Abstract
Transcription of a proto-oncogene c-fos is induced rapidly to high levels by various extracellular stimuli. To explore the molecular mechanism of c-fos gene induction, we established a defined in vitro transcription system for the c-fos promoter that consists of purified activators (SRF, Elk-1, cAMP-responsive element-binding protein, and ATF1), general transcription factors, and RNA polymerase II. In this reconstituted transcription system, activation of c-fos transcription was highly dependent upon coactivators such as PC4 and Mediator, indicating a very weak activation potential of the activators in the context of an unaltered promoter structure. This heightened coactivator dependence, however, allowed us to identify from HeLa nuclear extract a coactivator-like activity termed transcriptional regulator of c-fos (TREF) that enhanced c-fos transcription but not GAL4-VP16-dependent transcription. TREF cooperated with Mediator to enhance c-fos transcription by ∼60-fold over its basal level and, like Mediator, stimulated activator-independent (basal) transcription as well. Further purification of TREF revealed that it consists of at least three distinct components, one of which was purified to near homogeneity and identified as heterogeneous nuclear ribonucleoprotein R. Recombinant heterogeneous nuclear ribonucleoprotein R enhanced transcription from the c-fos promoter and displayed cooperativity with PC4 and Mediator, thus demonstrating its direct transcriptional activity.
The c-fos promoter is one of the very well studied gene promoters and responds in a rapid and transient manner to a myriad of extracellular signals such as growth factors, cytokines, and cellular stress (1). These signals are transmitted via cascades of kinases including MAPKs2 to the nucleus, wherein the transcription factors bound on the inducible cis-elements are activated upon phosphorylation (2). Among the important cis-elements within the c-fos promoter are the serum response element (SRE), upon which a dimer of SRF and one molecule of Elk-1 form a ternary complex (3), and the cAMP-responsive elements (CREs), which are bound by a heterodimer of CREB and ATF1 (4). Although the signal transduction and the subsequent regulation of the activators are well characterized, much less is known about the mechanism by which the activators elicit dramatic c-fos induction.
Transcription of a typical protein-coding gene such as c-fos requires general transcription factors (GTFs), including TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, which form the preinitiation complex with RNA polymerase II (RNAP II) on the promoter (5). Preinitiation complex formation and the following steps of transcription are the ultimate targets of activators, whose activating signals are transduced to GTFs and RNAP II in a process that is poorly understood. This process, nonetheless, has been shown to require another class of cofactors termed coactivators or coregulators that serve as a physical and functional bridge between activators and GTFs (6, 7). In the case of the c-fos gene, well characterized p300/CBP serves as a coactivator for SRF (8), Elk-1 (9), and CREB (10, 11), and coactivates transcription not only as a bridging factor but also as a histone acetyltransferase (12). More recent studies indicate that another type of coactivator termed Mediator is also involved in c-fos transcription. The Med23 subunit of Mediator, first identified as an E1A-interacting protein (13), has been demonstrated to interact with Elk-1 and plays a role for SRE-mediated transcriptional activation (14). Indeed, Med23 (−/−) knock-out cells show reduced transcription of SRE-containing genes such as egr-1 and c-fos (14, 15).
Mediator was initially discovered by the genetic and biochemical analyses in yeast (16–18). Subsequent biochemical dissection of mammalian nuclear extracts by several independent groups lead to the purification of mammalian Mediator complexes (6), variously named as NAT (negative regulator of activated transcription), the mouse Mediator complex, TRAP (thyroid hormone receptor-associated proteins), SMCC (SRB-MED-containing cofactor), DRIP (vitamin D receptor-interacting proteins), ARC (activator-recruited cofactor), CRSP (cofactor required for Sp1 activation), and PC2 (positive cofactor 2). The identified complexes turned out to be highly related to each other and probably constitute the identical Mediator or its various subcomplexes (6).
Given the diversity of the employed biochemical systems, it is somewhat surprising that only the requirement of Mediator was eventually revealed. The apparent unity, however, could be due to the use of artificially constructed activators and templates that may alter or even obviate the requirement of some cofactors. Indeed, for producing sufficiently strong signals of transcript, in vitro transcription systems typically utilize a strong activation domain fused to a DNA-binding domain (often the DNA-binding domain of yeast GAL4) and an artificially constructed model promoter with tandem repeats of activator-binding sites. Because of the intrinsically strong activation potential of the activator and the optimized geometry of activator-binding sites, it is possible that these systems obscure the requirement for some classes of cofactors.
To identify a novel cofactor requirement in an unbiased manner, we chose the c-fos promoter as a model and initiated to recapitulate transcription from the naturally configured promoter with only purified components. We employed a defined in vitro transcription system using purified GTFs (19), together with SRF, Elk-1, CREB, and ATF1 to analyze transcriptional activation from the c-fos promoter that retains cis-elements in its original configuration. Unlike the GAL4-VP16-based model promoter, activation of the c-fos promoter by SRF, Elk-1, CREB, and ATF1 was much more dependent on coactivators such as PC4 and Mediator. This heightened coactivator dependence allowed us to identify from HeLa nuclear extract a novel coactivator-like activity termed transcriptional regulator for c-fos (TREF), which augments activated transcription from the c-fos promoter but not the GAL4-VP16-dependent transcription. Upon further purification, TREF was separated into three activities, one of which was identified as hnRNP R. Recombinant hnRNP R stimulated c-fos transcription and cooperated with PC4 and Mediator, evoking a potent activation (∼100-fold) of the c-fos promoter over its basal level. Because hnRNP R is a presumptive RNA-binding protein and is possibly involved in regulating mRNA stability, our results suggest a potential link between transcription and the subsequent post-transcriptional processes for rapid and transient c-fos induction.
EXPERIMENTAL PROCEDURES
Construction of Baculoviruses for Expressing FLAG-tagged Proteins
cDNAs of human SRF, Elk-1, CREB, ATF1, and hnRNP R were isolated from HeLa first strand cDNA by a PCR-based method. Because the 5′ region of SRF cDNA could not be amplified by PCR because of a high GC content, ∼300 bp of the 5′ region was constructed by annealing synthetic oligonucleotides based on the published SRF DNA sequence. PCR was used to add NdeI and BamHI sites to the N- and C-terminal ends of each cDNA, and when necessary, internal NdeI and BamHI sites were mutated without altering the amino acid sequences. Each cDNA was sequenced in its entirety and then subcloned into a modified baculovirus transfer vector pTOF, which was constructed from its parental plasmid, pVL1392, by mutating the unique NdeI site within the plasmid and adding FLAG-encoding DNA sequence as well as NdeI and BamHI sites. Recombinant baculoviruses were prepared by transfection of Sf9 cells with each transfer vector (1.0 μg), linearized baculoviral DNA (0.25 μg), and cellfectin (Invitrogen), and the obtained viruses were plaque-purified and amplified.
