Abstract
Endothelial cells rapidly respond to changes in oxygen homeostasis by regulating gene expression. Regulator of G protein signaling 5 (RGS5) is a negative regulator of G protein-mediated signaling that is strongly expressed in vessels during angiogenesis; however, the role of RGS5 in hypoxia has not been fully understood. Under hypoxic conditions, we found that the expression of RGS5, but not other RGS, was induced in human umbilical vein endothelial cells (HUVEC). RGS5 mRNA was increased when HUVEC were incubated with chemicals that stabilized hypoxia-inducible factor-1α (HIF-1α), whereas hypoxia-stimulated RGS5 promoter activity was absent in HIF-1β−/− cells. Vascular endothelial growth factor (VEGF), which is regulated by HIF-1, did not appear to be involved in hypoxia-induced RGS5 expression; however, VEGF-mediated activation of p38 but not ERK1/2 was increased by RGS5. Overexpression of RGS5 in HUVEC exhibited a reduced growth rate without affecting the cell proliferation. Annexin V assay revealed that RGS5 induced apoptosis with significantly increased activation of caspase-3 and the Bax/Bcl-2 ratio. Small interfering RNA-specific for RGS5, caspase-3 inhibitor, and p38 inhibitor resulted in an attenuation of RGS5-stimulated apoptosis. Matrigel assay proved that RGS5 significantly impaired the angiogenic effect of VEGF and stimulated apoptosis in vivo. We concluded that RGS5 is a novel HIF-1-dependent, hypoxia-induced gene that is involved in the induction of endothelial apoptosis. Moreover, RGS5 antagonizes the angiogenic effect of VEGF by increasing the activation of p38 signaling, suggesting that RGS5 could be an important target for apoptotic therapy.
Hypoxia is a common pathophysiological phenomenon that has a profound impact on endothelial cell properties during many cardiovascular disease processes and tumorigenesis. It has been extensively suggested that cell response to hypoxia is mainly regulated by hypoxia inducible factor-1α (HIF-1α)2, which is rapidly ubiquitinated and degraded in normal conditions but can be stabilized by hypoxia (1, 2). Under hypoxic conditions, HIF-1 activates diverse genes involved in both cell growth and cell death (3). In endothelial cells, hypoxia stimulates the secretion of vascular endothelial growth factor (VEGF) and other angiogenic factors and receptors through transcriptional regulation by HIF-1, which leads to neovascularization and protection against ischemic injury (4–7). Conversely, HIF-1 has been shown to be a factor mediating hypoxia-induced apoptosis, growth of tumors with HIF-1α−/− was not retarded but accelerated because of decreased hypoxia-induced apoptosis (3).
Regulator of G protein signaling (RGS) proteins are responsible for the rapid turnoff of G protein-coupled receptor signaling pathways via the GTPase-stimulating protein activity of their RGS domain (8, 9). More than 25 RGS proteins have been identified, and there are indications that each will specifically regulate a particular G protein-coupled receptor pathway (10, 11). RGS5 belongs to the R4 subfamily of RGS proteins and is enriched in cardiovascular tissues, especially in pericytes and endothelial cells (12–14) but not in cultured vascular smooth muscle cells (15). RGS5 mRNA expression was found markedly decreased in a model of three-dimensional capillary morphogenesis (16). However, it is also reported that RGS5 mRNA is highly expressed in endothelial cells in the tumor vasculature of human renal cell carcinoma (17) and astrocytomas but not in HIF-1α deficient tumors (18). RGS5 is responsible for the abnormal tumor vascular morphology in mice. Loss of RGS5 results in pericyte maturation, vascular normalization, and consequent marked reductions in tumor hypoxia and vessel leakiness (19). These findings argue for an important role of RGS5 in endothelial function.
G protein-coupled receptor (GPCR) signaling pathways are involved in cellular responses to many extracellular stimuli and are major targets for drug discovery. RGS5 is a negative modulator of the angiotensin AT1a receptor. The inhibition of RGS5 expression enhanced angiotensin-stimulated inositol phosphate release (20). In addition, RGS5 attenuated endothelin-1-, sphingosine-1-phosphate-, and platelet-derived growth factor-induced ERK phosphorylation, which indicates that RGS5 exerts control over the platelet-derived growth factor receptor β and GPCR-mediated signaling pathways (14).
We conducted the present study to elucidate the molecular mechanisms controlling the regulation of RGS5 in hypoxic endothelium and the function of RGS5 in endothelial cells. The results presented here demonstrated that hypoxia increased RGS5 expression, which was mediated by HIF-1. Increased RGS5 expression induced apoptosis in human endothelial cells via increase of p38 MAPK activation. We report here, for the first time, that RGS5 is a novel hypoxia-inducible molecule involved in the regulation of endothelial cell behavior, indicating that RGS5 plays an important role in homeostasis in hypoxic endothelial cells.
EXPERIMENTAL PROCEDURES
Cell Culture
Human umbilical vein endothelial cells (HUVEC) were obtained from Clonetics (San Diego, CA) and cultured following the manufacturer's instructions. Human dermal microvascular endothelial cells-1 (HMEC-1; Center of Disease Control) were cultured in MCDB-131 (Invitrogen) containing 10% fetal bovine serum, 10 ng/ml epidermal growth factor, 1 μg/ml hydrocortisone, and 2 mm l-glutamine. C4-B13NBii1 (HIF-1β−/−) and vT2 (HIF-1β+/+) cell lines were obtained from American Type Culture Collection and cultured according to the manufacturer's instructions. HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum.
Hypoxia Incubation
Hypoxic exposure was performed using a molecular incubator chamber (Billups-Rothenberg) flushed with 5% CO2 and 95% N2. The concentration of oxygen (∼1%) was determined before and after incubation by using an oxygen analyzer (Vascular Technology). Other reagents used to mimic hypoxia included dimethyloxalylglycine (DMOG) (BIOMOL), cobalt chloride (CoCl2), and 3,4-dihydroxybenzoate (3,4-DHB) (Sigma).
