Abstract
Extracellular signal-regulated kinases (ERKs) play important physiological roles in proliferation, differentiation, and gene expression. ERK5 is approximately twice the size of ERK1/2, and its amino-terminal half contains the kinase domain that shares homology with ERK1/2 and TEY activation motif, whereas the carboxyl-terminal half is unique. In this study, we examined a physiological role of ERK5 in rat pheochromocytoma cells (PC12), comparing it with ERK1/2. Nerve growth factor (NGF) induced phosphorylation of both ERK5 and ERK1/2, whereas the cAMP analog dibutyryl cAMP (Bt2cAMP) caused only ERK1/2 phosphorylation. U0126, at 30 μm, that blocks ERK1/2 signaling selectively attenuated neurite outgrowth induced by NGF and Bt2cAMP, but BIX02188 and BIX02189, at 30 μm, that block ERK5 signaling and an ERK5 dominant-negative mutant suppressed only NGF-induced neurite outgrowth. Next, we examined the expression of tyrosine hydroxylase, a rate-limiting enzyme of catecholamine biosynthesis. Both NGF and Bt2cAMP increased tyrosine hydroxylase gene promoter activity in an ERK1/2-dependent manner but was ERK5-independent. However, when both ERK5 and ERK1/2 signalings were inhibited, tyrosine hydroxylase protein up-regulation by NGF and Bt2cAMP was abolished, because of the loss of stabilization of tyrosine hydroxylase protein by ERK5. Taking these results together, ERK5 is involved in neurite outgrowth and stabilization of tyrosine hydroxylase in PC12 cells, and ERK5, along with ERK1/2, plays essential roles in the neural differentiation process.
ERKs2 or MAPKs are involved in proliferation, differentiation, migration, and gene expression. ERK1/2 is activated by variety of stimuli, and the signaling pathway leading to ERK1/2 activation has been well characterized (1–3). ERK5 is approximately double the molecular size of ERK1/2. The kinase domain is encoded by its amino-terminal half and shares ∼50% homology with ERK1/2, whereas its unique carboxyl terminus encodes two proline-rich regions, a nuclear export domain and a nuclear localization domain (4, 5). Recently, it was reported that the autophosphorylated carboxyl terminus of ERK5 plays a critical role in activating transcription (6). The threonine and tyrosine residues on ERK5 are phosphorylated by MEK5 but not MEK1/2. In contrast, ERK1/2 is not phosphorylated by MEK5 but is phosphorylated by MEK1/2 (7, 8).
Several physiological roles of ERK5 have been reported (4, 5, 9). For example, ERK5 regulates S-phase entry by epidermal growth factor (EGF) in HeLa cells (10). In dorsal root ganglia, ERK5 is activated by nerve growth factor (NGF) during retrograde transport of TrkA, and this action prevents apoptosis (11). In developing cortical neurons, ERK5 plays a critical role in survival promoted by brain-derived neurotrophic factor (BDNF) (12). Neuronal differentiation is reduced by ERK5 knockdown using antisense morpholino oligonucleotides in Xenopus laevis (13). Also, ERK5 is required for generation of neurons from cortical progenitors (14). In animal models, ERK5 gene knockout is lethal at E9.5–10.5 because of cardiovascular defects, indicating the involvement of ERK5 in heart development (15). Pathophysiological roles for ERK5 have been proposed for tumor development and cardiac hypertrophy (4). Thus, ERK5 plays essential physiological roles in addition to ERK1/2.
In rat pheochromocytoma cells (PC12), EGF and NGF activate ERK5 via the small G-protein Ras (16). But it has been shown that another neurotrophin, BDNF, activates ERK5 through Rap1 in cortical neurons (17). In the mink lung epithelial cell line, interaction of Lck-associated adapter with MEKK2 is involved in activation of ERK5 (18), whereas in bone marrow-derived mast cells, protein kinase C mediates FcϵRI-induced ERK5 activation and cytokine production (19). Heterotrimeric G-protein-coupled receptor (GPCR) signals also activate ERK5. For example, GPCR agonists such as carbachol and thrombin activate ERK5 via Gαq/11 and Gα12/13 families of heterotrimeric G-proteins (1, 20). The ERK5 activation by carbachol and thrombin is not blocked by dominant-negative mutants of Ras and Rho nor the C3 toxin that inactivates Rho-mediated functions (20). In contrast, prostaglandin E2 and forskolin, both of which promote cAMP production, attenuate ERK5 phosphorylation induced by EGF via protein kinase A (21). In addition, we recently reported that βγ subunits of Gi/o activated by lysophosphatidic acid blocked ERK5 activation by EGF and NGF in PC12 cells (22). Thus, the mechanism of ERK5 regulation is complex, and the detailed mechanisms remain unclear.
PC12 cells are a good in vitro model of immature neurons. In response to NGF, they differentiate toward sympathetic neurons (23). We have shown that NGF and cAMP promote ERK1/2 phosphorylation via Ras or Rap1 in PC12 cells, and ERK1/2 activation is essential for neurite outgrowth by NGF or cAMP (24). Although the role of ERK5 in neural differentiation is suggested (13, 14), the detailed molecular mechanism in neuronal differentiation remains unclear. Therefore, in this study, we attempted to clarify the physiological role of ERK5 in the neural differentiation process, especially focusing on neurite outgrowth and neuron-specific gene expression such as tyrosine hydroxylase.
EXPERIMENTAL PROCEDURES
Materials
NGF, Bt2cAMP, D-luciferin, Hoechst-33258, and antibodies against FLAG M2 and β-actin were purchased from Sigma. EGF was purchased from PeproTech EC. (London, UK). Cycloheximide and SB203580 were purchased from Wako Pure Chemicals (Tokyo, Japan). U0126, PD98059, antibodies against phospho-ERK1/2, ERK1/2, phospho-ERK5, ERK5, and tyrosine hydroxylase and horseradish peroxidase (HRP)-conjugated anti-rabbit IgG secondary antibody were purchased from Cell Signaling (Beverly, MA). This phospho-specific ERK5 antibody also recognizes phospho-ERK1/2. Antibodies against ERK2, Rap1 and C3G, were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Enhanced chemiluminescence assay kit and HRP-conjugated anti-mouse IgG were purchased from GE Healthcare. Lipofectamine 2000 was purchased from Invitrogen. Phosphotyrosine hydroxylase (Ser-31) antibody was from Chemicon (Temecula, CA). BIX02188 and BIX02189 were kindly provided from Boehringer Ingelheim (Ridgefield, CT). DNA plasmid encoding tyrosine hydroxylase promoter-driven luciferase was kindly provided by Dr. Bistra Nankova (New York Medical College). DNA plasmids for measuring transcriptional activity of myocyte-enhancer factor (MEF) 2 (MEF2C/Gal4 and Gal4/Luciferase) were kindly given by Dr. Zhengui Xia (University of Washington) (25). DNA plasmid encoding mycERK5, MEK5A (S311A/T315V), and MEK5D (S311D/T315D) were from Dr. Eisuke Nishida (Kyoto University). mycERK5 kinase-dead mutant (K83M) was created from mycERK5 in this laboratory by using the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX). Adenovirus encoding RasS17N (RasN17) was provided by Dr. Dennis Stacey (Cleveland Clinic Institute). Adenovirus encoding FLAG-Rap1GAP1 was constructed using FLAG-Rap1GAP1 (26). C3G siRNA (sense, GGACUUUGAUGUUGAAUGUtt; antisense, ACAUUCAACAUCAAAGUCCtg) was purchased from Ambion (Austin, TX) (27).
Cell Culture
PC12 cells were obtained from the Japanese Cancer Research Bank (Tokyo, Japan). The cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (Cell Culture Laboratory, Cleveland, OH), 5% horse serum (Invitrogen), penicillin (50 units/ml), and streptomycin (50 μg/ml) in an incubator containing 5% CO2 at 37 °C. PC12 cells that stably overexpress dominant-negative MEK5 were cultured in the presence of G418 (Invitrogen). To obtain PC12 cells that stably overexpress EGFP-tagged Rap1GAP (28), the cells were transfected with EGFP-tagged Rap1GAP and cultured in the presence of G418. Then the EGFP-positive and -negative cells were selected by cell sorting using FACSAria (BD Biosciences). HEK293 cells were grown in DMEM supplemented with 10% fetal calf serum and the above antibiotics.