Preparation of High Five Cell Extract
High Five cells were infected with each amplified baculovirus and cultured at 27 °C for 50 h. The collected cells were washed once with phosphate-buffered saline; resuspended in one packed cell volume of hypotonic buffer (10 mm Tris-HCl, pH 7.3, at 4 °C, 1.5 mm MgCl2, 10 mm KCl) containing 0.5 mm PMSF, 1 mm dithiothreitol, 100 nm MG132, 10 mm β-glycerophosphate, and protease inhibitor mixture (Sigma); and then homogenized mildly in a Dounce homogenizer. After centrifugation, the supernatant was saved as the cytoplasmic (S100) fraction, and the pellet (crude nuclei) was suspended in one packed cell volume of low salt buffer (10 mm Tris-HCl, pH 7.3, at 4 °C, 25% glycerol, 0.2 mm EDTA, 1.5 mm MgCl2, and 10 mm KCl) and extracted by the addition of one packed cell volume of high salt buffer (10 mm Tris-HCl, pH 7.3, at 4 °C, 25% glycerol, 0.2 mm EDTA, 1.5 mm MgCl2, and 1.2 m KCl). The nuclear suspension was fractionated by centrifugation into the nuclear extract and nuclear pellet fractions.
Purification of F:SRF, F:Elk-1, F:CREB, F:ATF1, and F:hnRNP R
FLAG-tagged SRF (F:SRF)-containing S100 fraction was loaded onto HiTrap heparin (GE Healthcare) equilibrated with BC(100) containing 0.5 mm PMSF and 1 mm dithiothreitol. BC buffer consists of 20 mm Hepes-KOH, pH 7.9, 1 mm EDTA, and 10% glycerol, and the number in parentheses shows its KCl concentration (in mm). The bound proteins were eluted with a 10-column volume linear gradient from 100 to 1000 mm KCl. F:SRF bound to HiTrap heparin was eluted at 300–500 mm KCl. The fractions containing F:SRF were adjusted to 100 mm with BC(0) and loaded onto HiTrap Q (GE Healthcare), and the bound proteins were eluted with a 10-column volume linear gradient from 100 to 1000 mm KCl. F:SRF was eluted at 200–300 mm KCl, and the fractions containing F:SRF were loaded onto Mono S. F:SRF bound to Mono S was eluted at 150–250 mm KCl. F:Elk-1 was purified with the same columns as those used for F:SRF (HiTrap heparin, HiTrap Q, and Mono S). F:Elk-1 was eluted at 200–300 mm KCl from HiTrap heparin and was in the flow-through fractions from both HiTrap Q and Mono S. The High Five S100 fraction containing recombinant F:CREB (CREBα isoform) was loaded onto HiTrap SP (GE Healthcare) equilibrated with BC(100) containing 0.5 mm PMSF. F:CREB bound to HiTrap SP was eluted stepwise with BC(700). The eluate was dialyzed against BC(100) containing 0.5 mm PMSF and 0.1% Triton X-100, and F:CREB was further purified with anti-FLAG M2-agarose. The same method was also used for purifying F:ATF1. F:hnRNP R was purified similarly by SP Sepharose and Q Sepharose columns, followed by affinity purification using anti-FLAG M2-agarose.
GST Pull-down Assays
GST pull-down assays were performed essentially as described (19) using a FLAG-tagged PC4 expressed in Escherichia coli.
In Vitro Transcription
The promoter region of the c-fos gene (from −380 to −10) was isolated from human genomic DNA by PCR using the following primers: 5′-ggccgaattcgcactgcaccctcggtgttg-3′ and 5′-ggccgtcgacggccgcgccgcagccactgcttttataac-3′.
After disruption of the EagI site at −121, the c-fos promoter fragment was subcloned between the EcoRI and SalI sites of pUC19, and the G-less cassette from pG5HMC2AT (20) was fused to the c-fos promoter by using an EagI site introduced at −8. In vitro transcription assays were performed as described (21).
Purification of Mediator
To produce a 3×FLAG-Nut2-expressing cell line, pLNCX2–3×FLAG-Nut2 was introduced into AmphoPack-293 cells by Lipofectin (Invitrogen), and the produced viruses were used to infect HeLa S3 cells, which were then selected in 0.8 mg/ml G418 medium containing 10% fetal bovine serum and 1% penicillin-streptomycin solution (Invitrogen). The G418-resistant colony that expressed the highest level of 3×FLAG-Nut2 was expanded for nuclear extract preparation. The nuclear extract was fractionated on a P11 column, and the resulting 0.85 m KCl eluate was adjusted to BC(300) containing 0.1% Triton X-100 and mixed with M2-agarose (Sigma) at 4 °C for 1 h. After extensive washes with the same buffer, bound proteins were eluted with the buffer containing 0.1 mg/ml 3×FLAG peptide.
HeLa NE Fractionation
Fifty ml of HeLa nuclear extract (∼10 mg/ml protein) was adjusted to BC(100) and loaded onto a 50-ml P11 column, and the flow-through fraction was collected as fraction A. The bound proteins were then eluted stepwise in BC(300), BC(500), and BC(850), and the eluates were collected as fractions B, C, and D, respectively. Each fraction was then adjusted to BC(100) by dialysis and loaded onto an appropriate volume (∼10 ml/100 mg of protein) of DE52 column, and the flow-through fraction and the BC(300) eluate were collected.
RESULTS
Expression and Purification of Natural Activators for the c-fos Gene
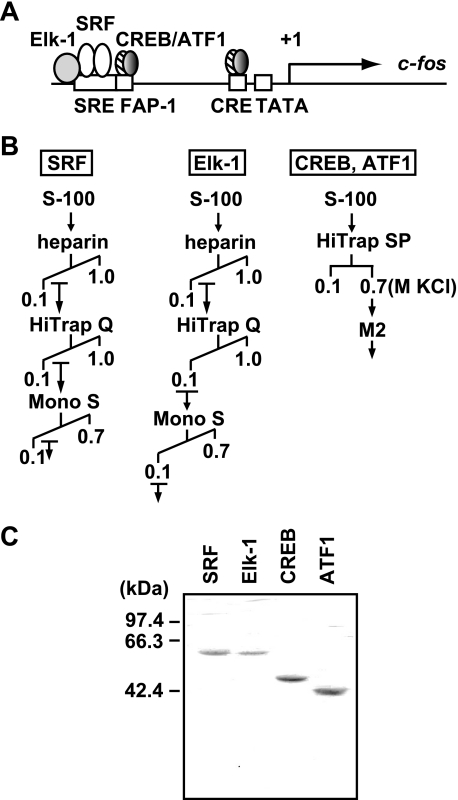
The c-fos promoter contains binding sites for several transcription factors, including SRF, Elk-1, CREB, and ATF1. SRF binds the SRE as a dimer and forms a ternary complex with Elk-1, and CREB binds the c-fos AP-1 site (FAP-1) and CRE either as a homodimer or as a heterodimer with ATF1 (Fig. 1A). These transcription factors play important roles for c-fos transcription, with each transcription factor differentially regulated by extracellular signals (22). The activities of SRF, Elk-1, CREB, and ATF1 are enhanced upon phosphorylation by MAPKs or their downstream effector kinases such as the mitogen- and stress-activated kinases and the ribosomal S6 kinases (23).
