Abstract
One of the mechanisms that minimize the aberrant cross-talk between cAMP- and cGMP-dependent signaling pathways relies on the selectivity of cAMP binding domains (CBDs). For instance, the CBDs of two critical eukaryotic cAMP receptors, i.e. protein kinase A (PKA) and the exchange protein activated by cAMP (EPAC), are both selectively activated by cAMP. However, the mechanisms underlying their cAMP versus cGMP selectivity are quite distinct. In PKA this selectivity is controlled mainly at the level of ligand affinity, whereas in EPAC it is mostly determined at the level of allostery. Currently, the molecular basis for these different selectivity mechanisms is not fully understood. We have therefore comparatively analyzed by NMR the cGMP-bound states of the essential CBDs of PKA and EPAC, revealing key differences between them. Specifically, cGMP binds PKA preserving the same syn base orientation as cAMP at the price of local steric clashes, which lead to a reduced affinity for cGMP. Unlike PKA, cGMP is recognized by EPAC in an anti conformation and generates several short and long range perturbations. Although these effects do not alter significantly the structure of the EPAC CBD investigated, remarkable differences in dynamics between the cAMP- and cGMP-bound states are detected for the ionic latch region. These observations suggest that one of the determinants of cGMP antagonism in EPAC is the modulation of the entropic control of inhibitory interactions and illustrate the pivotal role of allostery in determining signaling selectivity as a function of dynamic changes, even in the absence of significant affinity variations.
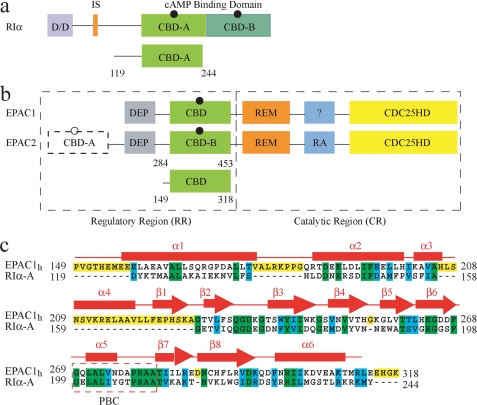
In eukaryotes, protein kinase A (PKA)2 and the exchange protein directly activated by cAMP (EPAC) are two major receptors for the cAMP second messenger (1–4). The activities of both PKA and EPAC are modulated in a cAMP-dependent manner through cAMP binding domains (CBDs) (1–4). In all isoforms of PKA, two tandem CBDs, denoted as CBD-A and CBD-B, are part of the regulatory subunit (R), in which they are preceded by an N-terminal dimerization docking module and a linker region (Fig. 1a) (1, 3). In the inactive state PKA exists as a tetrameric holo-enzyme complex, including two regulatory (R) subunits and two catalytic (C) subunits (1, 3). Binding of cAMP to the CBDs of the R subunits results in the release of the C subunits and in the activation of the kinase function (1, 3).
FIGURE 1.
Schematic representation of the domain organization in the regulatory subunit I-α of PKA (a) and in EPAC (b).The black circles indicate cAMP. a, D/D is the dimerization docking domain; the inhibitory site is shown in orange, and the two tandem cAMP binding domains, CBD-A and CBD-B, are highlighted in different shades of green. b, DEP, disheveled-egl-10-pleckstrin domain; REM, Ras exchange motif; RA, Ras-associated module, and the CDC25HD catalytic domain are represented in gray, green, orange, blue, and yellow, respectively. The black dashed line and the empty cAMP circle in EPAC2 indicate that this domain is not strictly necessary for the cAMP-dependent GEF activity. The module with question mark in EPAC1 denotes an unknown function for this domain. a and b, the CBD in light green shown below the full-length protein represents the construct used for the NMR studies. c, sequence alignment of the CBDs of bovine RIα- domain A and human EPAC1. Fully conserved residues are highlighted in green; cyan denotes conservation for the functional group only, and yellow indicates the residues present only in one of the CBDs. The secondary structure of apo-EPAC2m (PDB ID 1O7F) is shown in red.