SDS-PAGE and Western Blotting
Electrophoresis was performed on 8–11% acrylamide gels. Proteins were transferred electrically from the gel onto polyvinylidene difluoride membrane (Millipore, Bedford, MA) by the semi-dry blotting method. The blots were blocked for 1 h with 5% low fat milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) at room temperature and incubated with primary antibodies overnight at 4 °C. The blots were washed several times and incubated with HRP-conjugated anti-rabbit or anti-mouse IgG antibody as a secondary antibody in TBST containing 5% low fat milk at room temperature for 2 h. After rinsing with TBST, blots were developed using a chemiluminescence assay kit and visualized by the exposing the chemiluminescence from the membrane to the Hyperfilm enhanced chemiluminescence. The densities of the bands corresponding to tyrosine hydroxylase were analyzed by densitometry (ImageJ 1.36b, National Institutes of Health).
Rap1 Activation Assay
Activated Rap1 was isolated from cell lysates using a protocol adapted from that of Franke et al. (29). Treated cells were lysed in 300 μl of ice-cold lysis buffer (10% glycerol, 1% Nonidet P-40, 50 mm Tris-HCl (pH 8.0), 200 mm NaCl, 5 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, 1 μm leupeptin, 10 μg/ml soybean trypsin inhibitor, 10 mm NaF, 0.5 mm aprotinin, 1 mm Na3VO4). Lysates were clarified by low speed centrifugation, and supernatants containing 500–1000 μg of total protein were incubated with 40 μg of GST-RalGDS fusion protein (gift of Dr. J. L. Bos, Utrecht University, Utrecht, The Netherlands) for 40 min, followed by incubation with glutathione-agarose beads for an additional 40 min at 4 °C. Beads were rinsed three times with lysis buffer, and protein was eluted from the beads with Laemmli sample loading buffer. The amount of Rap1 bound to the beads was detected by Western blotting using antibody to Rap1.
Assay for Neurite Outgrowth
The neurite extension from PC12 cells was utilized as an index of neuronal differentiation. The cells were fixed with 4% paraformaldehyde and then the nuclei were stained with Hoechst-33258. The photographs were taken with CELAVIEW-RS100 (Olympus, Tokyo, Japan). The number of nuclei and total length of neurites were calculated with the CELAVIEW software (Olympus, Tokyo, Japan), and then the value of total neurite length divided by nucleus number was expressed as neurite length per cell (μm/cell). For transfection experiments (Figs. 6 and 7), the number of nuclei and total neurite length in GFP-positive cells were selectively counted by using the software. Data are expressed as means ± S.E. of the values of three wells (30).
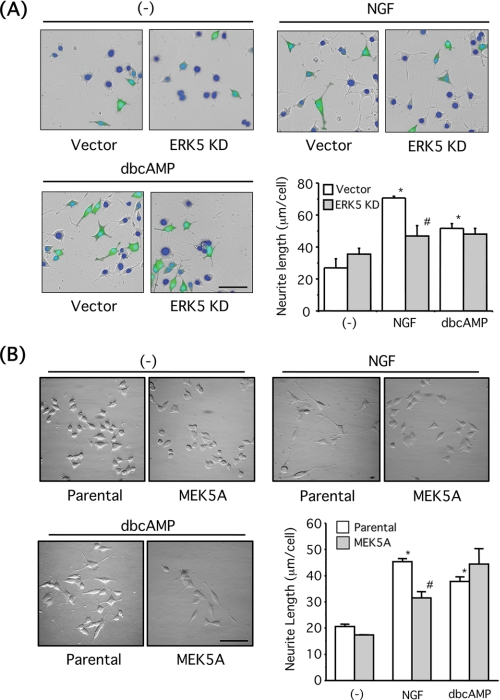
FIGURE 6.
Neurite outgrowth induced by NGF is blocked by overexpression of dominant-negative ERK5 and MEK5A mutants. A, PC12 cells were co-transfected with GFP and empty vector or ERK5KD. The cells were incubated with NGF (100 ng/ml) or Bt2cAMP (dbcAMP) (0.5 mm) for a day. The nuclei were stained with Hoechst-33258 (blue), and morphological changes of GFP-positive cells were observed under a fluorescence microscope. Scale bar, 100 μm. Neurite length of GFP-positive PC12 cells was measured as described under “Experimental Procedures.” Values represent the means ± S.E. (n = 3). NGF and Bt2cAMP significantly increased neurite length (*, p < 0.05), and the effect of NGF was reversed by ERK5KD (#, p < 0.05). B, parental PC12 cells or PC12 cells stably overexpressing MEK5A mutant were incubated with NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for a day. The nuclei were stained with Hoechst-33258, and the morphological changes were observed. Scale bar, 100 μm. Neurite length of PC12 cells was measured as described under “Experimental Procedures.” Values represent the means ± S.E. (n = 3). NGF and Bt2cAMP significantly increased neurite length (*, p < 0.05), and the effect of NGF was reversed by MEK5A overexpression (#, p < 0.05).
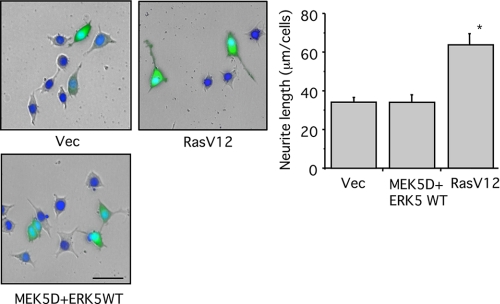
FIGURE 7.
ERK5 is not sufficient for neurite outgrowth in PC12 cells. PC12 cells were co-transfected with GFP and empty vector (Vec), MEK5D + ERK5WT, or RasV12. Then the cells were incubated for 2 days. The nuclei were stained with Hoechst-33258 (blue), and morphological changes of GFP-positive cells were observed under a fluorescence microscope. Scale bar, 50 μm. Neurite length of GFP-positive PC12 cells was measured as described under “Experimental Procedures.” Values represent the means ± S.E. (n = 3). RasV12 significantly increased neurite length (*, p < 0.05).
Luciferase Assay
The DNA plasmids were transfected into PC12 cells using the transfection reagent Lipofectamine 2000. Briefly, the cells were seeded onto 24-well plates at 1 × 105 (cells/well) and cultivated for a day. The DNA plasmids ((0.25 μg of Elk-1/Gal, 0.25 μg of 5× Gal4-E1B/Luciferase, 0.2 μg of β-galactosidase, and 0.3 μg of ERK5/MEK5 mutants or 0.1 μg of MEF2C/Gal, 0.1 μg of Gal-luciferase, 0.1 μg of β-galactosidase, and 1.2 μg of ERK5 mutants or 0.4 μg of tyrosine hydroxylase promoter/luciferase, 0.4 μg of ERK5 or MEK5 mutants, and 0.2 μg of β-galactosidase)) and transfection reagent (1 μl/tube) were mixed gently in DMEM (10 μl/tube) and incubated for 20 min at room temperature. After the addition of DMEM (40 μl/tube), this entire mixture was transferred to the cultured media (50 μl/well), which had been replaced with serum-free DMEM (200 μl). The cells were incubated for 4 h at 37 °C, and then the media were replaced with growth medium (500 μl) containing 10% fetal calf serum and 5% horse serum. For reporter gene assays, the cells were incubated with drugs at 37 °C for 6–8 h after serum starvation and subjected to luciferase assay. Cells were lysed in lysis buffer (1% Triton X-100, 110 mm K2HPO4, 15 mm KH2PO4 (pH 7.8)) (100 μl/well). Then after centrifuging the lysates to remove the cell debris, the supernatant (50 μl/tube) was mixed with 300 μl of assay buffer (25 mm Gly-Gly, 15 mm MgSO4, 5 mm ATP, 10 mm NaOH). The luciferase reaction was started by adding 100 μl of luciferin solution (150 μm), and luciferase activity was measured using a luminometer (GENE LIGHT 55, Microtech Nition, Funabashi, Japan). As an internal control, β-actin promoter-driven β-galactosidase activity was measured in the lysates to normalize for the transfection efficiency.
Adenoviral Infection
PC12 cells were infected with adenoviruses encoding dominant-negative RasN17 and FLAG-Rap1GAP1 that block endogenous activities of Ras and Rap1, respectively (31). The infection was carried out for 3 days at 50 m.o.i. for RasN17 adenovirus.
FLAG-Rap1GAP1 adenovirus at 50 m.o.i. was applied to cells together with transactivating virus at 250 m.o.i. Control cells were infected with transactivating virus alone. After infection for 3 days, the medium was replaced with serum-free medium, and the cells were stimulated with drugs.
Statistics
Data were expressed as the mean values ± S.E., and significant differences were analyzed using Tukey's method.