FIGURE 1.
Expression and purification of the full-length activators for the c-fos gene. A, schematic diagram of the c-fos gene. The regulatory cis-elements (SRE, FAP-1, and CRE) in the c-fos promoter and their cognate activators (SRF, Elk-1, CREB, and ATF1) are depicted in the diagram. B, purification schemes of SRF, Elk-1, CREB, and ATF1. Recombinant FLAG-tagged proteins were expressed in insect (High Five) cells, and their extracts were used for the following purification. Three columns (HiTrap heparin, HiTrap Q, and Mono S) were used for purification of SRF and Elk-1, whereas HiTrap SP and anti-FLAG M2 affinity resin were used for purification of CREB and ATF1. C, SDS-PAGE analysis of purified activators. Purified activators (∼1.0 μg) were resolved on a 10% SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue.
As a first step to better understand the regulatory mechanisms of c-fos transcription in vitro, it was essential to obtain intact SRF, Elk-1, CREB, and ATF1. For this purpose, we chose the baculovirus expression system, which can provide post-translational modifications as opposed to the E. coli system, because these transcription factors are subject to multiple post-translational modifications, including phosphorylation, glycosylation, and sumoylation that regulate their activity and stability (24, 25). Moreover, to prevent rapid degradation of the expressed proteins, we found that the inclusion of a proteasome inhibitor MG132 during purification was crucial, presumably because of the ubiquitination and proteasome-mediated protein degradation (26). As shown in Fig. 1B, the expressed activators were purified through combinations of conventional and affinity columns. Specifically, SRF and Elk-1 were purified by HiTrap heparin, HiTrap Q, and Mono S columns, whereas CREB and ATF1 were purified by a HiTrap SP column and then by anti-FLAG M2 affinity purification. The SDS-PAGE analysis showed that the obtained activators were essentially homogeneous with no apparent degradation (Fig. 1C).
The purified transcription factors were also tested for their DNA binding using gel shift (supplemental Fig. S1) and DNase I footprinting assays (supplemental Fig. S2). As expected, SRF and Elk-1 bound specifically to the SRE, and CREB and ATF1 bound to both the FAP-1 and CRE. Elk-1 binding to the SRE was completely dependent upon the presence of SRF. Both CREB and ATF1 bound to the FAP-1 and CRE as either homodimer or heterodimer; however, a heterodimer of CREB and ATF1 appeared to show the strongest binding. Together, the purified recombinant SRF, Elk-1, CREB, and ATF1 are intact and retain the binding activity to their cognate cis-elements.
SRF, Elk-1, CREB, and ATF1 Fail to Activate Transcription in the Reconstituted in Vitro Transcription System
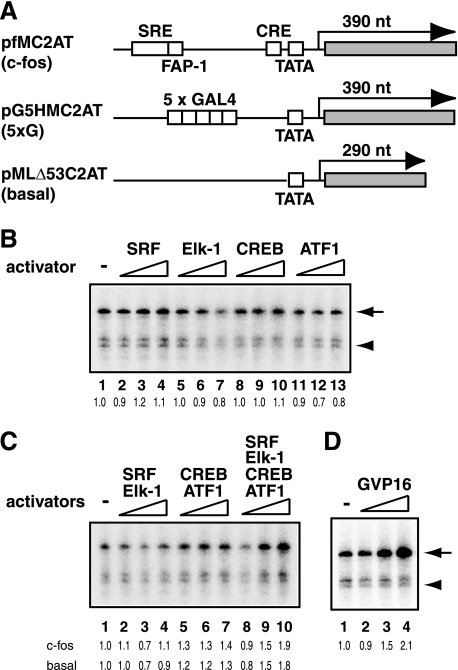
Having confirmed the DNA binding activities of recombinant SRF, Elk-1, CREB, and ATF1, we then tested their transcriptional activities. First, to reveal the intrinsic activation potential of these cellular activators, we employed an in vitro transcription system reconstituted only with RNAP II and six GTFs (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH). The reconstituted in vitro transcription system was devoid of any known coactivators but capable of supporting activated transcription by GAL4-VP16 (19). A c-fos template, pfMC2AT, was constructed by replacing the upstream GAL4-binding sites and the TATA box of pG5HMC2AT with the c-fos promoter region (Fig. 2A). Thus, the pfMC2AT template encompasses from −380 to −10 of the human c-fos gene, including the SRE, FAP-1, CRE, and TATA box, which allows simultaneous binding of seven activators, namely one Elk-1 and two SRFs to the SRE and each CREB/ATF1 heterodimer to the FAP-1 and CRE (Fig. 1A).
FIGURE 2.
Transcriptional activities of SRF, Elk-1, CREB, and ATF1 in the reconstituted in vitro transcription system. A, schematic diagram of the templates used for in vitro transcription assays. The c-fos template, pfMC2AT, contains the SRE, FAP-1, CRE, and the TATA box, which are derived from the human c-fos promoter, together with a 390-nucleotide (nt) G-less cassette. The 5×G template, pG5HMC2AT, contains five GAL4-binding sites, the HIV-1 TATA box, and a 390-nucleotide G-less cassette. The control plasmid, pMLΔ53C2AT, containing the Ad2MLP-derived TATA box and a 290-nucleotide G-less cassette, was used for measuring basal transcription as a control. B, in vitro transcription assays in the absence of coactivators. Transcription reactions were performed in a reconstituted system consisting of GTFs and RNAP II with the indicated recombinant activator. The amounts of activators were 5, 20, or 80 ng for SRF, CREB, and ATF1 and 2.5, 10, or 40 ng for Elk-1. The arrow and arrowhead indicate the positions of the transcripts from activator-dependent (pfMC2AT) and basal transcription (pMLΔ53C2AT), respectively, and the relative levels of activator-dependent transcription are shown below each lane. C, in vitro transcription assays were done with various combinations of the activators. The relative levels of activator-dependent and basal transcription are shown below each lane. D, GAL4-VP16-dependent transcription using pG5HMC2AT. The reactions contained 1.6, 6.3, or 25 ng of GAL4-VP16. The arrow indicates the transcripts from activator-dependent transcription (pG5HMC2AT).