Retroviral Transduction
Full-length RGS5 cDNA was obtained using reverse transcription-PCR from HUVEC and confirmed to be correct sequences corresponding to GenbankTM (NM_003617). RGS5 cDNA was cloned into pBMN-GFP vectors (Orbigen). In 10-cm dishes, 6 × 106 293T cells were transfected with pBMN-GFP-RGS5, pMD-VSVG, pJK3, and pCMV-tat using Polyethylenimine (Polysciences). Forty-eight h post transfection, virus-containing medium was collected, filtered through a 0.45-μm filter, and used to transduce HUVEC. Overexpression of RGS5 was confirmed by Western blotting.
Small Interfering RNA (siRNA) Transfection
siRNA targeting human RGS5 were synthesized by Genpharma, Inc. (ZhangJiang). Two duplexes of siRNA (siRNA-1: 5′-AGAUGGCUGAGAAGGCAAATT-3′, 5′-UUUGCCUUCUCAGCCAUCUTG-3′ and siRNA-2: 5′-GCGUGAUUCCCUGGACAAATT-3′, 5′-UUUGUCCAGGGAAUCACGCCA-3′) were confirmed to have knockdown ability by Northern and Western blotting. Another duplex of RNA that is not targeted to any human genes was used as a control. HUVEC were transfected with siRNA at a final concentration of 50 nm using Lipofectamine 2000 (Invitrogen).
Northern Blot Analysis
Total RNA was extracted from cells by TRIzol (Invitrogen). Fifteen μg of RNA were loaded per lane, separated on a 1.3% formaldehyde-agarose gel, and transferred to a nylon membrane (Millipore). The membrane was then UV-cross-linked. Northern blot was performed by using a digoxin Northern starter kit (Roche Applied Science) as per the manufacturer's instruction. An RNA probe for human RGS5 containing digoxin-labeled dUTP was used for hybridization.
Western Blot Analysis
Cells were lysed in radioimmune precipitation assay buffer (Boston BioProducts), and proteins were resolved by SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Millipore). After being blocked for 1 h in Tris-buffered saline-Tween 20 with 5% nonfat milk, the polyvinylidene difluoride membrane was then probed with primary antibodies (RGS5 polyclonal antiserum was obtained by immunized rabbits, 1:2,000; RGS2 and RGS4 antibodies were from Santa Cruz Biotechnology, 1:1,000; Bax, Bcl-2, and p53 antibodies were from BD Bioscience, 1:1,000; and total and cleaved caspase-3, total and phosphorylated p38, and ERK antibodies were from Cell Signaling, 1:1,000) overnight at 4 °C and then with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. The blots were detected with SuperSignal chemiluminescent substrate (Pierce Biotechnology).
Promoter Activity
A series of fragments encompassing the 5′-flanking region of the human RGS5 gene (GenBankTM NT_004487) were obtained by PCR from human genomic DNA and cloned into PGL-3 vector (Promega). The cultured cells including HMEC-1, HIF-1β−/−, and HIF-1β+/+ were transfected with the constructs using Lipofectamine 2000. Twenty-four h after transfection, cells were exposed to CoCl2 or hypoxia for 5 h, and luciferase activity was determined using the Dual-Luciferase assay system (Promega).
Matrigel Analysis
Matrigel assays were carried out as described (21). SKMEL/VEGF cells (1 × 107) alone or mixed with 1 × 107 Phoenix cells infected with retrovirus expressing RGS5 were suspended in 0.5 ml of growth factor-reduced matrigel (BD Biosciences) and injected subcutaneously into nu/nu mice. Tissues were harvested, photographed, and fixed with 4% paraformaldehyde for terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) and immunostaining. Three individual experiments were performed, n = 4 per group in each experiment.
Immunostaining Analysis
Frozen sections of the matrigel blocks were washed and incubated with anti-CD31 antibody (BD Biosciences) and counterstained by hematoxylin as described previously (22). For immunofluorescent CD31 and TUNEL double staining, sections were incubated with anti-CD31 antibody for 1 h at room temperature followed by incubation with a Texas Red-conjugated secondary antibody for 1 h. Subsequent TUNEL staining was based on instruction of an apoptosis detection kit (Roche Applied Science). All slides were imaged on a Leica DM IRB fluorescent microscope.
Cell Growth and Migration Analysis
104 cells were plated in each well of 12-well plates. At 5 h after plating (day 1) and at 2, 3, and 4 days after plating, the cells were fixed in 100% ethanol and subsequently stained with 0.1% crystal violet dissolved in 10% ethanol. After staining and thorough washing, the dye was extracted with 10% acetic acid, and absorbance was measured at 590 nm. Caspase inhibitor, cabobenzoxy-valyl-alanyl-aspartyl-(O-methyl)-fluoromethylketone (Z-VAD-FMK) (BIOMOL) was used in the cell growth measurements. To measure cell proliferation, HUVEC transduced with RGS5 or control were seeded in 12-well plates and incubated with 1 μCi/ml 3H[thymidine] at 37 °C for 4 h. Medium was then removed, and the wells were washed three times with phosphate-buffered saline. Radioactivity was extracted with 1 n NaOH and added to a scintillation vial containing 4 ml of ScintiVerse II (Fisher) solution. Thymidine incorporation was measured using a scintillation counter. Wells that contained no cells were labeled and counted to provide background counts. The cell migration rate was measured by wound healing assay. Equal numbers of cells (1 × 105) were plated in 12-well plates. The monolayer of cells were wounded by manual scratching with a pipette tip and then photographed in phase contrast with a Nikon microscope (0 h point) and placed into complete growth medium. Matching wound regions were photographed after 6, 12, and 24 h.