RESULTS
NGF, but Not cAMP, Induces ERK5 Phosphorylation in PC12 Cells
To examine whether NGF or cAMP can activate ERK5 in PC12 cells, PC12 cells were stimulated with NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for 3–360 min. Then phosphorylation of ERK5 and ERK1/2 was observed by Western blotting (Fig. 1). As shown previously, NGF induced the sustained phosphorylation of ERK5 and ERK1/2 (24, 32). In contrast, although Bt2cAMP induced ERK1/2 phosphorylation, it did not induce ERK5 phosphorylation.
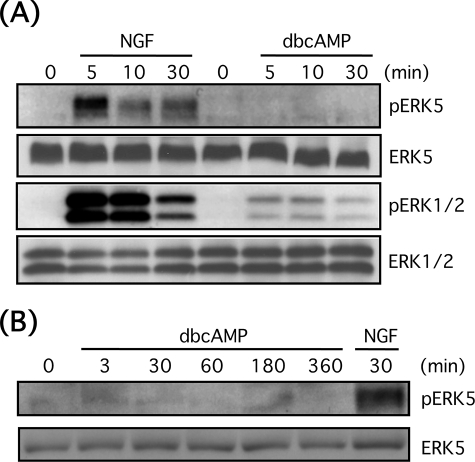
FIGURE 1.
NGF, but not Bt2cAMP (dbcAMP) causes ERK5 phosphorylation in PC12 cells. A, PC12 cells were stimulated with NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for 5–30 min. Then phospho-ERK5, total ERK5, phospho-ERK1/2, and total ERK1/2 were examined by Western blotting. B, PC12 cells were stimulated with Bt2cAMP (0.5 mm) for 3–360 min or NGF (100 ng/ml) for 30 min. Then phospho-ERK5 and total ERK5 were examined by Western blotting.
Whereas it has been reported that MEK inhibitors, such as PD98059 and U0126, block MEK5 as well as MEK1/2 (16), it has been shown that these inhibitors do not block ERK5 signaling efficiently at low doses (33–35). To examine the selectivity of these MEK inhibitors in our system, PC12 cells were preincubated with U0126 (10 or 30 μm) for 30 min and then further stimulated with NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for 10 min. Although ERK1/2 phosphorylation by both reagents was abolished by pretreatment with U0126, ERK5 phosphorylation was not blocked at all (Fig. 2A). Another MEK inhibitor, PD98059, also suppressed ERK1/2 phosphorylation by NGF and Bt2cAMP but did not suppress ERK5 phosphorylation by NGF (Fig. 2B). Judging from these results, we used U0126 as a selective inhibitor for ERK1/2 signaling in this study. Recently, BIX02188 and BIX 02189 were developed as selective pharmacological inhibitors of the MEK5/ERK5 pathway (36). PC12 cells were pretreated with or without BIX02188 and BIX02189 (3–30 μm) for 30 min, and the cells were stimulated with NGF (100 ng/ml) for 5 min. Both inhibitors blocked ERK5 phosphorylation in a concentration-dependent manner, whereas phosphorylation levels of ERK1/2 were unaffected (Fig. 2C). Therefore, we used BIX02188 and BIX02189 as selective pharmacological inhibitors for ERK5 signaling in this study.
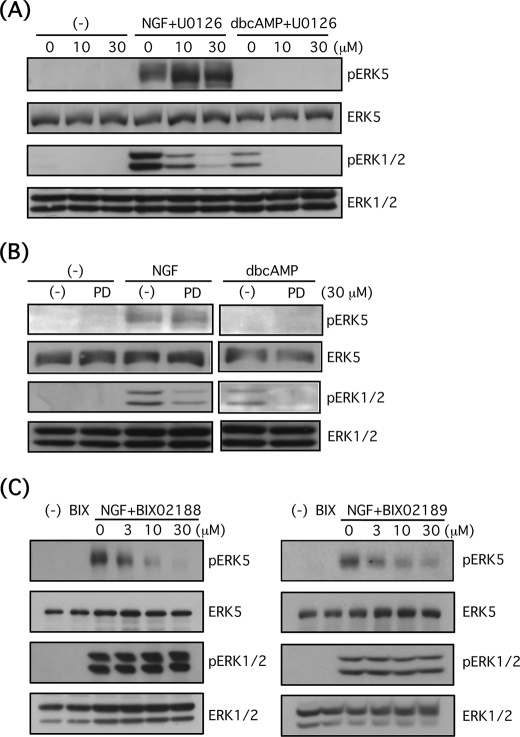
FIGURE 2.
U0126 and PD98059 selectively block ERK1/2 signaling, and BIX02188 and BIX02189 selectively inhibit ERK5 signaling in PC12 cells. A, after PC12 cells were pretreated with or without U0126 (10 or 30 μm) for 30 min, the cells were stimulated with NGF (100 ng/ml) or Bt2cAMP (dbcAMP) (0.5 mm) for 10 min. Then phospho-ERK5, total ERK5, phospho-ERK1/2, and total ERK1/2 were examined by Western blotting. B, after PC12 cells were pretreated with or without PD98059 (PD) (30 μm) for 30 min, the cells were stimulated with NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for 10 min. Then phospho-ERK5, total ERK5, phospho-ERK1/2, and total ERK1/2 were examined by Western blotting. C, after PC12 cells were pretreated with or without BIX02188 and BIX02189 (BIX) (3–30 μm) for 30 min, the cells were stimulated with NGF (100 ng/ml) for 5 min. Then phospho-ERK5, total ERK5, phospho-ERK1/2, and total ERK1/2 were examined by Western blotting.
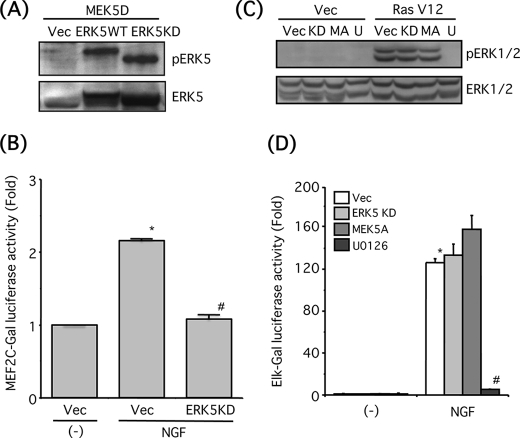
Next, we attempted to confirm the selectivity of dominant-negative mutants of ERK5 or MEK5. HEK293 cells were co-transfected with the constitutively active MEK5 mutant (MEK5D) and empty vector, ERK5 wild-type (ERK5WT), or dominant-negative ERK5 mutant (ERK5KD). MEK5D caused phosphorylation of both ERK5WT and ERK5KD, but the band shift of ERK5KD was smaller because ERK5KD lacks its kinase activity and subsequent autophosphorylation (Fig. 3A). It has been established that ERK5 and p38 MAPK also phosphorylate MEF2C at Ser-387 that is critical for transcriptional activation. In addition, p90 ribosomal S6 kinase 2 activated by ERK1/2 phosphorylates MEF2C on Ser-192 that is also required for transcriptional activation (37). To confirm the inhibitory effect of ERK5KD on endogenous ERK5 activity, MEF2C-dependent transcription was examined. PC12 cells were stimulated by NGF (100 ng/ml) for 8 h in the presence of U0126 (10 μm) and a p38 MAPK inhibitor SB203580 (1 μm). NGF significantly promoted MEF2C-dependent transcription, and this effect was significantly blocked by ERK5KD (Fig. 3B). Because constitutively active Ras oncogenic mutant (RasV12) is a strong activator for ERK1/2 signaling (38), we examined whether ERK5KD or dominant-negative MEK5 mutant (MEK5A) interferes with ERK1/2 phosphorylation induced by RasV12 in PC12 cells. Although U0126 completely inhibited ERK1/2 phosphorylation, the phosphorylation was not affected by ERK5KD or MEK5A (Fig. 3C). Luciferase expression resulting from Elk1 phosphorylation by ERK1/2 was often regarded as an index of in vivo ERK1/2 kinase activity, and it has been shown that ERK5 does not phosphorylate Elk1 (16, 25). NGF (100 ng/ml, 6 h) robustly promoted ERK1/2 activity, and this effect was completely blocked by U0126 but not affected by ERK5KD or MEK5A (Fig. 3D). These results suggest that ERK5KD or MEK5A does not affect ERK1/2 signaling.
FIGURE 3.