Consistent with the previous results, the reconstituted transcription system allowed GAL4-VP16 to activate transcription from its cognate template with five GAL4-binding sites (pG5HMC2AT) in the absence of any coactivator, achieving ∼2–3-fold activation at its saturating amount (Fig. 2D) (19). The same transcription system, however, failed to activate transcription when one of the c-fos gene activators (SRF, Elk-1, CREB, and ATF1) was added individually (Fig. 2B). Moreover, even when various combinations of the activators (SRF/Elk-1, CREB/ATF1, or SRF/Elk-1/CREB/ATF1) were tested, minimal levels of activation were observed (Fig. 2C). At the highest amounts of the four activators, where c-fos transcription increased apparently, basal (i.e. activator-independent) transcription also increased to similar extents, indicating no net effect on activator-dependent transcription under these conditions (Fig. 2C, lanes 9 and 10). Given that a single c-fos promoter binds seven activators (Fig. 1A), with each activator possessing at least one activation domain, it is unlikely that the number of activators or activation domains is insufficient for c-fos promoter activation. Thus, unlike viral activators (e.g. VP16), which can activate transcription in the absence of any coactivator, the tested activators (SRF, Elk-1, CREB, and ATF1) alone are insufficient to evoke activated transcription, suggesting a higher degree of dependence on coactivators.
A Coactivator PC4 Permits Transcriptional Activation of the c-fos Promoter by SRF, Elk-1, CREB, and ATF1
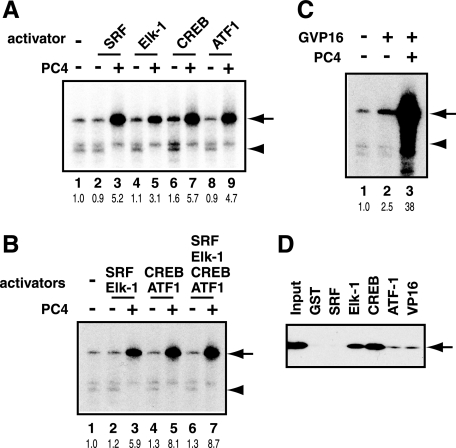
As an initial step to explore their coactivator requirement for activating the c-fos promoter, we tested the effect of PC4, a well characterized coactivator that augments in vitro transcription for a wide variety of activators (27, 28). We added PC4 to in vitro transcription reactions, in which various combinations of the c-fos activators were examined. Consistent with our earlier study (19), PC4 enhanced GAL4-VP16-dependent transcription from 2.5- to 38-fold, giving rise to additional increase (over 10-fold) in activation (Fig. 3C). Similarly, PC4 enabled the c-fos activators individually (Fig. 3A) or as combinations of SRF/Elk-1, CREB/ATF1, or SRF/Elk-1/CREB/ATF1 (Fig. 3B) to activate transcription by ∼5–9-fold. Thus, SRF, Elk-1, CREB, and AFT1 are indeed capable of activating transcription, although in a markedly PC4-dependent manner.
FIGURE 3.
Effect of coactivator PC4 on transcriptional activation by SRF, Elk-1, CREB, and ATF1. A, PC4 (200 ng) was added to the in vitro transcription reactions containing one of the activators (SRF, Elk-1, CREB, and ATF1). The positions of the transcripts from activator-dependent (arrow) and basal (arrowhead) transcription are indicated on the right. B, PC4 (200 ng) was added to the in vitro transcription reactions with various combinations of SRF, Elk-1, CREB, and ATF1 as activators. C, PC4 (200 ng) was added to in vitro transcription reactions from pG5HMC2AT with GAL4-VP16 as an activator. The arrow indicates activator-dependent transcription from pG5HMC2AT. D, interactions of PC4 with SRF, Elk-1, CREB, ATF1, and VP16. GST-fused activators were immobilized on glutathione-Sepharose 4B and incubated with FLAG-PC4. The bound proteins were eluted in high salt buffer and analyzed by immunoblot analysis using anti-FLAG M2 antibody. The position of FLAG-PC4 is indicated by an arrow on the right.
PC4 exerts its coactivator activity by mediating interactions between activators and GTFs such as TFIIA (27), TFIIB (29), and TFIIH (19). Given the ability of PC4 to enhance the transcriptional activities of SRF, Elk-1, CREB, and ATF1 (Fig. 3, A–C), we wished to know whether PC4 could interact with these activators. To determine this, we performed GST pull-down assays using GST-fused activators and FLAG-tagged PC4. Fig. 3D shows that PC4 interacted strongly with CREB and Elk-1, moderately with ATF1, and very weakly with SRF. The results indicate that these interactions may, at least in part, provide a mechanistic basis for PC4-enhanced transcriptional activities of SRF, Elk-1, CREB, and ATF1.
A Novel Activity Enhances in Vitro Transcription from the c-fos Promoter
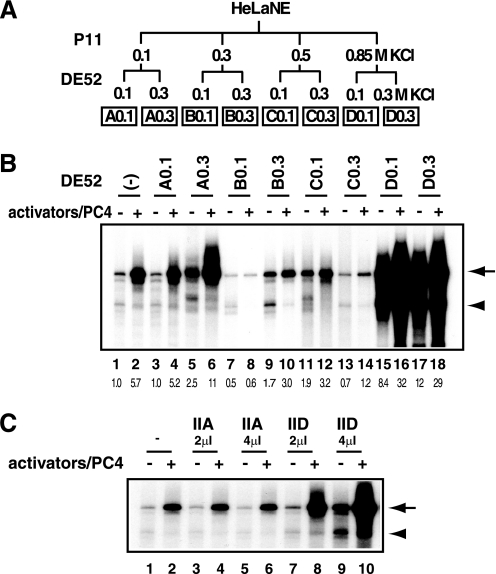
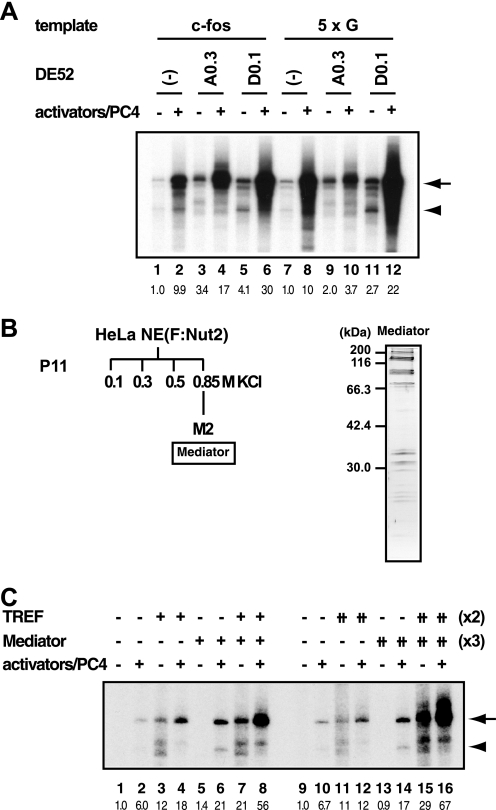
Numerous in vivo studies have shown that various extracellular stimuli elicit c-fos gene activation that typically reaches well over 100-fold. Thus, even in the presence of PC4, the observed activation of the c-fos promoter in vitro (Fig. 2, A and B) was unremarkable as compared with that generally observed within cells. To further explore the coactivator requirement for the c-fos promoter in an unbiased manner, we fractionated the nuclear extract from rapidly growing HeLa cells into eight fractions (Fig. 4A) and examined their effects on c-fos transcription in vitro.