Unlike PKA, EPAC is a single-chain protein that functions as a guanine nucleotide-exchange factor (GEF) for the small GTPase Rap1 and Rap2 (2, 4). The domain organization of EPAC includes an N-terminal regulatory region (RR) and a C-terminal catalytic region (CR) (Fig. 1b). There are two known homologous isoforms of EPAC, i.e. EPAC1 and EPAC2. One of the key differences between EPAC1 and EPAC2 is that in the former there is only a single CBD, whereas in the latter there are two noncontiguous CBDs, i.e. CBD-A and CBD-B. However, CBD-A has been shown not to be strictly necessary for the cAMP-dependent activation of EPAC (2, 4).
Both PKA and EPAC are critical for the regulation of a wide range of cAMP-dependent physiological processes (1–4), and impaired activity of these cAMP sensors has been implicated in cardiovascular pathology, diabetes, and Alzheimer disease (1–4). Therefore, PKA and EPAC represent attractive therapeutic targets. However, the design and development of specific drug leads targeting the CBDs of either of these two eukaryotic protein systems require an in depth analysis of how PKA and EPAC selectively recognize and allosterically respond to diverse cNMPs. For instance, the cAMP and cGMP second messengers control distinct groups of essential signaling pathways (1–6). It is therefore critical to minimize the cross-talk between the cAMP- and cGMP-dependent cellular responses. Although in vivo the selective control of the cAMP- and/or cGMP-dependent signaling pathways is a complex process that depends on multiple factors, including the modulation of cNMP synthesis, degradation, and compartmentalization (5), one of the key mechanisms to reduce the cAMP/cGMP cross-talk relies on the ability of both PKA and EPAC CBDs to sense selectively cAMP as opposed to cGMP.
Despite the fact that both PKA and EPAC CBDs are cAMP-selective sensors, these two signaling systems adopt different mechanisms to implement their cAMP-selective response. Specifically, in the PKA system cGMP is an agonist of cAMP, i.e. cGMP is able to activate PKA once it binds the R subunit. However, the affinity of cGMP for the PKA R subunit is significantly lower than that of cAMP, resulting in an activation constant that is 2 orders of magnitude higher than that of cAMP (i.e. Ka of 21 ± 2 nm for cAMP and of 4100 ± 20 nm for cGMP) (7, 8). Unlike PKA, EPAC preserves approximate micromolar affinities for both cAMP and cGMP, but in EPAC the latter cNMP is an antagonist of cAMP (9, 10), i.e. cGMP, like other N6-substituted cAMP analogs, binds effectively to the CBD of EPAC but fails to fully activate its GEF activity (9, 10).
The molecular basis for the cGMP antagonism selectively observed in EPAC but not in PKA is currently not fully understood. An initial hypothesis to explain the antagonist function of cGMP in EPAC has been recently proposed based on the structure of the ternary (Sp)-cAMPS-EPAC2m-Rap1 complex (11), which shows that the N6 of the (Sp)-cAMPS agonist forms an hydrogen-bond with the backbone carbonyl oxygen of Lys-450 located in the lid region (supplemental Table S1) (11). The disruption of this hydrogen bond by cGMP has been hypothesized to result in the inhibition of EPAC activation (11). However, a similar backbone hydrogen bond between the N6 of cAMP and the backbone carbonyl oxygen of Arg-632 has been observed also in the CBDs of the hyperpolarization-activated cyclic nucleotide-modulated channels (HCN) (supplemental Table S1), for which cGMP, like in PKA, is not an antagonist (12). This observation suggests that the elimination of the cAMP N6 hydrogen bond alone may not be sufficient to fully explain why cGMP is a cAMP antagonist with respect to the activation of EPAC. Furthermore, the previously proposed hypothesis based on the simple disruption by cGMP of the N6 hydrogen bond does not consider the possibility that cGMP may adopt an EPAC-bound conformation different from that of cAMP and/or that cGMP may affect also the inhibitory interactions between the ionic latch residues of the EPAC RR and the CDC25HD catalytic domain. These RR/CR salt bridges stabilize the EPAC system in an overall “closed” topology, whereby the RR sterically occludes access of Rap1 into the catalytic domain of EPAC (13). cAMP binding results in increased picosecond to nanosecond and millisecond to microsecond dynamics at the ionic latch region, which in turn leads to an increased entropic penalty for the inhibitory interactions mediated by the ionic latch (14). In other words, cAMP is able to weaken the inhibitory interactions between the regulatory and catalytic regions of EPAC by increasing the entropic cost associated with the formation of the cluster of ionic latch salt bridges between these two functional segments. This dynamically driven mechanism contributes to the observed cAMP-dependent shift toward active “open” conformations of EPAC, and we hypothesize that one of the effects of cGMP is to perturb the dynamic patterns of the EPAC CBD, thus altering the entropic control of the inhibitory interactions.