Apoptosis Analysis
The DNA ladder was examined in the infected HUVEC that were cultured in serum-free medium for 24 h. Cells were then removed from tissue culture plates by trypsin and resuspended in a lysis buffer (150 mm NaCl, 10 mm Tris-HCl, pH 8.0, 10 mm EDTA, 0.5% SDS, and 100 ng/ml proteinase K) for 4 h at 50 °C. DNA was extracted using phenol and chloroform followed by ethanol precipitation. The pellet was resuspended in Tris-EDTA buffer (10 mm Tris-HCl and 1 mm EDTA) and treated with RNase for 30 min at 37 °C. Ten micrograms of the DNA were fractionated by electrophoresis on a 2% agarose gel. Using an apoptosis assay kit (Invitrogen), annexin V binding assay was performed in confluent infected HUVEC in serum-free medium under hypoxic or normoxic conditions for 24 h and analyzed by flow cytometry using a FACScan (BD Biosciences). For each treatment, 10,000 cells were counted and then evaluated using Cell Quest software. Cells that were annexin V-positive and propidium iodide-negative were counted for early stages of apoptosis.
Statistical Analysis
Statistical significance was assessed by Student's t test, and p < 0.05 was considered statistically significant.
RESULTS
Hypoxia Increases RGS5 Expression in Endothelial Cells
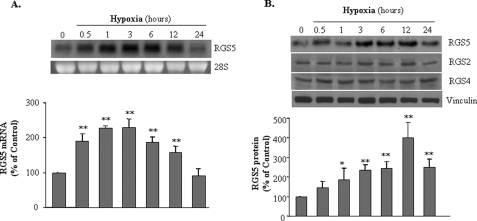
HUVEC were exposed to hypoxic conditions (O2<1%) for periods of 0.5, 1, 3, 6, 12, or 24 h. As compared with cultured HUVEC in normoxia (21% O2), RGS5 mRNA expression was induced by hypoxia immediately from 0.5 h, continuously increased up to 12 h, and peaked at 3 h (2.30 ± 0.25-fold). The increased RGS5 expression returned to baseline at 24 h (Fig. 1A). In addition to the increase of RGS5 mRNA, the protein level was also induced from 3 h to 12 h after hypoxia and declined at 24 h. RGS5 protein expression showed a time-dependent pattern, whereas the other two RGS family members that were also expressed in endothelial cells, RGS2 and RGS4, were not induced by hypoxia (Fig. 1B). These results suggest that hypoxia exclusively up-regulate the expression of RGS5 in endothelial cells.
FIGURE 1.
Induction of RGS5 expression by hypoxia in endothelial cells. A, RGS5 mRNA expression in HUVEC was assessed after various durations of exposure to hypoxia (1% oxygen from 0.5 to 24 h). Northern blot analysis (top) shows that hypoxia induces RGS5 mRNA expression in a time-dependent manner. The quantitative analysis based on three experiments (bottom) indicates that a significant increase of RGS5 mRNA is very rapid at 30 min with continual effects at 1, 3, 6, and 12 h (**, p < 0.01). B, immunoblot analysis demonstrated that RGS5 protein response to hypoxia was initiated at 1 h (*, p < 0.05), remained consistently high until 12 h, and declined at 24 h (**, p < 0.01). However, hypoxia did not change RGS2 and RGS4 at the protein levels. The results were quantified based on three experiments by ImageJ and are presented as mean ± S.D.
HIF-1α Is Involved in the Hypoxia-induced Expression of RGS5
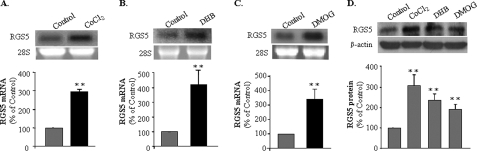
We next focused on which mechanism was involved in the regulation of RGS5 by hypoxia. CoCl2 is recognized as a hypoxia-mimicking compound that stabilizes HIF, a key transcriptional regulator activated only in hypoxic conditions. Incubation of HUVEC in the presence of 150 μm CoCl2 resulted in an induction of RGS5 mRNA (Fig. 2A) with the same time-dependent pattern as the hypoxia induction (data not shown). To further investigate whether the induction of RGS5 is HIF-1-regulated in endothelial cells, HUVEC were incubated in the presence of prolyl hydroxylases inhibitor, ethyl 3,4-DHB or DMOG. As shown in Fig. 2B, exposure of HUVEC to 200 μm DHB for 6 h resulted in a strong increase of RGS5 mRNA expression. Similarly, the cells incubated in the presence of 500 μm DMOG for 6 h, in which the HIF-1α degradation would be blocked for the same duration, showed a significant increase of RGS5 mRNA expression (Fig. 2C). In addition to the increase of RGS5 mRNA, the expression of RGS5 protein was also induced in HUVEC incubated with CoCl2, DHB, and DMOG respectively (Fig. 2D). Together, these results indicate that RGS5 mRNA and protein expression are controlled by changes in oxygen level and that HIF-1α is involved in the regulation of RGS5 in hypoxic endothelial cells.
FIGURE 2.
HIF-1α mediated the induction of RGS5 by hypoxia in endothelial cells. A, RGS5 mRNA expression was significantly increased by 3-fold in the HUVEC that were incubated with 150 μm of CoCl2 (black bar) for 6 h. B and C, the treatment of HUVEC with chemicals that could stabilize HIF-1α, 200 μm of DHB (B) and 500 μm of DMOG (C), results in the significant increase of RGS5 mRNA expression by 4- and 3.5-fold, respectively. D, the protein level of RGS5 was also induced by CoCl2, DHB, and DMOG, respectively. The data were quantified based on three experiments by ImageJ and presented as mean ± S.D. (**, p < 0.01).
HIF-1 Increases the Promoter Activity of RGS5
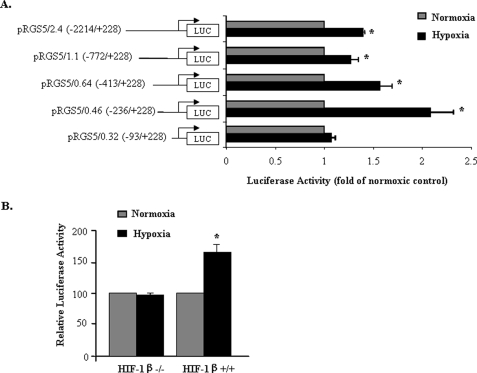
To evaluate whether RGS5 is a target gene for HIF-1α, promoter regions of RGS5 were cloned and constructed. In HMEC-1, the activities of the luciferase construct of RGS5 promoter were measured. As shown in Fig. 3A, exposure of HMEC-1 transfected with RGS5 promoter of various length sequences including 2.4 kb (down to −2214); 1.1 kb (to −772); 0.64 kb (to −413); and 0.46 kb (to −236), but not 0.32 kb (to −93) to CoCl2 (150 μm, 6 h) led to an increase in luciferase activity when compared with HMEC-1 not incubated with CoCl2. In addition, when the construct of RGS5 promoter (2.4 kb) was transfected into the cells lacking HIF-1β, with which HIF-1α forms obligate heterodimers (23), and then exposed to hypoxia, no induction of RGS5 promoter activity was detected, whereas induction was observed (1.6 ± 0.11-fold) in the control cells normally expressing HIF-1β (Fig. 3B). These results imply that the HIF pathway is required for hypoxia-regulated RGS5 expression.