FIGURE 4.
A novel activity enhances transcription from the c-fos promoter. A, fractionation scheme of HeLa nuclear extract. HeLa nuclear extract was fractionated stepwise by P11 (0.1, 0.3, 0.5, and 0.85 m KCl), and the derived fraction was further fractionated by DE52 (0.1 and 0.3 m KCl) into eight fractions. The names of the DE52-derived fractions are as indicated at the bottom. B, in vitro transcription assays. Each fraction was added to the in vitro transcription reactions using the c-fos template, pfMC2AT, in the presence or absence of the activators (SRF, Elk-1, CREB, and ATF1). PC4 was also added with the activators. C, effect of varying amounts of purified TFIIA or TFIID on c-fos transcription.
As expected, the D0.3 fraction, which contains TFIID (supplemental Fig. S3), increased both basal and activated transcription (Fig. 4B, lanes 17 and 18), because the amount of TFIID in the current transcription system was set below the saturating amount to reveal the effect of activators (Fig. 4C, lanes 7–10). Furthermore, the D0.1 fraction showed an activity reminiscent of Mediator (Fig. 4B, lanes 15 and 16) (30), indicating that the D0.1 activity is, at least in part, attributable to Mediator, although the presence of additional coactivators cannot be excluded. Intriguingly, the A0.3 fraction, in which no biochemically defined coactivator had been reported, stimulated transcription from the c-fos promoter (Fig. 4B, lanes 5 and 6). Although this fraction includes TFIIA (31), which is important for activated transcription in vitro (19), further addition of TFIIA did not affect the level of c-fos transcription at all (Fig. 4C, lanes 3–6), thus excluding the possibility that the observed stimulatory effect was due to TFIIA. Collectively, our biochemical analyses identified a novel activity, which we termed TREF, that can stimulate in vitro transcription from the c-fos promoter.
TREF Enhances Transcription from the c-fos but Not the 5×G Promoter and Displays Cooperativity with Mediator
We then addressed whether or not TREF was specific to the c-fos promoter. As shown in Fig. 5A, TREF increased both basal transcription (lane 3) and activator-dependent transcription (lane 4) from the c-fos promoter, eliciting a ∼17-fold net increase of transcription from the basal level (compare lanes 1 and 4). However, whereas TREF increased basal transcription from the 5×G promoter, it somewhat diminished GAL4-VP16-dependent transcription from the same promoter (Fig. 5A, lanes 9 and 10). Thus, although TREF enhances transcription through the core promoter as well as the upstream regulatory elements, the stimulatory effect of TREF through the upstream elements appears to have some activator specificity. By contrast, the D0.1 fraction, which principally reflects the activity of Mediator, stimulated both basal and activator-dependent transcription from the c-fos (Fig. 5A, lanes 5 and 6) and 5×G promoters (Fig. 5A, lanes 11 and 12) in a similar manner. Finally, the fully phosphorylated activators and its unphosphorylated (i.e. phosphorylation-defective) mutants displayed essentially indistinguishable response to TREF, indicating that phosphorylation of the activators do not affect the TREF activity (data not shown).
FIGURE 5.
Promoter specificity of TREF and its cooperativity with Mediator. A, a novel coactivator activity, TREF, is specific to the c-fos promoter. In vitro transcription was performed using the c-fos template (pfMC2AT) or 5×G template (pG5HMC2AT) in the presence or absence of the activators (SRF, Elk-1, CREB, ATF1, and PC4 for pfMC2AT; GAL4-VP16 and PC4 for pG5HMC2AT). Either A0.3 or D0.1 fraction was added to the indicated reactions. The arrow indicates the transcripts from pfMC2AT (lanes 1–6) or pG5HMC2AT (lanes 7–12). B, purification of Mediator. The nuclear extract from the HeLa cell line that expresses FLAG-tagged Nut2 (F:Nut2) was fractionated by P11, and Mediator was purified from the 0.85 m KCl fraction using anti-FLAG M2 affinity gel. C, TREF cooperates with Mediator in activating transcription from the c-fos promoter. TREF and Mediator were added to the transcription reactions, and the positions of transcripts from pfMC2AT and pMLΔ53C2AT are indicated by the arrow and arrowhead, respectively.
Next, we tested the functional cooperativity of TREF and Mediator, a major coactivator component of the D0.1 fraction, in stimulating c-fos transcription. As shown in Fig. 5C, both TREF and Mediator, which was isolated from HeLa cell line stably expressing FLAG-Nut2 (Fig. 5B), enhanced activator-dependent transcription from the c-fos promoter (lanes 4, 6, 12 and 14). When TREF and Mediator were added simultaneously to the transcription reactions, they displayed further enhancement of transcription from the c-fos promoter, resulting in ∼60-fold net activation (Fig. 5C, lanes 8 and 16) over the basal levels (Fig. 5C, lanes 1 and 9). Importantly, this cooperativity between TREF and Mediator was observed in the range of amounts where a 2-fold increase of TREF (Fig. 5C, compare lanes 3 and 4 versus lanes 11 and 12) or a 3-fold increase of Mediator (Fig. 5C, compare lanes 5 and 6 versus lanes 13 and 14) had little additional effect on transcription individually. Thus, TREF not only stimulates c-fos transcription through the core promoter and the upstream regulatory elements but also displays cooperativity with Mediator for enhancing c-fos transcription, indicating that the c-fos gene may require multiple coactivators for its dramatic level of transcriptional activation observed in vivo.