To test our hypotheses on cGMP agonism/antagonism and to further understand the molecular mechanisms underlying the different signaling responses of PKA and EPAC to the two endogenous second messengers cAMP and cGMP, here we present a comparative NMR analysis of cGMP binding and allostery for the critical CBDs of both PKA and EPAC. These results were also compared with the data on the same domains in their apo- and cAMP-bound states (14–20). All these studies rely on the RIα-(119–244) and the related EPAC1h-(149–318) constructs (Fig. 1), which have been previously validated as models of the essential CBDs of PKA and EPAC, respectively (14–20).
Our comparative NMR analysis has revealed that the structure, dynamics, and allosteric activation pathways of the PKA CBD-A are not significantly altered when cAMP is replaced by cGMP. However, significant differences between these two cNMPs are found for the EPAC1h CBD at the level of both ligand recognition and modulation of dynamic modes, leading to a mechanism in which cGMP shifts the activation equilibrium of EPAC toward the auto-inhibited state, thus accounting for its antagonistic function.
EXPERIMENTAL PROCEDURES
Expression, Purification, and NMR Sample Preparation of RIα-(119–244)
The regulatory subunit of PKA was expressed and purified as described before (15–19). In brief, singly uniformly 15N-labeled samples were prepared by growing the bacteria in 15NH4Cl containing M9 minimal media. The bacteria were grown at 37 °C before induction with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside at a 0.8 A600 nm for 20 h at 22 °C. The cells were then lysed by three cycles of French press after resuspending them in the lysis buffer (20 mm MES, pH 6.5, 100 mm NaCl, 2 mm EGTA, 2 mm EDTA, and 5 mm dithiothreitol). The cell debris was removed by centrifugation, and the protein was fractionated by a 40% ammonium sulfate cut. The precipitant was again resuspended with lysis buffer and incubated overnight with cAMP-Sepharose beads at 4 °C. The resins were then thoroughly washed with high salt buffer (lysis buffer containing 700 mm NaCl) before eluting the protein with 25, 35, and 40 mm cAMP (Sigma). The protein was further purified through a Superdex S75 gel filtration column, which was pre-equilibrated with 50 mm MES, pH 6.5, 100 mm NaCl. The cAMP sample was prepared by adding 1 mm cAMP and 5% D2O to the dilute sample and then concentrating to a protein concentration of 0.5 mm. The cGMP-bound RIα-(119–244) sample was prepared by eluting with cGMP rather than cAMP.
The sample for the Nz exchange experiments was prepared following a previously explained denaturation/renaturation protocol (15, 16). cAMP was removed by extensive dialysis in 8 m urea, and the protein was refolded to its native conformation by a stepwise removal of urea. The correct refolding of the protein was validated by comparing the HSQC spectra of the original cAMP bound and the refolded sample to which cAMP was re-added as a control. The 50:50 cAMP and cGMP sample was prepared by adding equal amounts of both ligands to the cAMP-free protein.