Dominant-negative mutants of ERK5 and MEK5 do not suppress ERK1/2 signaling. A, HEK293 cells were co-transfected with a constitutively active mutant of MEK5 (MEK5D) and empty vector (Vec), ERK5 wild-type (WT), or ERK5 kinase-dead mutant (KD). Then phospho-ERK5 and total ERK5 were examined by Western blotting. B, PC12 cells were co-transfected with MEF2C/Gal, 5× Gal4-E1B/Luciferase, and β-galactosidase and empty vector (Vec) or ERK5KD. Two days after transfection, the cells were incubated with NGF (100 ng/ml) for 8 h in the presence of U0126 (10 μm) and SB203580 (1 μm). MEF2C-Gal-dependent transcription activity was monitored by measuring luciferase activity as described under “Experimental Procedures.” MEF2C-gal-dependent transcription activity was expressed as the fold over the control (no drug). Values represent the means ± S.E. (n = 3). NGF significantly increased the MEF2C-gal-dependent transcription activity (*, p < 0.05) and ERK5KD significantly reduced the transcription activity (#, p < 0.05). C, PC12 cells were co-transfected with Ras oncogenic mutant (RasV12) and empty vector (Vec), ERK5 KD (KD), or dominant-negative mutant of MEK5 (MEK5A, MA). The cells were incubated with or without U0126 (U) (30 μm) for 30 min, and then phospho-ERK1/2 and total ERK1/2 were examined by Western blotting. D, PC12 cells were co-transfected with Elk-1/Gal, 5× Gal4-E1B/Luciferase, and β-galactosidase and empty vector, ERK5KD or MEK5A. Two days after transfection, the cells were pretreated with or without U0126 (30 μm) for 30 min and incubated with NGF (100 ng/ml) for 6 h. ERK1/2 kinase activity was monitored by measuring luciferase activity as described under “Experimental Procedures.” Transfection efficiency was normalized by measuring β-galactosidase activity. ERK1/2 kinase activity was expressed as the fold over the control (no drug). Values represent the means ± S.E. (n = 3). NGF significantly increased ERK1/2 kinase activity (*, p < 0.05), and U0126 abolished the kinase activity (#, p < 0.05).
Neither Ras Nor Rap1 Mediates ERK5 Phosphorylation by NGF in PC12 Cells
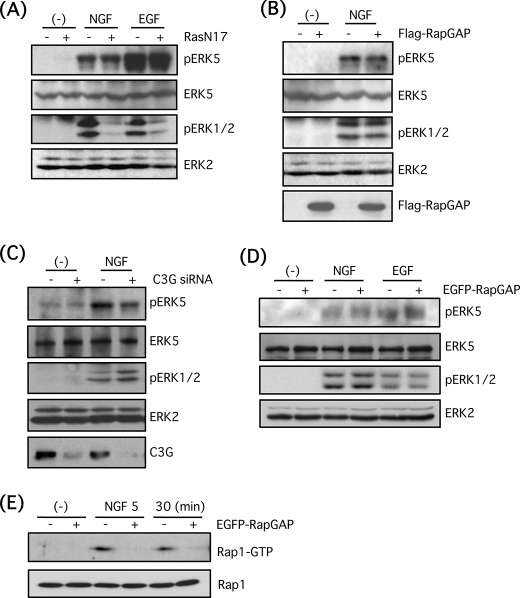
Whereas it has been shown that Ras regulates ERK5 activity in PC12 cells and COS7 cells (16), there are reports demonstrating that Ras was not involved in ERK5 activation by growth factors or GPCR agonists such as carbachol and thrombin (10, 20). On the other hand, Rap1, another small G-protein, is necessary for ERK5 activation by BDNF in cortical neurons (17). Other studies examining Rap1 have shown contrasting results that Rap1 activated by exchange protein activated by cAMP inhibits ERK5 activity within protein complexes containing protein kinase A anchoring protein (39). Thus, the involvement of Ras and Rap1 in ERK5 activation is controversial. Therefore, we attempted to examine the involvement of these small G-proteins in ERK5 activation in PC12 cells. PC12 cells were infected with adenovirus encoding dominant-negative mutant of Ras (RasN17) for 3 days at 50 m.o.i. Then the cells were stimulated with NGF (100 ng/ml) or EGF (100 ng/ml) for 5 min. Although the Ras-dependent ERK1/2 phosphorylation by NGF and EGF was largely reduced by RasN17, ERK5 phosphorylation by these growth factors was resistant to RasN17, suggesting that Ras is not involved in ERK5 phosphorylation by NGF and EGF in PC12 cells (Fig. 4A). Similarly, FLAG-Rap1GAP1 that inactivates Rap1 activity was overexpressed by adenoviral infection at 50 m.o.i. for 3 days, and then PC12 cells were stimulated with NGF (100 ng/ml) for 5 min. ERK5 phosphorylation was resistant to Rap1GAP1 as shown in the case of RasN17 (Fig. 4B). In addition, Rap1 activity was completely blocked by infection of adenovirus encoding FLAG-Rap1GAP1 (data not shown). Also, a major Rap1 guanine nucleotide exchange factor, C3G, is known to mediate Rap1 activation by NGF (32, 40). Therefore, we attempted to block Rap1 activation by C3G knockdown. In the condition that C3G expression is diminished by siRNA, ERK5 phosphorylation was not largely reduced (Fig. 4C). Furthermore, ERK5 phosphorylation by NGF or EGF was examined in PC12 cells that stably overexpress EGFP-Rap1GAP. However, ERK5 phosphorylation was not blocked again by NGF (100 ng/ml) or EGF (100 ng/ml) (Fig. 4D). In EGFP-Rap1GAP cells, Rap1 activation by NGF was efficiently reduced, as determined by pulldown assay with GST-RalGDS (Fig. 4E). Taking these results together, ERK5 phosphorylation by NGF is Ras- and Rap1-independent.
FIGURE 4.
Small G-proteins, Ras and Rap1, are not involved in ERK5 phosphorylation by NGF in PC12 cells. A, PC12 cells were infected with adenovirus encoding the dominant-negative mutant of Ras (RasN17) for 3 days at 50 m.o.i. Then the cells were stimulated with NGF (100 ng/ml) or EGF (100 ng/ml) for 5 min, and phospho-ERK5, total ERK5, phospho-ERK1/2, and total ERK2 were examined by Western blotting. B, PC12 cells were infected with adenovirus encoding FLAG-Rap1GAP1 for 3 days at 50 m.o.i. in the presence of transactivating virus (250 m.o.i.). Then the cells were stimulated with NGF (100 ng/ml) for 5 min, and phospho-ERK5, total ERK5, phospho-ERK1/2, total ERK2, and FLAG-Rap1GAP1 were examined by Western blotting. C, PC12 cells were transfected with double-stranded siRNA for C3G. After 3 days, the cells were stimulated with NGF (100 ng/ml) for 5 min, and phospho-ERK5, total ERK5, phospho-ERK1/2, total ERK2, and C3G were examined by Western blotting. D, EGFP-Rap1GAP-positive or -negative PC12 cells were stimulated with NGF (100 ng/ml) or EGF (100 ng/ml) for 5 min, and phospho-ERK5, total ERK5, phospho-ERK1/2, and total ERK2 were examined by Western blotting. E, EGFP-Rap1GAP-positive or -negative PC12 cells were stimulated with NGF (100 ng/ml) for 5 and 30 min, and Rap1 pulldown was performed as described under “Experimental Procedures.” Rap1-GTP and total Rap1 were examined by Western blotting.
ERK5 Activity Is Required for Neurite Outgrowth Induced by NGF in PC12 Cells
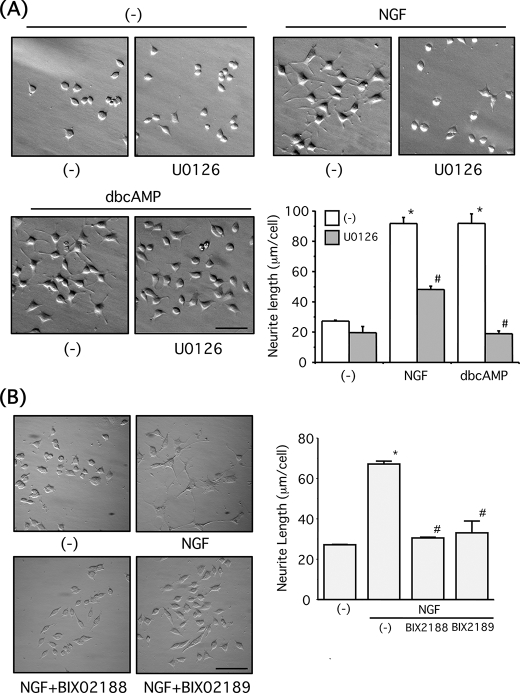
PC12 cells extend neurites in response to NGF or Bt2cAMP in an ERK1/2-dependent manner (24). As shown previously, we confirmed that U0126 significantly attenuated the neurite outgrowth by both NGF and Bt2cAMP (Fig. 5A). Furthermore, both BIX02188 and BIX02189 (30 μm) significantly reduced neurite extension by NGF (100 ng/ml, 1 day) in PC12 cells (Fig. 5B), indicating ERK5 is required for the NGF-induced neurite outgrowth. Next, PC12 cells were co-transfected with GFP and empty vector or ERK5KD and then further stimulated with NGF (100 ng/ml, 1 day) or Bt2cAMP (0.5 mm, 1 day). When GFP-positive cells were examined, ERK5KD significantly reduced the NGF-induced neurite extension (Fig. 6A). However, the neurite outgrowth induced by Bt2cAMP was not attenuated by ERK5KD overexpression (Fig. 6A). Similarly, the neurite outgrowth induced by NGF (100 ng/ml, 1 day) but not Bt2cAMP (0.5 mm, 1 day) was significantly inhibited in PC12 cells that stably overexpress MEK5A (Fig. 6B). These biochemical and pharmacological experiments strongly indicate that ERK5 activity is required for the NGF-induced neurite outgrowth in PC12 cells. To investigate whether ERK5 is sufficient for neurite outgrowth, the cells were co-transfected with GFP and empty vector or MEK5D + ERK5. RasV12 was used as a positive control. Whereas RasV12 caused neurite outgrowth, MEK5D + ERK5 did not induce significant neurite outgrowth in PC12 cells (Fig. 7), suggesting ERK5 is necessary but not sufficient for neurite outgrowth.