hnRNP R Is an Active Component of TREF
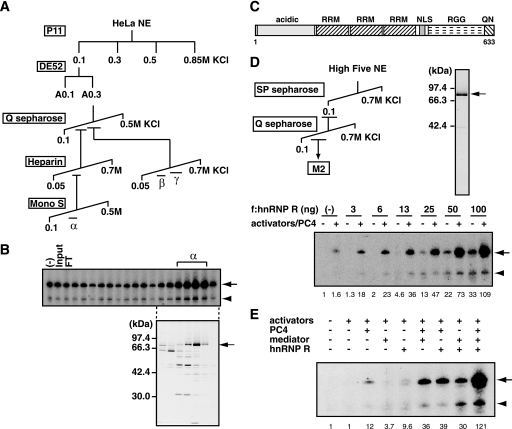
To further characterize the TREF activity, we purified the A0.3 fraction using c-fos transcription assays with SRF, Elk-1, CREB, and ATF1 as activators and PC4 as a coactivator. Our initial attempt to purify TREF resulted in a dramatic reduction in the TREF activity; however, we realized that the reduced activity was due to the separation of the original TREF activity into different fractions. Indeed, as shown in Fig. 6A, purification with a larger amount of the A0.3 fraction by Q Sepharose and heparin-Sepharose showed that the TREF activity could be separated into at least three activities that were tentatively designated as TREFα, TREFβ, and TREFγ. TREFα was purified further to near homogeneity by a Mono S column, and SDS-PAGE analysis revealed that active fractions contained a ∼80-kDa protein as a major polypeptide that correlated with the TREFα activity (Fig. 6B). Mass spectrometric analysis identified this 80-kDa polypeptide as hnRNP R, a putative RNA-binding protein, whose function remains ill defined. hnRNP R is a 633-amino acid protein with three RRMs and one arginine-glycine-glycine rich (RGG) box and belongs to a group of proteins termed hnRNPs (32) (Fig. 6C).
FIGURE 6.
hnRNP R stimulates transcription from the c-fos promoter. A, purification scheme of TREF activities. The TREF activity (A0.3) was further purified by Q Sepharose, heparin-Sepharose, and Mono S columns and separated into three chromatographically distinct activities designated TREFα, TREFβ, and TREFγ. B, transcriptional activities of the Mono S fractions of TREFα were assayed using pfMC2AT in the presence of SRF, Elk-1, CREB, ATF1, and PC4. The corresponding fractions were analyzed by SDS-PAGE and silver staining. The position of a ∼80-kDa protein that correlated with the transcriptional activity is indicated on the right. C, structural domains of hnRNP R. RRM, NLS, and RGG indicate the RNA recognition motif, nuclear localization signal, and arginine-glycine-glycine-rich domain, respectively. The regions rich in acidic amino acids or in glutamine and asparagine is indicated by acidic or QN, respectively. D, purification of recombinant hnRNP R. FLAG-tagged hnRNP R was expressed in insect cells and purified by SP Sepharose, Q Sepharose, and anti-FLAG M2-agarose. The purified hnRNP R was analyzed by SDS-PAGE. Transcriptional activity of hnRNP R was tested in the transcription assays using pfMC2AT, in which the presence of SRF, Elk-1, CREB, ATF1, and PC4 is indicated by +. Relative levels of activator-dependent transcription from pfMC2AT are indicated below the panel. E, hnRNP R, PC4, and Mediator were added in various combinations to the transcription reactions containing SRF, Elk-1, CREB, and ATF1 as activators. Relative levels of activator-dependent transcription from pfMC2AT are indicated below the panel.
To demonstrate that hnRNP R possesses the TREFα activity, we expressed recombinant hnRNP R by a baculovirus expression system and purified it to homogeneity (Fig. 6D). As shown in Fig. 6D, recombinant hnRNP R displayed a coactivator activity in a dose-dependent manner in the presence of PC4. The effect of hnRNP R on GAL4-VP16-dependent transcription, however, was minuscule (supplemental Fig. S4), indicating that its coactivator activity varies depending upon the used activators. When PC4, Mediator, and hnRNP R were tested individually or in combination, each coactivator stimulated c-fos transcription by 4–12-fold. hnRNP R and Mediator also stimulated activator-independent transcription to similar extents, indicating that both of them act directly to the core promoter in the absence of activators. Similar to the activity of a more crude TREF fraction (i.e. A0.3), hnRNP R also showed cooperativity with PC4 and Mediator (Fig. 6D), and the combined effect of the three coactivators on activator-dependent c-fos transcription reached well over 100-fold (Fig. 6E). Together, our results indicate that hnRNP R serves as a transcriptional coactivator for c-fos transcription.
DISCUSSION
In this study, we have established an in vitro transcription system that utilizes unaltered cellular activators (SRF, Elk-1, CREB, and ATF1) and the natural c-fos promoter to explore its coactivator requirement in an unbiased manner. We have identified from HeLa nuclear extract a novel coactivator activity, termed TREF, that specifically augments transcription from the c-fos promoter and cooperates with Mediator. TREF appears to be composed of at least three activities, one of which was identified as hnRNP R. Recombinant hnRNP R stimulated transcription from the c-fos promoter and cooperated with PC4 and Mediator to elicit a high level of c-fos activation.
Functional Distinction between Viral and Cellular Transcription Factors
Our transcription assays showed that the tested cellular activators fail to activate transcription in the absence of any coactivator, a condition under which the archetypical transcriptional activator GAL4-VP16 can still activate transcription. Viral activators such as VP16, E1A, Tax, and pX typically do not bind DNA directly but piggyback on DNA-bound cellular activators to enhance gene expression (33). For example, a prototypical viral protein VP16 becomes tethered to DNA by forming a complex with Oct-1 and HCF and then interacts with GTFs (TBP, TAFs, TFIIB, and TFIIH) (34). Similarly, another viral protein pX is recruited to DNA via its interactions with transcriptional activators and then activates transcription through interactions with TBP, TFIIB, and TFIIH (35, 36). Given these properties, viral activators resemble a coactivator rather than a mere transcriptional activator.