Expression, Purification, and NMR Sample Preparation of EPAC1h-(149–318)
The 15N- and 13C/15N-labeled EPAC1h-(149–318) samples were expressed as glutathione S-transferase fusion proteins (11, 13, 14, 20). The bacteria were induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside at a 0.6 A600 nm for 3 h at 37 °C (14, 20). The cells were then resuspended in lysis buffer (phosphate-buffered saline, 10 mm EDTA, 10% v/v glycerol, supplemented with protease inhibitors) and lysed by French press. The supernatant obtained after removing the cell debris was incubated with GSH-Sepharose beads at 4 °C for 4 h. The beads were then thoroughly washed with wash buffer (0.5 m NaCl and 50 mm Tris, pH 8). The glutathione S-transferase tag was removed from EPAC by using biotinylated thrombin (Novagen) for 16 h at 4 °C followed by thrombin removal via streptavidin beads. The protein was further purified through a Q column by using a 0–1 m NaCl gradient in 50 mm Tris, pH 7.6 buffer. Finally the protein was dialyzed against the NMR buffer (50 mm Tris, pH 7.6, with MES, 50 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine, 0.02% w/v NaN3, 5% v/v D2O) and concentrated to 0.5 mm, unless otherwise specified. To obtain cAMP- or cGMP-bound EPAC1h-(149–318), 2 mm cAMP or cGMP was added to the final purified apoprotein.
NMR Spectroscopy
All NMR spectra were acquired with a Bruker AV 700 spectrometer equipped with a TCI cryoprobe at 306 K. Unless otherwise mentioned, all spectra were recorded with 128 complex points and a 31.8 ppm spectral width for the 15N dimension and 1024 complex points and a 14.2 ppm spectral width for the 1H dimension. The carrier frequencies for the 1H, 13C, and 15N were positioned at 4.7, 176.0, and 119.0 ppm, respectively. All two-dimensional spectra were processed using Xwinnmr (Bruker, Inc.) and/or NMRPipe (21). The 1H chemical shifts were calibrated with 2,2-dimethyl-2-silapentanesulfonic acid, and the 15N/13C chemical shifts were calibrated indirectly using their respective gyromagnetic ratios (22). The cross-peak fit heights were measured by implementing Gaussian line fitting through the program Sparky version 3.111 (23).
The Nz exchange experiments (24–26) for the assignment of cGMP-bound RIα-(119–244) were acquired with a recycle delay of 2 s and an Nz mixing time of 250 ms. The data were processed using linear prediction, and the compounded chemical shifts between the cAMP- and cGMP-bound states of RIα-(119–244) were computed using the following formula: Δδcompound = ((Δδ1H)2 + (Δδ15N/6.5)2)1/2, where Δδ1H and Δδ15N are the differences between the proton and nitrogen chemical shifts, respectively (27). The chemical shift changes between the apo- and the ligand-bound states were calculated according to δω = 0.2Δδ15N + Δδ1H (28).
The assignment of cGMP-bound EPAC1h-(149–318) was obtained through standard triple resonance experiments (i.e. HNCO, HNCA, HN(CO)CA, CBCA(CO)NH, and HNCACB), as explained previously (14, 20, 29). The triple resonance data were analyzed through Sparky version 3.111 (23), and the PECAN software was used to determine the probabilities of secondary structural elements by using the secondary chemical shifts (30). Compounded chemical shifts between the cAMP- and cGMP-bound states of EPAC1h-(149–318) and the chemical shift changes between the apo- and the ligand-bound forms were calculated as described above for RIα-(119–244).
syn/anti Base Orientation of Free cAMP and cGMP
0.5 mm cAMP and cGMP solutions were prepared in a 100% D2O buffer (20 mm phosphate buffer, pH 7.5, and 50 mm NaCl). To confirm the assignment of the cAMP and cGMP one-dimensional 1H spectra, two-dimensional homo-nuclear (1H-1H) total correlation spectroscopy spectra were acquired with spectral widths of 8389 Hz for both the directly and indirectly detected dimensions using 2048 and 512 complex points, respectively. The total correlation spectroscopy spin-lock was set at 60 ms with a 10 kHz strength. The resonance of the adenine H8 proton was distinguished from that of the partially overlapped adenine H2 proton based on the one-dimensional spectra of d8-cAMP. The d8-cAMP sample was prepared by substituting the H8 proton with a deuteron by boiling the sample overnight in D2O (31). The conformation of free cNMPs was probed through off-resonance ROESY experiments (32). The data were acquired with a mixing time of 158 ms and a spin-lock strength of 8.5 kHz. The t1 and t2 complex points were set at 2048 and 700, respectively, with a spectral width of 8389 Hz for both dimensions.