FIGURE 5.
Pharmacological inhibitors for ERK1/2 and ERK5 block neurite outgrowth induced by NGF in PC12 cells. A, PC12 cells were pretreated with or without U0126 (30 μm) for 30 min. After incubation with NGF (100 ng/ml) or Bt2cAMP (dbcAMP) (0.5 mm) for a day, the nuclei were stained with Hoechst-33258, and the morphological changes were observed. Scale bar, 100 μm. Neurite length of PC12 cells was measured as described under “Experimental Procedures.” Values represent the means ± S.E. (n = 3). NGF and Bt2cAMP significantly increased neurite length (*, p < 0.05), and these effects were reversed by U0126 (#, p < 0.05). B, PC12 cells were incubated with NGF (100 ng/ml) for a day in the presence of BIX02188 or BIX02189 (30 μm) for a day. The nuclei were stained with Hoechst-33258, and the morphological changes were observed. Scale bar, 100 μm. Neurite length of PC12 cells was measured as described under “Experimental Procedures.” Values represent the means ± S.E. (n = 3). NGF significantly increased neurite length (*, p < 0.05), and these effects were reversed by BIX02188 and BIX02189 (#, p < 0.05).
ERK5 Activity Is Required for Stabilization of Tyrosine Hydroxylase in PC12 Cells
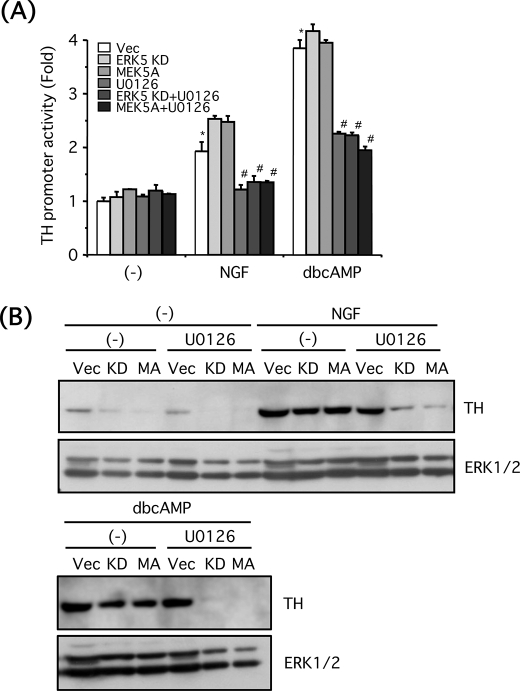
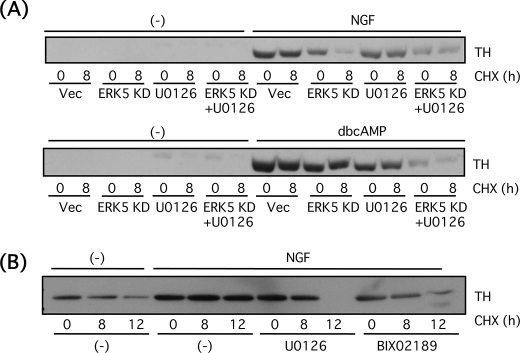
Catecholamines such as dopamine and norepinephrine are major neurotransmitters (hormones) secreted from PC12 cells, and tyrosine hydroxylase is one of the rate-limiting enzymes for the catecholamine biosynthesis. Because ERK5 is required for morphological differentiation of PC12 cells by NGF, we next investigated whether ERK5 is involved in functional differentiation of PC12 cells by determining tyrosine hydroxylase expression. After PC12 cells were incubated in the presence of NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for a day, tyrosine hydroxylase protein was up-regulated (Fig. 8B). Tyrosine hydroxylase gene promoter contains multiple transcription factor binding sites (41), and some of the transcription factors such as AP-1 (activator protein-1) and cAMP response element-binding protein (CREB) involved in the tyrosine hydroxylase transcription are thought to be regulated by ERK5 (6, 9, 11). To test this, PC12 cells were transfected with DNA plasmid encoding the tyrosine hydroxylase gene promoter linked to the luciferase gene thus allowing us to measure the promoter activity. Both NGF (100 ng/ml, 6 h) and Bt2cAMP (0.5 mm, 6 h) increased the promoter activity, and these effects were largely inhibited by U0126 (30 μm). However, ERK5KD or MEK5A did not affect the promoter activity in the presence or absence of U0126 (Fig. 8A). This result suggests ERK1/2 but not ERK5 is required for activation of tyrosine hydroxylase promoter. The tyrosine hydroxylase protein expression level was also investigated. Basal tyrosine hydroxylase expression level was reduced by blocking ERK5 signaling (Fig. 8B). When tyrosine hydroxylase was up-regulated by NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for 24 h, the inhibitory effect of ERK5KD or MEK5A alone was small. However, tyrosine hydroxylase protein expression was largely blocked when ERK5 and ERK1/2 signaling was inhibited by the interfering mutants and U0126 (30 μm). NGF-induced tyrosine hydroxylase levels were reduced by 81% in the presence of ERK5KD + U0126 and by 90% in the presence of MEK5A + U0126. Bt2cAMP-induced tyrosine hydroxylase levels were reduced by 94% in the presence of ERK5KD + U0126 and by 97% in the presence of MEK5A + U0126 (Fig. 8B). Taking these results together, it is suggested that ERK5 regulates either translational or post-translational modification of tyrosine hydroxylase, in conjunction with ERK1/2. To clarify the role of ERK5 in regulation of tyrosine hydroxylase protein level in more detail, PC12 cells transfected with empty vector or ERK5KD were incubated with NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for a day in the presence or absence of U0126 (30 μm) and then treated for 8 h with cycloheximide (20 μg/ml) to block translation. Despite cycloheximide treatment, tyrosine hydroxylase protein expression continued to be inhibited by the combined blockade of both ERK5 and ERK1/2 again as seen in Fig. 8B (70% reduction compared with that of NGF alone and 79% reduction compared with Bt2cAMP alone) (Fig. 9A). Even after treatment with cycloheximide for 8 h, tyrosine hydroxylase protein up-regulated by NGF or Bt2cAMP was stable and not altered by U0126. However, in case of NGF, but not Bt2cAMP, the tyrosine hydroxylase protein was unstable in the presence of ERK5KD. In the presence of cycloheximide, ERK5KD induced a 78% reduction in tyrosine hydroxylase levels (Fig. 9A). The similar experiment was performed by using BIX02189 (30 μm) to block MEK5 signaling. Again, tyrosine hydroxylase was degraded in a time-dependent manner in the presence of cycloheximide (20 μg/ml) (21% reduction at 8 h and 60% reduction at 12 h), and tyrosine hydroxylase up-regulated by NGF was stabilized and resistant to cycloheximide up to at least 12 h (Fig. 9B). However, in the presence of U0126 (30 μm), tyrosine hydroxylase 8 h after cycloheximide treatment was stable (14% reduction), as seen in Fig. 9A, but was degraded after 12 h (97% reduction). On the other hand, in the presence of BIX02189 (30 μm), tyrosine hydroxylase stability was reduced (33% reduction at 8 h and 68% reduction at 12 h) (Fig. 9B). These results indicate that ERK5 is involved in stabilization of tyrosine hydroxylase in the case of NGF.
FIGURE 8.