Besides acting as a coactivator, viral activators may have usurped part of the TFIID function as well. For example, the SV40 large T antigen (37), the hepatitis B viral protein pX (38), and the human cytomegalovirus IE proteins (39) can functionally obviate the requirement of TAF1, a TFIID subunit that functions as a coactivator. Thus, viral activators in general may depend less on coactivators than the cellular activators. This would allow viral activators to maximize their transcriptional potency in a manner detached from cellular regulatory networks. Cellular activators, by contrast, may retain their regulatory potential at the cost of activation potency, possibly necessitating a greater dependence on coactivators.
hnRNP R Possesses a Coactivator Activity for the c-fos Promoter
The current study has identified a novel type of coactivator, termed TREF, that stimulates transcription from the c-fos promoter in vitro and is apparently distinct from the more generally required Mediator (40). The TREF activity increases activator-dependent as well as basal transcription from the c-fos promoter in a manner similar to that of Mediator (18, 41, 42). Unlike Mediator, however, TREF does not stimulate GAL4-VP16-dependent transcription in vitro. Together with its cooperativity with Mediator in stimulating transcription from the c-fos promoter and its differing chromatographic behavior from Mediator (30, 43), TREF appears to be a novel activity distinct from the well characterized Mediator. Indeed, several steps of chromatography revealed that TREF is separated into three distinct components, one of which, TREFα, turned out to be hnRNP R. As it is, TREFα is also distinct from the reported coactivators for SRF or CREB, including CoS (coactivator for SRF-activated transcription) (44), MRTFs (myocardin-related transcription factors) (45, 46), CBP (CREB binding protein)/p300 (47), and TORC (transducer of regulated CREB) (48, 49).
hnRNP R belongs to a group of diverse RNA-binding proteins termed hnRNPs, which are characterized by RNA-binding motifs such as the RGG box, RRM, and K homology domains (50). hnRNPs are involved in a wide array of RNA metabolism including transcription, splicing, RNA export, RNA stability, and translation (50). No specific function, however, has been assigned to hnRNP R except for a recent report that suggests its role in ARE-mediated RNA degradation (51). Most of hnRNPs that have been analyzed so far do not appear to affect transcription (50); however, several lines of evidence indicate that hnRNP K serves as a transcription factor. Indeed, hnRNP K binds to a C-rich DNA sequence termed the CT element, which is located upstream of the c-myc gene and interacts directly with TBP (52). Moreover, hnRNP K has been demonstrated to activate transcription in vitro and in vivo (52). Although hnRNP R is structurally unrelated to hnRNP K, which has three KH domains but no RRM or RGG box (53), our purification and functional analyses of TREFα indicate that hnRNP R is another member of hnRNPs that directly stimulates transcription in addition to its possible role in regulating mRNA stability (51).
Recent studies have revealed that in addition to stimulating transcription, some coactivators participate actively in post-transcriptional processes. For example, the thermogenic coactivator PGC-1 regulates alternative splicing of the transcript from the gene promoter on which it is loaded (54). This coupling of transcription and splicing requires the RRM and serine- and arginine-rich domain within PGC-1 (54), indicating the importance of RNA-binding domains for the coupling. A coactivator for nuclear receptors, CoAA, also influences alternative splicing of the transcript in a steroid hormone-specific manner (55). CoAA contains two RRMs and is structurally an hnRNP-like protein (56). Thus, a subset of diverse proteins with RNA-binding domains such as the RRM, KH domain, serine- and arginine-rich domain, and/or RGG box may be actively involved in both transcription and post-transcriptional processes.
The mechanism by which hnRNP R stimulates c-fos transcription awaits future studies. However, given that hnRNP R is found among the proteins associated with phosphorylated C-terminal domain (57) and thereby presumed to be complexed with elongating RNAP II, one attractive hypothesis is that hnRNP R stimulates RNAP II elongation, perhaps through its interactions with phosphorylated C-terminal domain and with transcribed RNA. If so, C-terminal domain phosphorylation by TFIIH and/or p-TEFb might regulate the binding of hnRNP R to the C-terminal domain and alter its coactivator activity. All in all, our results support the emerging view that transcription is tightly coupled to the subsequent RNA metabolism.
Supplementary Material
Acknowledgments
We gratefully acknowledge the technical assistance of Michiyo Takeuchi. We thank Robert G. Roeder for plasmids and Sohail Malik for antibodies and cell lines.
This work was supported in part by Grants-in-aid for Scientific Research (to A. F. and K. H.) and Special Coordination Funds for Promoting Science and Technology (to A. F.) of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and supplemental text.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and supplemental text.
- MAPK
- mitogen-activated protein kinase
- GTF
- general transcription factor
- TREF
- transcriptional regulator of c-fos
- RNAP II
- RNA polymerase II
- hnRNP R
- heterogeneous nuclear ribonucleoprotein R
- RRM
- RNA recognition motif
- RGG
- arginine-glycine-glycine rich
- CRE
- cAMP-responsive element
- CREB
- CRE-binding protein
- SRE
- serum response element
- PMSF
- phenylmethylsulfonyl fluoride
- GST
- glutathione S-transferase
- SRF
- serum response factor
- Elk-1
- Ets-related transcription factor
- ATF1
- activating transcription factor 1
- FAP-1
- c-fos AP-1 site.
REFERENCES
- 1.Treisman R. (1995) EMBO J. 14,4905–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cahill M. A., Janknecht R., Nordheim A. (1996) Curr. Biol. 6,16–19 [DOI] [PubMed] [Google Scholar]
- 3.Price M. A., Hill C., Treisman R. (1996) Philos. Trans. R. Soc. Lond. B Biol. Sci. 351,551–559 [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Prywes R. (2000) Oncogene 19,1379–1385 [DOI] [PubMed] [Google Scholar]
- 5.Roeder R. G. (1996) Trends Biochem. Sci. 21,327–335 [PubMed] [Google Scholar]
- 6.Malik S., Roeder R. G. (2000) Trends Biochem. Sci. 25,277–283 [DOI] [PubMed] [Google Scholar]
- 7.Lewis B. A., Reinberg D. (2003) J. Cell Sci. 116,3667–3675 [DOI] [PubMed] [Google Scholar]