syn/anti Base Orientation of EPAC1h-(149–318)-bound cAMP and cGMP
The orientation of the adenine base in EPAC1h-(149–318)-bound cAMP and cGMP was determined from the transfer NOEs observed in a two-dimensional 13C/15N double-filtered nuclear Overhauser effect spectroscopy experiment using a 0.5 mm double-labeled 13C/15N EPAC1h-(149–318) and 0.7 mm cNMP ligand (cAMP or cGMP) (33, 34). Samples were prepared in a D2O solution composed of 20 mm phosphate buffer at pH 7.5 and 50 mm NaCl. The spectra were measured with 2048 and 420 complex points and spectral widths of 8389 × 8389 Hz, respectively. The NOE mixing time was set at 80 ms. Cross-peaks arising from cross-relaxation were differentiated from exchange nuclear Overhauser effect spectroscopy peaks by a two-dimensional-ROESY experiment (35) recorded with a spectral width of 11 ppm for both dimensions and with 1024 and 200 complex points, respectively. The ROESY mixing time was set at 15 ms and the continuous wave spin-lock strength at 2.5 kHz.
Hydrogen/Deuterium Exchange Experiments
Amide proton H/D exchange rates were determined from the cross-peak intensities in a series of 15N HSQC spectra acquired as function of time. The cGMP-bound RIα-(119–244) was exchanged to D2O by passing it through a gel filtration column pre-equilibrated with a 100% D2O buffer (1 mm cGMP, 50 mm MES, pH 6.5, 100 mm NaCl), as described previously (15). cGMP- and cAMP-bound EPAC1h-(149–318) were exchanged to D2O by 10-fold dilution in a 100% D2O buffer (2 mm cGMP or cAMP, 50 mm Tris, pH 7.6, 50 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine). The apo EPAC sample was exchanged with D2O in a 100% D2O buffer without cGMP or cAMP. A total of 84 HSQC spectra was acquired. For the first 30 spectra two scans were acquired, whereas the rest of the spectra were recorded with four scans. The time between the first exposure to D2O and the first HSQC spectra (dead time) was ∼20 min. The intensities of the slowly exchanging amide peaks were monitored for several days after exposure to D2O. The HSQC cross-peak intensities were analyzed using NMRPipe (21), and the exchange rates were determined by using the Curvefit software by implementing Levenberg-Marquardt nonlinear least square exponential fitting (36). The protection factors were calculated using the ratio of intrinsic exchange rates (computed based on polyalanine through the program SPHERE (37)) to the experimental H/D exchange rates, assuming an EX2 exchanging mechanism as explained previously (15). Details on the correlation between H/D protection factors and dissociation constants are available in the supplemental material.
15N NMR Relaxation Measurements
The spin-lattice (R1) and spin-spin (R2) relaxation rates and the hetero-nuclear steady state {1H}-15N NOEs were measured for a 0.1 mm sample of cGMP-bound EPAC1h-(149–318). The R1 relaxation rates were measured with water flip back and sensitivity-enhanced pulse sequences using relaxation delays of 100 (two times), 200, 300, 400 (two times), 500, 600, 800, and 1000 ms (38, 39). The R2 measurements were implemented using CPMG relaxation delays of 8.48, 16.96, 25.44, 33.92, 42.4, 50.88, 59.36, 76.32, and 93.28 ms with an offset and duty cycle compensated 15N R2 CPMG pulse sequence, where νCPMG was set at 472 Hz (40). Both R1 and R2 experiments were acquired with recycle delays of 1.5 s. The steady state {1H}-{15N} NOEs were determined from the ratio of spectra collected either with a 5-s 1H saturation or without 1H saturation and 10 s of recycle delay (41). The entire series of R1 and R2 relaxation rates were collected in triplicate, and the NOE spectra were collected as a set of 10 replicas. The acquired spectra were then co-added in the time domain prior to Fourier transformation. The R1 and R2 relaxation rates were determined by using the program Sparky (23) using the cross-peak fit heights. The errors on the R1 and R2 rates were estimated from the Gaussian distributed random noise. The error on the NOE values were gauged based on the standard deviation between fit heights in the replicate spectra. The effects of diffusional anisotropy and the overall tumbling of the protein on the relaxation rates and the reduced spectral densities were evaluated as described previously by using the HYDRONMR program (14, 42, 43).