FIGURE 3.
Up-regulation of the RGS5 promoter by hypoxia is HIF-1 dependent. A, transient transfection assays in HMEC-1 using a set of truncated RGS5 promoter luciferase (LUC) constructs in normal condition (gray bar) or in the presence of 150 μm of CoCl2 (black bar) for 6 h. pRL-CMV was cotransfected as an internal control to normalize the transfection efficiency. The hypoxia-activated RGS5 promoter is located at the sequence between RGS5 0.46 and RGS5 0.32. The data are expressed as the mean ± S.D. of the ratio of activity between luciferase and pRL-CMV from three separate experiments. B, a construct encompassing the RGS5 promoter (2.4 kb) was transfected into HIF-1β−/− and HIF-1β+/+ cell lines followed by deprivation of oxygen for 3 h. Luciferase activity was normalized by pRL-CMV in normoxic (gray bar) and hypoxic (black bar) conditions. The significant induction of hypoxia-induced RGS5 promoter activity was observed only in HIF-1β+/+ cells. The results were quantified based on three experiments and are presented as mean ± S.D. (*, p < 0.05).
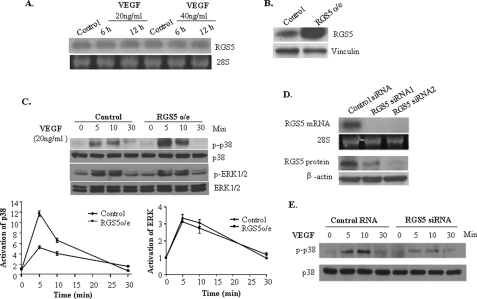
RGS5 Induction Is Associated with VEGF Signaling Pathway Effects
VEGF is an important gene regulated by HIF-1α under hypoxic conditions. VEGF signaling has broad effects on the growth of endothelial cells. To examine the relationship between these two HIF-dependent genes, RGS5 and VEGF, HUVEC were treated with VEGF by different doses and duration. The results showed that VEGF did not regulate RGS5 expression (Fig. 4A). However, in HUVEC with overexpressing RGS5 (RGS5 o/e) (Fig. 4B), VEGF-induced p38 activation was significantly increased, but ERK1/2 activation was not changed (Fig. 4C). A decreased phosphorylation level of p38 was observed in the HUVEC transfected with RGS5 siRNA (Fig. 4, D and E). These results suggest that HIF-1 independently regulates VEGF and RGS5, but RGS5 can affect the VEGF pathway.
FIGURE 4.
RGS5 induction is associated with VEGF signaling pathway effects. A, HUVEC were treated with VEGF (20 ng/ml and 40 ng/ml) for 6 and 12 h. Northern blot analysis indicated that RGS5 mRNA was not regulated by VEGF. B, Western blot showed that RGS5 was successfully expressed at high levels in HUVEC by retroviral transduction. C, RGS5 o/e and control HUVEC were treated with 20 ng/ml of VEGF for 5–30 min. Immunoblotting (top) was performed to detect the phosphorylation and total p38 and ERK. Quantitative analysis of -fold activation of p38 (bottom left panel) and ERK (bottom right panel) showed that VEGF-stimulated phosphorylation of p38 was significantly enhanced by RGS5, whereas the phosphorylation of ERK1/2 did not show a difference. The data are presented as mean ± S.D. based on three independent experiments. D, two siRNA duplexes of RGS5 showed knockdown efficiency over 90% of RGS5 expression by Northern and Western blotting. E, HUVEC transfected with control siRNA or RGS5 siRNA were treated with 20 ng/ml of VEGF for 5–30 min. An impaired level of phosphorylated p38 (p-p38) was detected in RGS5 knockdown cells by Western blotting.
RGS5 Attenuates Endothelial Growth without Suppressing Endothelial Proliferation and Migration
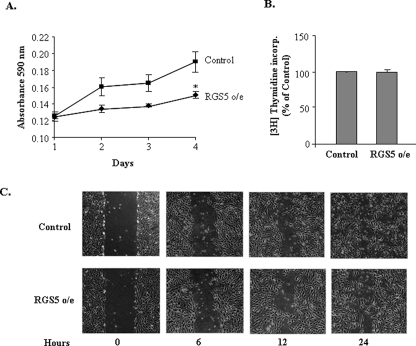
Hypoxia-induced genes could lead to endothelial cells to undergo opposing processes, either angiogenesis or apoptosis; however, little is known regarding the functional properties of RGS5 in endothelial cells under hypoxia. To test the consequence of up-regulation of RGS5 in hypoxia, functional properties of RGS5 in endothelial cells were examined in HUVEC with RGS5 o/e. As shown in Fig. 5A, a growth curve demonstrated that the HUVEC with RGS5 o/e grew significantly slower than the control cells that were infected with empty vectors. However, this growth defect did not present in parallel to the proliferative rates that were measured by thymidine incorporation (Fig. 5B), which indicated that the ability of RGS5 might not be correlated to hypoxia-induced angiogenesis. Wound healing scratch assays were performed to determine whether stable RGS5 expression would also affect the endothelial migration (Fig. 5C). Both the control and the RGS5 o/e cells completed the wound healing within 24 h. No significant difference was observed during the process.
FIGURE 5.