ERK5 is involved in up-regulation of tyrosine hydroxylase protein in PC12 cells. A, PC12 cells were co-transfected with tyrosine hydroxylase promoter/luciferase and β-galactosidase and empty vector (Vec), ERK5KD, MEK5D, or MEK5A. Two days after transfection, the cells were pretreated with or without U0126 (30 μm) for 30 min and incubated with NGF (100 ng/ml) or Bt2cAMP (dbcAMP) (0.5 mm) for 6 h. Tyrosine hydroxylase gene promoter activity was expressed as the fold over the control (no drug). Values represent the means ± S.E. (n = 3). NGF and Bt2cAMP significantly increased the promoter activity (*, p < 0.05), and U0126 abolished the promoter activity (#, p < 0.05). B, PC12 cells were transfected with empty vector, ERK5KD (KD), or MEK5A (MA). Two days after the transfection, the cells were pretreated with or without U0126 (30 μm) for 30 min and incubated with NGF (100 ng/ml) or Bt2cAMP (0.5 mm) for 24 h. Then tyrosine hydroxylase and ERK1/2 were examined by Western blotting.
FIGURE 9.
ERK5 stabilizes tyrosine hydroxylase protein in PC12 cells. A, PC12 cells were transfected with empty vector (Vec) or ERK5KD (KD). Two days after transfection, the cells were pretreated with or without U0126 (30 μm) for 30 min and incubated with NGF (100 ng/ml) or Bt2cAMP (dbcAMP) (0.5 mm) for 24 h. After incubating with cycloheximide (CHX; 20 μg/ml) for 8 h, tyrosine hydroxylase was examined by Western blotting. B, PC12 cells were pretreated with or without U0126 (30 μm) or BIX02189 (30 μm) for 30 min and incubated with NGF (100 ng/ml) for 24 h. After incubating with cycloheximide (CHX) (20 μg/ml) for 8 or 12 h, tyrosine hydroxylase (TH) was examined by Western blotting.
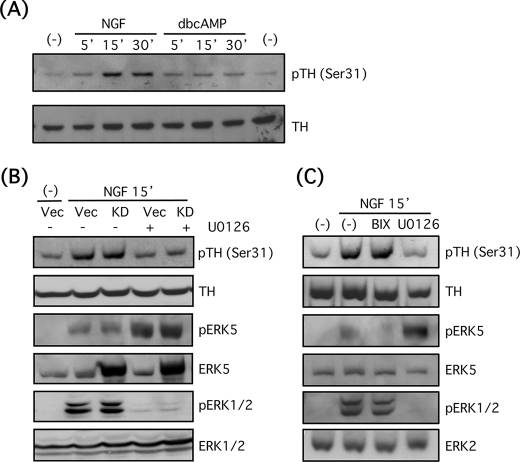
Tyrosine hydroxylase has at least four phosphorylation sites at Ser-8, -19, -31, and -40, and the phosphorylation was induced by multiple kinases such as protein kinase A (42). It has been demonstrated that tyrosine hydroxylase at Ser-31 is phosphorylated by ERK1/2 or its downstream effector, cyclin-dependent kinase 5, and this phosphorylation site is important for stabilization of tyrosine hydroxylase (42). For this reason, we attempted to test the hypothesis that stability of tyrosine hydroxylase protein is also regulated by Ser-31 phosphorylation by ERK5 or its downstream effector. As expected, NGF (100 ng/ml) induced phosphorylation of tyrosine hydroxylase at Ser-31 in a time-dependent manner (2.3-fold at 5 min, 5.7-fold at 15 min, and 4.3-fold at 30 min) (Fig. 10A). On the other hand, the effect of Bt2cAMP (0.5 mm) was minimal (2.0-fold at 5 min, 2.2-fold at 15 min, and 2.2-fold at 30 min) (Fig. 10A). Next, we examined whether ERK5 was required for the NGF-induced phosphorylation of tyrosine hydroxylase. Although the NGF-induced phosphorylation was not altered at all by overexpression of the dominant-negative ERK5 mutant, U0126 (30 μm) attenuated this phosphorylation (72% inhibition) (Fig. 10B). In addition, BIX02189 (30 μm) showed no inhibitory effect on the NGF-induced phosphorylation of tyrosine hydroxylase (Fig. 10C). These results suggest that ERK5 is essential for stabilization of tyrosine hydroxylase but that this is independent of the phosphorylation status at Ser-31.
FIGURE 10.
ERK1/2 but not ERK5 promotes tyrosine hydroxylase phosphorylation at Ser-31 in PC12 cells. A, PC12 cells were incubated with NGF (100 ng/ml) or Bt2cAMP (dbcAMP) (0.5 mm) for 5, 15, and 30 min. Then phosphotyrosine hydroxylase (Ser-31) and tyrosine hydroxylase (TH) were examined by Western blotting. B, PC12 cells were transfected with empty vector (Vec) or ERK5KD (KD). Two days after transfection, the cells were pretreated with or without U0126 (30 μm) for 30 min and incubated with NGF (100 ng/ml) for 15 min. Then phosphotyrosine hydroxylase (pTH) (Ser-31), tyrosine hydroxylase, and phospho-ERK5, total ERK5, phospho-ERK1/2, and ERK1/2 were examined by Western blotting. C, PC12 cells were pretreated with or without BIX02189 (BIX; 30 μm) or U0126 (30 μm) for 30 min and incubated with NGF (100 ng/ml) for 15 min. Then phosphotyrosine hydroxylase (Ser-31), tyrosine hydroxylase, and phospho-ERK5, total ERK5, phospho-ERK1/2, and ERK2 were examined by Western blotting.
DISCUSSION
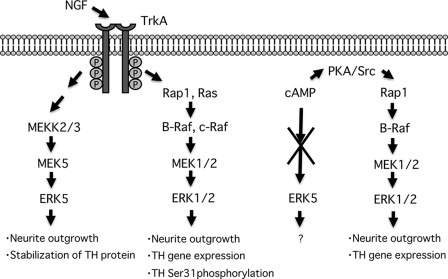
In this study, we investigated the physiological role of ERK5 in differentiation of PC12 cells using both biochemical and pharmacological approaches. We found for the first time that ERK5 activation was required for NGF-induced neurite outgrowth and stabilization of the neurotransmitter-synthesizing enzyme tyrosine hydroxylase, whereas ERK1/2 activity is required for neurite outgrowth, gene expression of tyrosine hydroxylase, and phosphorylation of the enzyme at Ser-31. cAMP induces only ERK1/2 phosphorylation that is required for neurite outgrowth and gene expression of tyrosine hydroxylase (Fig. 11).
FIGURE 11.
Putative signaling pathways and physiological roles of ERK5 and ERK1/2 in PC12 cells.
It has been demonstrated that conventional MEK inhibitors, including U0126 and PD98059, block MEK5 as well as MEK1/2 (16). Nevertheless, other studies have suggested that MEK5/ERK5 signaling is resistant to these inhibitors (34) or that extremely high concentrations (100 μm) are required to block ERK5 phosphorylation (33, 35). At the concentrations used in this study, we show that these inhibitors selectively blocked ERK1/2 signaling (Fig. 2, A and B). In contrast, we confirmed that BIX02188 and BIX02189, new MEK5/ERK5 inhibitors, blocked ERK5 phosphorylation efficiently without affecting ERK1/2 phosphorylation status (Fig. 2C), which enabled us to distinguish the role of ERK5 from ERK1/2. Whereas NGF and EGF induced the phosphorylations of both ERK5 and ERK1/2, cAMP only induced ERK1/2 phosphorylation (Figs. 1 and 2). This is consistent with the previous reports that neither cAMP, Ca2+, nor glutamate can stimulate ERK5 activity in cortical neurons (25) and that cAMP negatively regulates ERK5 activity (21, 22). Thus, cAMP can be regarded as a selective ERK1/2 activator among the ERKs examined in this study.
ERK5 is required for neuronal survival by neurotrophin such as NGF and BDNF (9, 11) or proliferation by growth factors (10). However, the physiological role of ERK5 in neuronal differentiation by neurotrophin/growth factors has been poorly understood. Only a few studies have been reported that abnormal neuronal development was observed by ERK5 knockdown using antisense morpholino oligonucleotides in X. laevis (13) and that ERK5 is required for generation of neurons from cortical progenitors (14). In this study, we demonstrated for the first time that ERK5 is essential for neurite outgrowth induced by NGF in PC12 cells and stabilization of the neuron-specific gene such as tyrosine hydroxylase. ERK5 activity is not sufficient to induce neurite outgrowth in this study (Fig. 7), suggesting that other signaling pathways are also required. To promote neural differentiation, the role of ERK1/2 pathway is generally thought to involve the regulation of transcription of genes that are necessary for the differentiation (43). Similarly, it has been proposed that neuronal differentiation by ERK5 results from activation of transcription factors. Transcription factors, including Sap-1, AP-1, CREB, nuclear factor-κB, and MEF2C, are directly or indirectly activated by ERK5 (9, 11, 12, 16, 44–46). Thus, these transcription factors are candidates that mediate neuronal differentiation by ERK5.