Reduced Spectral Density Analyses
The R1 and R2 relaxation rates and hetero-nuclear steady state {1H}-{15N} NOEs were analyzed by reduced spectral density mapping (44, 45), under the assumption that the spectral density functions at high frequency are equal as follows: J(ωN + ωH) = J(ωH) = J(ωN − ωH). The J(0) values were derived from the R1, R2, and {1H}-{15N} NOE data, and the J(ωN + ωH) and J(ωN) values were computed from the R1 and {1H}-{15N} NOE data (44, 45). Errors in the reduced spectral density were evaluated from the propagation of the uncertainties in the measured 15N relaxation rates and {1H}-{15N} NOE data.
RESULTS
syn/anti Base Orientations in Free and Bound cAMP
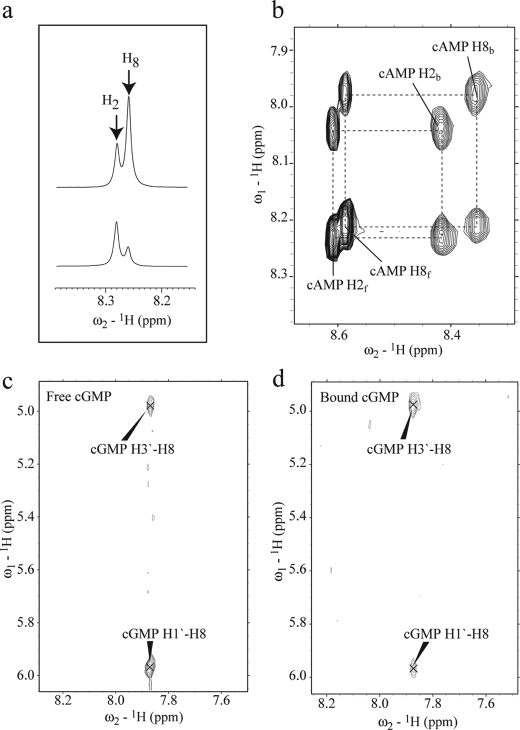
Analysis of the structures previously solved for different cyclic nucleotide-bound CBDs (supplemental Table S1) reveals that these ligands can dock either in a syn or in an anti conformation (11, 12, 46–49). For instance, cAMP binds both CBDs of PKA in a syn conformation, as shown previously by x-ray crystallography (46), whereas the same cNMP docks into the CBDs of cyclic nucleotide-gated ion channels in an anti orientation (48). The syn and anti conformations differ mainly for the orientation of the aromatic base with respect to the ribose moiety. In syn cNMPs the base is rotated over the ribose ring, although in anti-cNMPs the base is rotated away from the ribose ring. Using the bound cNMPs structures listed in supplemental Table S1 (11, 12, 46–49), it is possible to evaluate for both syn and anti conformations the expected distance ranges between NMR detectable protons, such as the H1′, H2′, H2, and H8 atoms of cAMP (supplemental Fig. S1), as summarized in supplemental Table S2. The supplemental Table S2 shows that, as expected, the H1′–H2′ distance does not change significantly with the change in orientation of the nucleotide base, and therefore this distance was selected as a reference in syn versus anti comparisons. The supplemental Table S2 also reveals that the syn and the anti conformations differ for the relative distances between H8 and the ribose protons, indicating that the NOEs involving H8 are sufficient to probe the orientation of the adenine base. For instance, in the syn conformation the H8–H1′ distance is significantly shorter than that in the anti conformation (supplemental Table S2), whereas the distance between H8–H3′ is longer in the syn conformation relative to the anti orientation (supplemental Table S2). Therefore, we expect for the syn conformation a strong NOE signal between H8 and H1′ along with a weak or absent NOE signal between H8 and H3′. On the other hand, the anti conformation will give rise to a medium NOE signal between H8 and H1′ and a stronger NOE signal between H8 and H3′. These NOE “signatures” for the syn and anti conformations were used to interpret the experimental distances measured for free and bound cyclic nucleotides (Fig. 2 and supplemental material).