RGS5 attenuates endothelial growth without suppressing endothelial proliferation. A, growth curves were performed by using HUVEC that overexpress RGS5 or control with blank vector. The absorbance at 590 nm reflects cell number and demonstrates that the HUVEC with RGS5 o/e significantly attenuates endothelial cell growth. *, p < 0.05. B, 3H[thymidine] incorporation (incorp.) showed no significant difference in endothelial cell proliferation in HUVEC with or without RGS5 overexpression. The data are expressed as percentages of RGS5 o/e cells versus control cells from three experiments and are presented as the mean ± S.D. C, phase contrast images were taken at different time points to assess cell migration in HUVEC culture plates that initially had an equal number of cells in control and RGS5 o/e. Measuring the distance of the gaps that were created by scratching with pipette tips showed that RGS5 overexpression did not influence cell migration.
RGS5 Enhances Apoptosis Associated with Increased Phosphorylation of p38 MAPK
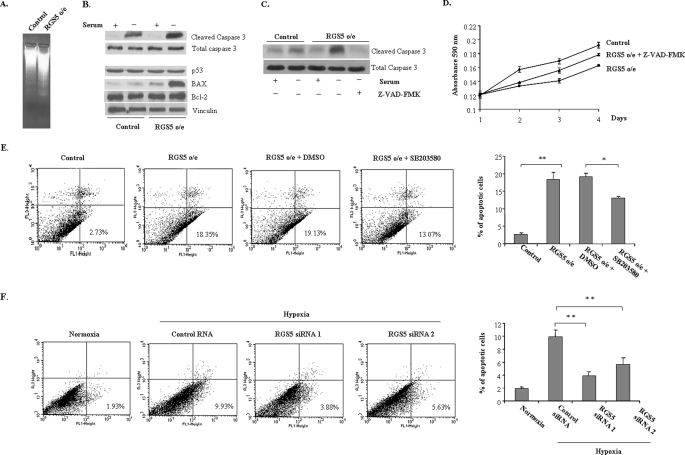
Because RGS5 attenuated the growth rate of endothelial cells but did not impact its proliferation, it has been considered that RGS5 may affect the apoptosis of endothelial cells. To test this possibility, apoptosis was induced by withdrawing serum and growth factors from HUVEC culture medium for 24 h. In Fig. 6A, a DNA laddering analysis illustrated a significant increase of apoptosis in HUVEC with RGS5 o/e compared with controls. In addition, an increase of activated caspase-3 was observed in HUVEC with RGS5 o/e (Fig. 6B), which could be blocked by application of Z-VAD-FMK (Fig. 6C), and consequently resulted in preventing the RGS5-attenuated endothelial cell growth (Fig. 6D). Moreover, in HUVEC with RGS5 o/e, pro-apoptotic protein Bax was significantly increased by nutrient deprivation whereas there was no difference in Bcl-2 expression between RGS5 o/e and control cells, indicating that the apoptosis occurred because of the change in the Bcl-2/Bax ratio (24) (Fig. 6B). In addition, p53 was not regulated in RGS5 o/e endothelial cells (Fig. 6B) although it was correlated with the induction of apoptosis in hypoxic endothelial cells (25). Other than changes of apoptotic factors, RGS5 o/e endothelial cells undergoing apoptosis was also confirmed by FACS analysis. Fig. 6E demonstrated an increase in the numbers of annexin V-positive and propidium iodide-negative cells in HUVEC with RGS5 o/e in comparison to controls after exposure to serum free medium for 24 h (18.35% versus 2.73%), whereas application of p38 inhibitor SB203580 (20 μm) to RGS5 o/e HUVEC resulted in a significant decrease in the number of apoptotic cells. Furthermore, with silencing of the RGS5 gene, HUVEC were rescued to a certain extent from hypoxia-induced apoptosis (Fig. 6F). These findings suggest that RGS5 was involved in the enhancement of apoptosis in association with p38 MAPK activation in endothelial cells.
FIGURE 6.
RGS5 enhances apoptosis of endothelial cells. A, DNA was extracted and fractionated by electrophoresis after HUVEC were cultured in serum-free medium for 24 h to induce apoptosis. A significant increase of DNA laddering was present in RGS5 o/e cells compared with control cells. B, immunoblot of cleaved caspase-3, total caspase-3, p53, Bax, and Bcl-2 were performed in either RGS5 o/e or control HUVEC after being incubated in serum-free medium for 24 h. The RGS5 o/e cells showed an increase of activation of caspase-3 and ratio of Bax versus Bcl-2, whereas no change was observed in p53 expression. C, a caspase inhibitor, Z-VAD-FMK (20 μm), significantly blocked the cleavage of caspase-3. D, application of Z-VAD-FMK prevented the effect of RGS5 on the growth of HUVEC. E, annexin V binding assay was performed in the HUVEC with or without RGS5 o/e at 24 h after exposure to serum-free medium. The x axis indicates the density of annexin V staining, and the y axis indicates the density of propidium iodide staining. The numbers in the lower right corner indicate the percentage of apoptotic cells. Values represent the average of three experiments. Inhibitor of p38, SB203580 (20 μm), was used to block the activation of p38 in RGS5 o/e HUVEC. The number of apoptotic cells was decreased in the presence of SB203580. F, knockdown RGS5 by two different sequences of siRNA duplexes decreased the number of apoptotic cells after exposed to hypoxia for 24 h. DMSO, dimethyl sulfoxide. *, p < 0.05; **, p < 0.01.
