J. 273,3625–3637 [DOI] [PubMed] [Google Scholar]
- 25.Hemschemeier A., Fouchard S., Cournac L., Peltier G., Happe T. (2008) Planta 227,397–407 [DOI] [PubMed] [Google Scholar]
- 26.Posewitz M. C., Smolinski S. L., Kanakagiri S., Melis A., Seibert M., Ghirardi M. L. (2004) Plant Cell 16,2151–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruse O., Rupprecht J., Bader K. P., Thomas-Hall S., Schenk P. M., Finazzi G., Hankamer B. (2005) J. Biol. Chem. 280,34170–34177 [DOI] [PubMed] [Google Scholar]
- 28.Menegus F., Cattaruzza L., Mattana M., Beffagna N., Ragg E. (1991) Plant Physiol. 95,760–767 [DOI] [PMC free article] [PubMed] [Google Scholar]