- 8.Ramirez S., Ait-Si-Ali S., Robin P., Trouche D., Harel-Bellan A. (1997) J. Biol. Chem. 272,31016–31021 [DOI] [PubMed] [Google Scholar]
- 9.Janknecht R., Nordheim A. (1996) Biochem. Biophys. Res. Commun. 228,831–837 [DOI] [PubMed] [Google Scholar]
- 10.Kwok R. P., Lundblad J. R., Chrivia J. C., Richards J. P., Bächinger H. P., Brennan R. G., Roberts S. G., Green M. R., Goodman R. H. (1994) Nature 370,223–226 [DOI] [PubMed] [Google Scholar]
- 11.Arany Z., Sellers W. R., Livingston D. M., Eckner R. (1994) Cell 77,799–800 [DOI] [PubMed] [Google Scholar]
- 12.Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996) Cell 87,953–959 [DOI] [PubMed] [Google Scholar]
- 13.Boyer T. G., Martin M. E., Lees E., Ricciardi R. P., Berk A. J. (1999) Nature 399,276–279 [DOI] [PubMed] [Google Scholar]
- 14.Stevens J. L., Cantin G. T., Wang G., Shevchenko A., Berk A. J. (2002) Science 296,755–758 [DOI] [PubMed] [Google Scholar]
- 15.Wang G., Balamotis M. A., Stevens J. L., Yamaguchi Y., Handa H., Berk A. J. (2005) Mol. Cell 17,683–694 [DOI] [PubMed] [Google Scholar]
- 16.Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D. (1990) Cell 61,1209–1215 [DOI] [PubMed] [Google Scholar]
- 17.Flanagan P. M., Kelleher R. J., 3rd, Sayre M. H., Tschochner H., Kornberg R. D. (1991) Nature 350,436–438 [DOI] [PubMed] [Google Scholar]
- 18.Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. (1994) Cell 77,599–608 [DOI] [PubMed] [Google Scholar]
- 19.Fukuda A., Nakadai T., Shimada M., Tsukui T., Matsumoto M., Nogi Y., Meisterernst M., Hisatake K. (2004) Mol. Cell Biol. 24,6525–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuda A., Nogi Y., Hisatake K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,1206–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukuda A., Yamauchi J., Wu S. Y., Chiang C. M., Muramatsu M., Hisatake K. (2001) Genes Cells 6,707–719 [DOI] [PubMed] [Google Scholar]
- 22.Hill C. S., Treisman R. (1995) EMBO J. 14,5037–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn K. L., Espino P. S., Drobic B., He S., Davie J. R. (2005) Biochem. Cell Biol. 83,1–14 [DOI] [PubMed] [Google Scholar]
- 24.Reason A. J., Morris H. R., Panico M., Marais R., Treisman R. H., Haltiwanger R. S., Hart G. W., Kelly W. G., Dell A. (1992) J. Biol. Chem. 267,16911–16921 [PubMed] [Google Scholar]
- 25.Comerford K. M., Leonard M. O., Karhausen J., Carey R., Colgan S. P., Taylor C. T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipford J. R., Deshaies R. J. (2003) Nat. Cell Biol. 5,845–850 [DOI] [PubMed] [Google Scholar]
- 27.Ge H., Roeder R. G. (1994) Cell 78,513–523 [DOI] [PubMed] [Google Scholar]
- 28.Kretzschmar M., Kaiser K., Lottspeich F., Meisterernst M. (1994) Cell 78,525–534 [DOI] [PubMed] [Google Scholar]
- 29.Henry N. L., Bushnell D. A., Kornberg R. D. (1996) J. Biol. Chem. 271,21842–21847 [DOI] [PubMed] [Google Scholar]
- 30.Malik S., Gu W., Wu W., Qin J., Roeder R. G. (2000) Mol. Cell 5,753–760 [DOI] [PubMed] [Google Scholar]
- 31.Reinberg D., Horikoshi M., Roeder R. G. (1987) J. Biol. Chem. 262,3322–3330 [PubMed] [Google Scholar]
- 32.Hassfeld W., Chan E. K., Mathison D. A., Portman D., Dreyfuss G., Steiner G., Tan E. M. (1998) Nucleic Acids Res. 26,439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flint J., Shenk T. (1997) Annu. Rev. Genet. 31,177–212 [DOI] [PubMed] [Google Scholar]
- 34.Wysocka J., Herr W. (2003) Trends Biochem. Sci. 28,294–304 [DOI] [PubMed] [Google Scholar]
- 35.Haviv I., Vaizel D., Shaul Y. (1996) EMBO J. 15,3413–3420 [PMC free article] [PubMed] [Google Scholar]
- 36.Haviv I., Shamay M., Doitsh G., Shaul Y. (1998) Mol. Cell Biol. 18,1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damania B., Alwine J. C. (1996) Genes Dev. 10,1369–1381 [DOI] [PubMed] [Google Scholar]
- 38.Haviv I., Matza Y., Shaul Y. (1998) Genes Dev. 12,1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukac D. M., Harel N. Y., Tanese N., Alwine J. C. (1997) J. Virol. 71,7227–7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malik S., Roeder R. G. (2005) Trends Biochem. Sci. 30,256–263 [DOI] [PubMed] [Google Scholar]
- 41.Mittler G., Kremmer E., Timmers H. T., Meisterernst M. (2001) EMBO Rep. 2,808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baek H. J., Malik S., Qin J., Roeder R. G. (2002) Mol. Cell Biol. 22,2842–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Näär A. M., Beaurang P. A., Zhou S., Abraham S., Solomon W., Tjian R. (1999) Nature 398,828–832 [DOI] [PubMed] [Google Scholar]
- 44.Zhu H., Pyrwes R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89,5291–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105,851–862 [DOI] [PubMed] [Google Scholar]
- 46.Wang D. Z., Li S., Hockemeyer D., Sutherland L., Wang Z., Schratt G., Richardson J. A., Nordheim A., Olson E. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,14855–14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan H. M., La Thangue N. B. (2001) J. Cell Sci. 114,2363–2373 [DOI] [PubMed] [Google Scholar]
- 48.Iourgenko V., Zhang W., Mickanin C., Daly I., Jiang C., Hexham J. M., Orth A. P., Miraglia L., Meltzer J., Garza D., Chirn G. W., McWhinnie E., Cohen D., Skelton J., Terry R., Yu Y., Bodian D., Buxton F. P., Zhu J., Song C., Labow M. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,12147–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conkright M. D., Canettieri G., Screaton R., Guzman E., Miraglia L., Hogenesch J. B., Montminy M. (2003) Mol. Cell 12,413–423 [DOI] [PubMed] [Google Scholar]
- 50.Krecic A. M., Swanson M. S. (1999) Curr. Opin. Cell Biol. 11,363–371 [DOI] [PubMed] [Google Scholar]
- 51.Huang J., Li S. J., Chen X. H., Han Y., Xu P. (2008) Cell Mol. Biol. Lett. 13,303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michelotti E. F., Michelotti G. A., Aronsohn A. I., Levens D. (1996) Mol. Cell Biol. 16,2350–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bomsztyk K., Denisenko O., Ostrowski J. (2004) Bioessays 26,629–638 [DOI] [PubMed] [Google Scholar]
- 54.Monsalve M., Wu Z., Adelmant G., Puigserver P., Fan M., Spiegelman B. M. (2000) Mol. Cell 6,307–316 [DOI] [PubMed] [Google Scholar]
- 55.Auboeuf D., Dowhan D. H., Li X., Larkin K., Ko L., Berget S. M., O'Malley B. W. (2004) Mol. Cell Biol. 24,442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwasaki T., Chin W. W., Ko L. (2001) J. Biol. Chem. 276,33375–33383 [DOI] [PubMed] [Google Scholar]
- 57.Carty S. M., Greenleaf A. L. (2002) Mol. Cell Proteomics 1,598–610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.