ERK1/2 activity was required for tyrosine hydroxylase gene expression (Fig. 8A). The tyrosine hydroxylase promoter includes multiple regulation sites for transcription factors, including AP-1, SP-1, and CREB, and the activity of certain factors is also regulated by ERK5 in addition to ERK1/2. However, ERK5-dependent activation of the tyrosine hydroxylase promoter was not observed in this study (Fig. 8A). Surprisingly, ERK5 activity was essential for maintaining levels of tyrosine hydroxylase protein (Fig. 8B). This action also involved ERK1/2 because blocking both ERK5 and ERK1/2 signaling was required to abolish up-regulation of the tyrosine hydroxylase protein (Fig. 8B). A rationale for these effects is that ERK5 regulates the stability of tyrosine hydroxylase (Fig. 9). Tyrosine hydroxylase is the rate-limiting enzyme for catecholamine synthesis and is important for maintaining the catecholamine content. Therefore, we further investigated the mechanism of ERK5 to stabilize tyrosine hydroxylase protein. It has been demonstrated that tyrosine hydroxylase phosphorylation at Ser-31 is important for its stability and that ERK1/2 or its downstream effector, cyclin-dependent kinase 5, can phosphorylate this site (42). This phosphorylation triggers the conformational change of tyrosine hydroxylase to one that is more resistant to proteases or ubiquitination. Thus, we attempted to examine the role of ERK5 in phosphorylation of tyrosine hydroxylase. Although we found that NGF increased its phosphorylation at Ser-31, it was completely blocked only by blocking MEK1/2 but not MEK5, indicating that this phosphorylation was ERK1/2-dependent (Fig. 10). Hence, it is unlikely that stabilization of tyrosine hydroxylase by ERK5 depends on this Ser-31 phosphorylation status. However, we cannot rule out the possibility that ERK5 phosphorylates another site that influences its stability. We also observed that ERK5 regulated the stability of other neuron-specific proteins such as the neurofilament light chain,3 suggesting that the protein stabilization by ERK5 is assumed to be not tyrosine hydroxylase-specific but a more general phenomenon. Recently, it was demonstrated that glycogen synthase kinase-3β (GSK3β) was inactivated by phosphorylation at Ser-9 residue by ERK5 and that a product of the early response gene, Snail protein, was stabilized resulting from inactivation of GSK3β, as in case of β-catenin (47). Thus, negative regulation of GSK3β by ERK5 is an alternative explanation for the mechanism of ERK5 for the increase in protein stability.
In this study, cAMP did not, by itself, cause ERK5 phosphorylation (Figs. 1 and 2). However, tyrosine hydroxylase protein level was reduced by blocking both ERK5 and ERK1/2 signaling as in case of NGF (Fig. 8B). A possible assumption to explain this result is that basal ERK5 kinase activity is required for stabilization of tyrosine hydroxylase. Another possibility is that this reflects a novel cross-talk mechanism between ERK5 and ERK1/2. Recently, it was shown that the ERK5 carboxyl terminus was phosphorylated by the amino-terminal half of ERK5 that included the kinase domain (6). The amino-terminal half of ERK5 shares homology with ERK1/2. Hence, if the amino-terminal half of ERK5 can induce phosphorylation of the ERK5 carboxyl terminus, it may be possible that ERK1/2 phosphorylates the ERK5 carboxyl terminus, which influences ERK5 transcription activity independently of ERK5 kinase activation. In fact, endogenous ERK5 can form a complex with ERK2.3 cAMP may utilize the ERK5 signaling pathway by phosphorylation of the ERK5 carboxyl terminus by ERK1/2.
It has been shown that ERK5 activation by NGF and EGF in PC12 cells is largely blocked by the dominant-negative mutant of Ras (RasN17) (16). In addition, a constitutively active mutant, RasV12, caused ERK5 activation in PC12 cells (16), indicating Ras is responsible for growth factor signaling to ERK5. Nevertheless, EGF-induced ERK5 phosphorylation was not inhibited by RasN17, and RasV12 did not activate ERK5 in HeLa cells (10). In cortical neurons, ERK5 activation by BDNF was not blocked by RasN17 (17). In COS7 cells, signaling from thrombin PAR1 or muscarinic 1 receptors to ERK5 did not require Ras and Rho family GTPases (20). In our study, RasN17 blocked neither NGF nor EGF-induced ERK5 phosphorylation, although ERK1/2 phosphorylation in the same cell lysates was largely blocked RasN17 (Fig. 4A). Also, RasV12 was not sufficient for ERK5 phosphorylation although ERK1/2 phosphorylation was observed by this mutant (22). In one study showing that Ras was involved in ERK5 activation (16), the ERK5 kinase assay was carried out using myelin basic protein as a substrate after ERK5 immunoprecipitation. However, the myelin basic protein is a well known substrate for ERK1/2 as well. In our preliminary experiments, endogenous ERK5 forms a complex with ERK2.3 Therefore, it is possible that ERK1/2 was contaminated in their ERK5 immunoprecipitates, and the results were influenced by Ras-dependent ERK1/2 activity in their experiments. Taking these results together, we conclude that Ras is not responsible for NGF or EGF-induced ERK5 activation in PC12 cells.
In terms of Rap1, Rap1 instead of Ras was involved in BDNF activation of ERK5 in cortical neurons (17). However, Rap1 that is activated by exchange protein activated by cAMP inactivates ERK5 in protein kinase A anchoring protein-associated pools (39). In our previous studies, cAMP strongly activated Rap1 through protein kinase A and Src tyrosine kinase in PC12 cells (24). However, Rap1 activated by forskolin did not induce ERK5 phosphorylation (22). In this study, ERK5 was not activated by a cAMP analog capable of activating Rap1 (Fig. 1). Moreover, EGF activated ERK5 in PC12 cells (Fig. 4) (22) although EGF did not increase Rap1 activity in PC12 cells determined by pulldown assay (24). These published results strongly suggest that Rap1 is not a major transducer for ERK5 activation in PC12 cells. In this study, we took two distinct approaches as follows: 1) overexpression of Rap1GAP1, an inactivator of Rap1; and 2) siRNA knockdown of C3G GEF that mediates activation of Rap1 by NGF (32, 40). Our results indicate Rap1 is not responsible for ERK5 activation by NGF (Fig. 4), and it is assumed that the Rap1-dependent ERK5 activation in cortical neurons (17) may be cell type-specific. It has been demonstrated that a Rho GAP, NOMA-GAP, is essential for the sustained ERK5 activation by NGF in PC12 cells (48), suggesting it as a possible alternative signaling pathway other than Ras and Rap1 signaling. Thus, requirement of small G-proteins for ERK5 regulation in various cells should be clarified in more detail.
In summary, we found for the first time that ERK5 mediated neurite outgrowth and tyrosine hydroxylase stabilization in PC12 cells. The detailed mechanism of ERK5 activation, substrate specificity between ERK5 and ERK1/2, and physiological role of ERK5 in vivo should be subjects for future study.
Acknowledgments
We thank Dr. Yoshihiro Shimada, Dr. Tamiyo Kobayshi, Dr. Kosuke Takagi, and Dr. Yuichiro Matsuo (Olympus, Tokyo, Japan) for neurite outgrowth analysis with CELAVIEW-RS100. We thank Boehringer Ingelheim (Ridgefield, CT) for providing BIX02188 and BIX02189. We also thank Dr. Bistra Nankova (New York Medical College, New York), Dr. Zhengui Xia (University of Washington, Seattle), and Dr. Eisuke Nishida (Kyoto University, Kyoto, Japan) for providing DNA plasmids. PC12 cells that stably overexpress EGFP-Rap1GAP were selected with FACSAria at Biomedical Research Core of Tohoku University Graduate School of Medicine (Sendai, Japan).
This work was supported in part by Grants-in-aid from the Japan Society for the Promotion of Science 20790053 (to Y. O.) and 19659011 (to N. N.), Japan-United States Brain Research Cooperative Program (to Y. O.), and Exploratory Research Program for Young Scientists of Tohoku University (to Y. O.).
Y. Obara, A. Yamauchi, S. Takehara, and W. Nemoto, M. Takahashi, P. J. S. Stork, and N. Nakahata, unpublished observations.