RGS5 Stimulates Apoptosis and Impairs VEGF-induced Angiogenesis
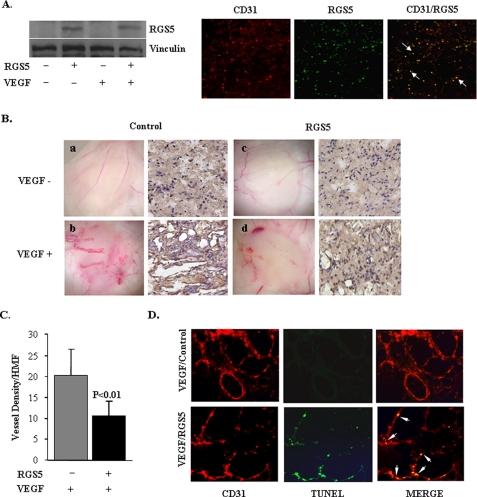
To further elucidate the function of RGS5 in the in vivo setting, matrigel assay was performed in nude mice. On day 3 after implantation of matrigel plugs that were incorporated with SK-MEL-2 melanoma cells expressing VEGF-A165 (SK-MEL/VEGF cells) and/or Phoenix cells packaging retroviruses expressing RGS5, expression of RGS5 in the vasculature of matrigel was confirmed by Western blotting and immunostaining (Fig. 7A). The angiogenic responses were evaluated by histology and immunohistochemistry for the endothelial cell marker CD31 (Fig. 7B). Strong angiogenesis was induced in plugs containing SK-MEL/VEGF cells (Fig. 7B, b). However, the matrigel plug that contained RGS5 significantly impaired the VEGF-induced angiogenesis (Fig. 7B, d) evidenced by the decreased vessel density accounted by CD31 positive cells (Fig. 7C). TUNEL and CD31 labeling were performed to evaluate endothelial cell apoptosis. As Fig. 7D illustrates, the concurrent TUNEL/CD31-positive cells were only observed in the matrigel plug that contained expressing RGS5.
FIGURE 7.
RGS5 impairs VEGF-induced angiogenesis in the matrigel assay in vivo. A, Western blotting (left) and immunostaining (right) for CD31 (red) and RGS5 (green) showed that RGS5 was expressed in the vasculature of matrigel. Arrows indicate the co-localization of CD31 and RGS5. B, shown is the macroscopic and immunohistochemical staining of implantation of matrigels that were mixed with Phoenix cells that packaged empty virus (a and b) or RGS5 virus (c and d). SKMEL/VEGF cells that secreted VEGF were added in matrigel plugs (b and d). VEGF-induced blood vessel growth was decreased when RGS5 was overexpressed. C, shown are the quantitative measurements of vessel density in matrigels from Bb and Bd as angiogenic responses. RGS5 significantly attenuated VEGF-induced angiogenesis. The results were quantified based on three individual experiments and are presented as mean ± S.D. (**, p < 0.01). D, immunofluorescent double staining for TUNEL (green) and CD31 (red). Arrows indicate the apoptotic endothelial cells with co-localization of CD31 and TUNEL (yellow) in the matrigel section with RGS5 overexpression.
DISCUSSION
Hypoxia and ischemia trigger a multitude of responses designed to compensate for reduced oxygen availability. In endothelial cells, these responses include increased expression of growth factors and their receptors to induce angiogenesis (5, 7). On the other hand, hypoxia-regulated genes can lead cells to apoptosis to maintain the homeostasis (3). Thus, a thorough understanding of the mechanisms that hypoxia-related genes use to regulate endothelial cell behavior may aid in the comprehension of vascular homeostasis.
In this report, we demonstrated that regulator of G protein signaling 5 is a novel hypoxia-induced gene in endothelial cells. Expression of RGS5 can be significantly induced by hypoxia at both mRNA and protein levels. Hypoxia induces expression of RGS5 but not RGS2 and RGS4, although these three RGS members have been reported to express in endothelial cells (12, 26). This suggests that hypoxia-induced endothelial RGS5 is specific within the RGS protein family.
We also demonstrated that hypoxia-induced RGS5 expression is mediated by the transcription factor HIF-1. The evidence in this report shows that the expression of RGS5 mRNA was up-regulated when the endothelial cells were incubated with CoCl2, a chemical that mimics hypoxia. The same induction was observed by treating cells with DMOG or DHB, which can stabilize HIF-1α by inhibiting prolyl hydroxylases. These data were supported by previous reports showing that RGS5 mRNA was highly expressed in endothelial cells in tumor vasculature (17) but not in HIF-1α-deficient tumors (18). In addition, RGS promoter activity increased in the cells that expressed HIF-1β but not in the cells in which HIF-1β was absent, indicating that HIF-1 is required for the induction of RGS5. However, by searching the transcriptional factor-binding elements, there is no typical HIF-1 binding site (hypoxia responsive element 5′-RCGTG-3′) found in the 5′-flanking region of RGS5. Despite that a hypoxia responsive element sequence was identified in the first intron of RGS5, the RGS5 promoter activity was not enhanced by the first intron of RGS5 (data not shown). Our data do not exclude the possibility that HIF-1 regulates RGS5 indirectly through other genes. Further investigation is needed to detail the mechanism.
RGS5 has been reported to respond to a variety of angiogenic signals in vascular cells. RGS5 expression is up-regulated in ovarian angiogenesis and in the granulation tissue of cutaneous wounds. RGS5 is also induced in the vasculature of premalignant lesions during the “angiogenic switch,” and its expression is further elevated in tumor vessels (13, 17, 18). It has been reported that RGS5 mRNA was markedly decreased in a three-dimensional capillary morphogenesis model (16), indicating that RGS5 is not a proangiogenic factor. Very few reports have explored the function of RGS5 in endothelial cells during hypoxia/ischemia. In our study, hypoxia-induced RGS5 can lead endothelial cells to undergo apoptosis. Our results showed that the increased RGS5 contributed to the attenuation of endothelial cell growth rate and stimulation of apoptotic genes, including cleaved caspase-3 and the ratio of Bax to Bcl-2, which presents complementary evidence to previous reports that G protein-coupled receptor (GPCR) signaling pathways are involved in cellular response to hypoxia via HIF-1α (27, 28). GPCR activation initiates rapid apoptosis on both receptor-mediated signaling and receptor phosphorylation (29). In cardiac myocytes, sustained or excessive activation of either Gq- or Gs-signaling pathways resulted in apoptotic loss of cardiomyocytes both in vitro and in vivo (30). However, as a negative regulator of G protein, RGS family proteins such as RGS3 were also involved in stimulating apoptosis (31). In Hela cells, steroid receptor-binding protein-RGS induced apoptosis in the presence or absence of the caspase inhibitor Z-VAD-FMK (32). Thus, an HIF-dependent increased level of RGS5 may regulate the GPCR signaling pathway in hypoxia-related apoptosis.