- ERK
- extracellular signal-regulated kinase
- PC12
- pheochromocytoma cells
- NGF
- nerve growth factor
- Bt2cAMP
- dibutyryl cAMP
- MAPK
- mitogen-activated protein kinase
- EGF
- epidermal growth factor
- BDNF
- brain-derived neurotrophic factor
- GPCR
- G-protein-coupled receptor
- HRP
- horseradish peroxidase
- DMEM
- Dulbecco's modified Eagle's medium
- CREB
- cAMP-response element-binding protein
- GSK3β
- glycogen synthase kinase-3β
- m.o.i.
- multiplicity of infection
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP.
REFERENCES
- 1.Goldsmith Z. G., Dhanasekaran D. N. (2007) Oncogene 26,3122–3142 [DOI] [PubMed] [Google Scholar]
- 2.Nishida E., Gotoh Y. (1993) Trends Biochem. Sci. 18,128–131 [DOI] [PubMed] [Google Scholar]
- 3.Robinson M. J., Cobb M. H. (1997) Curr. Opin. Cell Biol. 9,180–186 [DOI] [PubMed] [Google Scholar]
- 4.Wang X., Tournier C. (2006) Cell. Signal. 18,753–760 [DOI] [PubMed] [Google Scholar]
- 5.Nishimoto S., Nishida E. (2006) EMBO Rep. 7,782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morimoto H., Kondoh K., Nishimoto S., Terasawa K., Nishida E. (2007) J. Biol. Chem. 282,35449–35456 [DOI] [PubMed] [Google Scholar]
- 7.English J. M., Vanderbilt C. A., Xu S., Marcus S., Cobb M. H. (1995) J. Biol. Chem. 270,28897–28902 [DOI] [PubMed] [Google Scholar]
- 8.Zhou G., Bao Z. Q., Dixon J. E. (1995) J. Biol. Chem. 270,12665–12669 [DOI] [PubMed] [Google Scholar]
- 9.Cavanaugh J. E. (2004) Eur. J. Biochem. 271,2056–2059 [DOI] [PubMed] [Google Scholar]
- 10.Kato Y., Tapping R. I., Huang S., Watson M. H., Ulevitch R. J., Lee J. D. (1998) Nature 395,713–716 [DOI] [PubMed] [Google Scholar]
- 11.Watson F. L., Heerssen H. M., Bhattacharyya A., Klesse L., Lin M. Z., Segal R. A. (2001) Nat. Neurosci. 4,981–988 [DOI] [PubMed] [Google Scholar]
- 12.Liu L., Cavanaugh J. E., Wang Y., Sakagami H., Mao Z., Xia Z. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimoto S., Kusakabe M., Nishida E. (2005) EMBO Rep. 6,1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L., Cundiff P., Abel G., Wang Y., Faigle R., Sakagami H., Xu M., Xia Z. (2006) Proc. Natl. Acad. Sci. U.S.A. 103,9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi M., Lee J. D. (2004) J. Mol. Med. 82,800–808 [DOI] [PubMed] [Google Scholar]
- 16.Kamakura S., Moriguchi T., Nishida E. (1999) J. Biol. Chem. 274,26563–26571 [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Su B., Xia Z. (2006) J. Biol. Chem. 281,35965–35974 [DOI] [PubMed] [Google Scholar]
- 18.Sun W., Wei X., Kesavan K., Garrington T. P., Fan R., Mei J., Anderson S. M., Gelfand E. W., Johnson G. L. (2003) Mol. Cell. Biol. 23,2298–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G., Lucas J. J., Gelfand E. W. (2005) Cell. Immunol. 238,10–18 [DOI] [PubMed] [Google Scholar]
- 20.Fukuhara S., Marinissen M. J., Chiariello M., Gutkind J. S. (2000) J. Biol. Chem. 275,21730–21736 [DOI] [PubMed] [Google Scholar]
- 21.Pearson G. W., Earnest S., Cobb M. H. (2006) Mol. Cell. Biol. 26,3039–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obara Y., Okano Y., Ono S., Yamauchi A., Hoshino T., Kurose H., Nakahata N. (2008) Cell. Signal. 20,1275–1283 [DOI] [PubMed] [Google Scholar]
- 23.Greene L. A., Tischler A. S. (1976) Proc. Natl. Acad. Sci. U.S.A. 73,2424–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obara Y., Labudda K., Dillon T. J., Stork P. J. (2004) J. Cell Sci. 117,6085–6094 [DOI] [PubMed] [Google Scholar]
- 25.Cavanaugh J. E., Ham J., Hetman M., Poser S., Yan C., Xia Z. (2001) J. Neurosci. 21,434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carey K. D., Dillon T. J., Schmitt J. M., Baird A. M., Holdorf A. D., Straus D. B., Shaw A. S., Stork P. J. (2000) Mol. Cell. Biol. 20,8409–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Dillon T. J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P. J. (2006) Mol. Cell. Biol. 26,2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng J., Casey P. J. (2002) J. Biol. Chem. 277,43417–43424 [DOI] [PubMed] [Google Scholar]
- 29.Franke B., Akkerman J. W., Bos J. L. (1997) EMBO J. 16,252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obara Y., Aoki T., Kusano M., Ohizumi Y. (2002) J. Pharmacol. Exp. Ther. 301,803–811 [DOI] [PubMed] [Google Scholar]
- 31.Obara Y., Horgan A. M., Stork P. J. (2007) J. Neurochem. 101,470–482 [DOI] [PubMed] [Google Scholar]
- 32.York R. D., Yao H., Dillon T., Ellig C. L., Eckert S. P., McCleskey E. W., Stork P. J. (1998) Nature 392,622–626 [DOI] [PubMed] [Google Scholar]
- 33.Rovida E., Spinelli E., Sdelci S., Barbetti V., Morandi A., Giuntoli S., Dello Sbarba P. (2008) J. Immunol. 180,4166–4172 [DOI] [PubMed] [Google Scholar]
- 34.Pollack V., Sarközi R., Banki Z., Feifel E., Wehn S., Gstraunthaler G., Stoiber H., Mayer G., Montesano R., Strutz F., Schramek H. (2007) Am. J. Physiol. Renal Physiol. 293,F1714–F1726 [DOI] [PubMed] [Google Scholar]
- 35.Mody N., Leitch J., Armstrong C., Dixon J., Cohen P. (2001) FEBS Lett. 502,21–24 [DOI] [PubMed] [Google Scholar]
- 36.Tatake R. J., O'Neill M. M., Kennedy C. A., Wayne A. L., Jakes S., Wu D., Kugler S. Z., Jr., Kashem M. A., Kaplita P., Snow R. J. (2008) Biochem. Biophys. Res. Commun. 377,120–125 [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Liu L., Xia Z. (2007) J. Neurochem. 102,957–966 [DOI] [PubMed] [Google Scholar]
- 38.Vossler M. R., Yao H., York R. D., Pan M. G., Rim C. S., Stork P. J. (1997) Cell 89,73–82 [DOI] [PubMed] [Google Scholar]
- 39.Dodge-Kafka K. L., Soughayer J., Pare G. C., Carlisle Michel J. J., Langeberg L. K., Kapiloff M. S., Scott J. D. (2005) Nature 437,574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hisata S., Sakisaka T., Baba T., Yamada T., Aoki K., Matsuda M., Takai Y. (2007) J. Cell Biol. 178,843–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gueorguiev V. D., Cheng S. Y., Sabban E. L. (2006) J. Biol. Chem. 281,10188–10195 [DOI] [PubMed] [Google Scholar]
- 42.Moy L. Y., Tsai L. H. (2004) J. Biol. Chem. 279,54487–54493 [DOI] [PubMed] [Google Scholar]
- 43.Markus A., Patel T. D., Snider W. D. (2002) Curr. Opin. Neurobiol. 12,523–531 [DOI] [PubMed] [Google Scholar]
- 44.Garaude J., Cherni S., Kaminski S., Delepine E., Chable-Bessia C., Benkirane M., Borges J., Pandiella A., Iñiguez M. A., Fresno M., Hipskind R. A., Villalba M. (2006) J. Immunol. 177,7607–7617 [DOI] [PubMed] [Google Scholar]
- 45.Cude K., Wang Y., Choi H. J., Hsuan S. L., Zhang H., Wang C. Y., Xia Z. (2007) J. Cell Biol. 177,253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato Y., Kravchenko V. V., Tapping R. I., Han J., Ulevitch R. J., Lee J. D. (1997) EMBO J. 16,7054–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchetti A., Colletti M., Cozzolino A. M., Steindler C., Lunadei M., Mancone C., Tripodi M. (2008) Cell. Signal. 20,2113–2118 [DOI] [PubMed] [Google Scholar]
- 48.Rosário M., Franke R., Bednarski C., Birchmeier W. (2007) J. Cell Biol. 178,503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]