Hypoxia stabilizes HIF-1α to induce HIF-1 target genes such as VEGF (33) and GLUT-1 (glucose transporter-1) (34). The up-regulation of these molecules in endothelial cells can induce angiogenesis to restore blood-supplied nutrients and energy. However, activation of caspase-3 and Apaf-1-mediated caspase-9 have also been reported in several cell types under hypoxic conditions (5, 35), suggesting that hypoxia induces apoptosis as well, in which HIF-1 also plays an important role (3). The knowledge, however, of the mutual adjustment between these two hypoxia-related opposite pathways in endothelial cells is very limited. In the present study, the expression of RGS5 was not affected by VEGF stimulation, but RGS5 magnified VEGF-mediated activation of p38 MAPK. VEGF-induced vessel sprouting into the matrigel plug in mice was also impaired by introducing high expression of RGS5. In various cell types, it was well established that ERK is activated by several growth factors of consequence for cell proliferation and differentiation. In contrast, it is generally considered that c-Jun NH2-terminal kinase (JNK) and p38 are activated by environmental stress, bacterial lipopolysaccharide, and inflammatory cytokines (36). Inhibition of p38 enhanced FGF-2-induced tubular morphogenesis and VEGF-induced angiogenesis in vitro and in vivo by decreasing apoptosis (37, 38), suggesting that p38 signaling is important for cells undergoing apoptosis. Although p38 was suggested to be involved in cell migration (39), our data did not show that it affected the HUVEC migration in stable RGS5 overexpression cells. Therefore, the enhanced activation of p38 by RGS5 in endothelial cells may very well contribute to the apoptotic effect and play a negative regulatory role in VEGF-induced angiogenesis under hypoxic conditions.
It has been reported that pertussis toxin-sensitive G proteins are involved in Flt-1 (VEGFR1)-mediated down-regulation of HUVEC proliferation (40), and KDR (VEGFR2) and Flt-1 employ heterotrimeric G proteins G11α and Gi/o in their signaling pathways (41). In addition, VEGFR2-mediated p38 activation resulted in apoptosis (42) and inhibition of p38 activation is critical to VEGFR2-mediated endothelial cell survival (43). Thus, RGS5-stimulated VEGF-induced activation of p38 may be explained by G protein interaction with the VEGFR pathway. However, whether RGS5 directly regulates VEGFR activation or whether it occurs through other G protein-coupled receptors is to be determined in a future study.
The present study identifies RGS5 as a hypoxia-inducible regulator that promotes apoptosis in endothelial cells. Increased expression of RGS5 by HIF-1 unbalances VEGF-mediated MAPK pathways; thus, the cells are more apt to undergo apoptosis when suffering the stress of nutrient or oxygen deprivation. This study revealed a cross-talk between the HIF-1-induced angiogenesis and apoptosis pathways. Elucidating how G protein signaling controls different cellular responses in the VEGF signaling pathway will lead to an understanding of the precise mechanism of angiogenesis and apoptosis and, ultimately, the development of new therapeutic strategies.
Acknowledgments
We thank Drs. Gregory Robinson and Huiyan Zeng (Beth Israel Deaconess Medical Center/Harvard Medical School) for the gift of SKMEL/VEGF cells. We also very much appreciate Dr. Jincai Luo (Institute of Molecular Medicine, Peking University) for technical training and assistance.
This work was supported in part by National Institutes of Health Grant HLR01082837 (to J. L.). This work was also supported by American Heart Association Grant-in-aid 0265494T (to J. L.) and the China Scholarship Council (to J. Y. and X. A.).
- HIF-1α
- hypoxia-inducible factor-1α
- siRNA
- small interfering RNA
- VEGF
- vascular endothelial growth factor
- GPCR
- G protein-coupled receptor
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- HUVEC
- human umbilical vein endothelial cells
- DHB
- dihydroxybenzoate
- RGS5 o/e
- overexpressing RGS5
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun NH2-terminal kinase
- MAPK
- mitogen-activated protein kinase
- GFP
- green fluorescent protein
- Z-VAD-FMK
- cabobenzoxy-valyl-alanyl-aspartyl-(O-methyl)-fluoromethylketone.
REFERENCES
- 1.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292,464–468 [DOI] [PubMed] [Google Scholar]
- 2.Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Cell 107,43–54 [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P., Dor Y., Herbert J. M., Fukumura D., Brusselmans K., Dewerchin M., Neeman M., Bono F., Abramovitch R., Maxwell P., Koch C. J., Ratcliffe P., Moons L., Jain R. K., Collen D., Keshert E. (1998) Nature 394,485–490 [DOI] [PubMed] [Google Scholar]
- 4.Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. (1998) Genes Dev. 12,149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunelle J. K., Chandel N. S. (2002) Apoptosis 7,475–482 [DOI] [PubMed] [Google Scholar]
- 6.Liu Y., Cox S. R., Morita T., Kourembanas S. (1995) Circ. Res. 77,638–643 [DOI] [PubMed] [Google Scholar]
- 7.Mandriota S. J., Pyke C., Di Sanza C., Quinodoz P., Pittet B., Pepper M. S. (2000) Am. J. Pathol. 156,2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hepler J. R., Berman D. M., Gilman A. G., Kozasa T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94,428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman D. M., Kozasa T., Gilman A. G. (1996) J. Biol. Chem. 271,27209–27212 [DOI] [PubMed] [Google Scholar]
- 10.De Vries L., Gist Farquhar M. (1999) Trends Cell Biol. 9,138–144 [DOI] [PubMed] [Google Scholar]
- 11.Wieland T., Mittmann C. (2003) Pharmacol. Ther. 97,95–115 [DOI] [PubMed] [Google Scholar]
- 12.Kirsch T., Wellner M., Luft F. C., Haller H., Lippoldt A. (2001) Brain Res. 910,106–115 [DOI] [PubMed] [Google Scholar]
- 13.Adams L. D., Geary R. L., McManus B., Schwartz S. M. (2000) Circ. Res. 87,623–631 [DOI] [PubMed] [Google Scholar]
- 14.Cho H., Kozasa T., Bondjers C., Betsholtz C., Kehrl J. H. (2003) FASEB J. 17,440–442 [DOI] [PubMed] [Google Scholar]
- 15.Grant S. L., Lassègue B., Griendling K. K., Ushio-Fukai M., Lyons P. R., Alexander R. W. (2000) Mol. Pharmacol. 57,460–467 [DOI] [PubMed] [Google Scholar]
- 16.Bell S. E., Mavila A., Salazar R., Bayless K. J., Kanagala S., Maxwell S. A., Davis G. E. (2001) J. Cell Sci. 114,2755–2773 [DOI] [PubMed] [Google Scholar]
- 17.Furuya M., Nishiyama M., Kimura S., Suyama T., Naya Y., Ito H., Nikaido T., Ishikura H. (2004) J. Pathol. 203,551–558 [DOI] [PubMed] [Google Scholar]
- 18.Berger M., Bergers G., Arnold B., Hämmerling G. J., Ganss R. (2005) Blood 105,1094–1101 [DOI] [PubMed] [Google Scholar]
- 19.Hamzah J., Jugold M., Kiessling F., Rigby P., Manzur M., Marti H. H., Rabie T., Kaden S., Gröne H. J., Hämmerling G. J., Arnold B., Ganss R. (2008) Nature 453,410–414 [DOI] [PubMed] [Google Scholar]
- 20.Wang Q., Liu M., Mullah B., Siderovski D. P., Neubig R. R. (2002) J. Biol. Chem. 277,24949–24958 [DOI] [PubMed] [Google Scholar]
- 21.Zeng H., Qin L., Zhao D., Tan X., Manseau E. J., Van Hoang M., Senger D. R., Brown L. F., Nagy J. A., Dvorak H. F. (2006) J. Exp. Med. 203,719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettersson A., Nagy J. A., Brown L. F., Sundberg C., Morgan E., Jungles S., Carter R., Krieger J. E., Manseau E. J., Harvey V. S., Eckelhoefer I. A., Feng D., Dvorak A. M., Mulligan R. C., Dvorak H. F. (2000) Lab. Invest. 80,99–115 [DOI] [PubMed] [Google Scholar]
- 23.Wood S. M., Gleadle J. M., Pugh C. W., Hankinson O., Ratcliffe P. J. (1996) J. Biol. Chem. 271,15117–15123 [DOI] [PubMed] [Google Scholar]
- 24.Yang E., Korsmeyer S. J. (1996) Blood 88,386–401 [PubMed] [Google Scholar]
- 25.Stempien-Otero A., Karsan A., Cornejo C. J., Xiang H., Eunson T., Morrison R. S., Kay M., Winn R., Harlan J. (1999) J. Biol. Chem. 274,8039–8045 [DOI] [PubMed] [Google Scholar]
- 26.Cho H., Harrison K., Schwartz O., Kehrl J. H. (2003) Biochem. J. 371,973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sodhi A., Montaner S., Patel V., Zohar M., Bais C., Mesri E. A., Gutkind J. S. (2000) Cancer Res. 60,4873–4880 [PubMed] [Google Scholar]
- 28.Spinella F., Rosanò L., Di Castro V., Natali P. G., Bagnato A. (2002) J. Biol. Chem. 277,27850–27855 [DOI] [PubMed] [Google Scholar]
- 29.Revankar C. M., Vines C. M., Cimino D. F., Prossnitz E. R. (2004) J. Biol. Chem. 279,24578–24584 [DOI] [PubMed] [Google Scholar]
- 30.Adams J. W., Brown J. H. (2001) Oncogene 20,1626–1634 [DOI] [PubMed] [Google Scholar]
- 31.Dulin N. O., Pratt P., Tiruppathi C., Niu J., Voyno-Yasenetskaya T., Dunn M. J. (2000) J. Biol. Chem. 275,21317–21323 [DOI] [PubMed] [Google Scholar]
- 32.Bansal G., Druey K. M., Xie Z. (2007) Pharmacol. Ther. 116,473–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shweiki D., Itin A., Soffer D., Keshet E. (1992) Nature 359,843–845 [DOI] [PubMed] [Google Scholar]
- 34.Ebert B. L., Firth J. D., Ratcliffe P. J. (1995) J. Biol. Chem. 270,29083–29089 [DOI] [PubMed] [Google Scholar]
- 35.McClintock D. S., Santore M. T., Lee V. Y., Brunelle J., Budinger G. R., Zong W. X., Thompson C. B., Hay N., Chandel N. S. (2002) Mol. Cell. Biol. 22,94–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyriakis J. M., Avruch J. (1996) BioEssays 18,567–577 [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto T., Turesson I., Book M., Gerwins P., Claesson-Welsh L. (2002) J. Cell Biol. 156,149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Issbrücker K., Marti H. H., Hippenstiel S., Springmann G., Voswinckel R., Gaumann A., Breier G., Drexler H. C., Suttorp N., Clauss M. (2003) FASEB J. 17,262–264 [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto T., Yokote K., Tamura K., Takemoto M., Ueno H., Saito Y., Mori S. (1999) J. Biol. Chem. 274,13954–13960 [DOI] [PubMed] [Google Scholar]
- 40.Zeng H., Zhao D., Mukhopadhyay D. (2002) J. Biol. Chem. 277,4003–4009 Epub 2001 Nov 4028 [DOI] [PubMed] [Google Scholar]
- 41.Zeng H., Zhao D., Yang S., Datta K., Mukhopadhyay D. (2003) J. Biol. Chem. 278,20738–20745 [DOI] [PubMed] [Google Scholar]
- 42.Ferrari G., Pintucci G., Seghezzi G., Hyman K., Galloway A. C., Mignatti P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103,17260–17265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yilmaz A., Kliche S., Mayr-Beyrle U., Fellbrich G., Waltenberger J. (2003) Biochem. Biophys. Res. Commun. 306,730–736 [DOI] [PubMed] [Google Scholar]