FIGURE 2.
a, expansions from the one-dimensional spectra of cAMP (top) and of d8-cAMP (bottom). Replacement of the proton at the 8th position of the adenine base (supplemental Fig. S1) by deuterium significantly reduces the H8 signal in d8-cAMP. b, expansion from the ROESY spectrum of EPAC1h-(149–318)-bound cAMP. Black dashed lines connect the cross-peaks between free cAMP (suffixed with f) and EPAC-bound cAMP (suffixed with b). c, expansion of the off-resonance ROESY spectrum of free cGMP. d, same expansion as in c but for the transfer 13C15N double-filtered nuclear Overhauser effect spectroscopy spectrum of cGMP bound to EPAC1h-(149–318). The cross-peaks are labeled with the assigned proton pairs.
The normalized NOE and rotating frame nuclear Overhauser effect intensities measured for EPAC-bound and for free cAMP are summarized in Tables 1 and 2, respectively. For free cAMP, a medium NOE signal between H8 and H1′ was observed (Table 2), which is consistent with a predominantly anti conformation, as also supported by the weak NOE detected between the H2 and H1′ protons (Table 2 and supplemental Table S2). However, the strong H8–H1′ NOE signal observed for EPAC-bound cAMP (Table 1) suggests that cAMP binds to EPAC in a syn conformation, as also confirmed by the absence of the H2–H1′ NOE (Table 1, supplemental Table S2). This result is also independently confirmed by the syn conformation determined for (Sp)-cAMPS in the recently published (Sp)-cAMPS-EPAC2m-RAP ternary complex (11), validating the effectiveness of our NOE-based approach for distinguishing between syn and anti orientations.
TABLE 1.
Normalized NOEs for cAMP and cGMP bound to EPAC 1h-(149–318)
| H1′ |
H2a,b |
H8 |
|||
|---|---|---|---|---|---|
| cAMP | cGMP | cAMP | cAMP | cGMP | |
| H4′ | w | n | n | n | n |
| H3′ | ov | n | ov | ov | s |
| H2′ | s | s | w | w | w |
| H1′ | n | s | m | ||
| H5′ | w | w | n | n | n |
a In cGMP H2 is substituted with NH2; therefore, no NOE has been observed at this position.
b All NOEs are normalized against the intensity of the H1′– H2′NOE cross-peak because the distance between H1′ and H2′ does not change in the syn and anti conformations (see supplemental Tables S2 and S3). s denotes a strong NOE; m denotes a medium NOE; w denotes a weak NOE; n means no NOE observed; ov indicates peak overlap.
TABLE 2.
Normalized NOEs intensities for Free cAMP and cGMP
Data are as described in Table 1, and all symbols have the same meaning as in Table 1.
| H1′ |
H2 |
H8 |
|||
|---|---|---|---|---|---|
| cAMP | cGMP | cAMP | cAMP | cGMP | |
| H4′ | n | n | n | n | n |
| H3′ | ov | w | ov | ov | m |
| H2′ | s | s | w | s | w |
| H1′ | w | m | s | ||
| H5′ | s | s | w | s | w |
syn/anti Base Orientations in Free and Bound cGMP
The NOE method was then extended to free and EPAC-bound cGMP, for which no previous structural information is available (Tables 1 and 2 and supplemental Table S2). The strong H8–H1′ NOE observed for free cGMP (Fig. 2c and Table 2) suggests that cGMP exists predominantly in a syn conformation in solution. However, the medium H8–H1′ NOE and the strong H8–H3′ NOE measured for EPAC-bound cGMP (Fig. 3d and Table 1) indicate that upon binding to EPAC1h-(149–318) there is an equilibrium shift, whereby the state with the highest population corresponds to cGMP in an anti conformation. This result defines a distinct difference between cGMP bound to EPAC and to PKA, as in the latter x-ray crystallography has shown that cGMP prefers a syn conformation (supplemental Table S1) (47). The conclusions of these syn/anti conformational analyses are summarized in Table 3.
FIGURE 3.
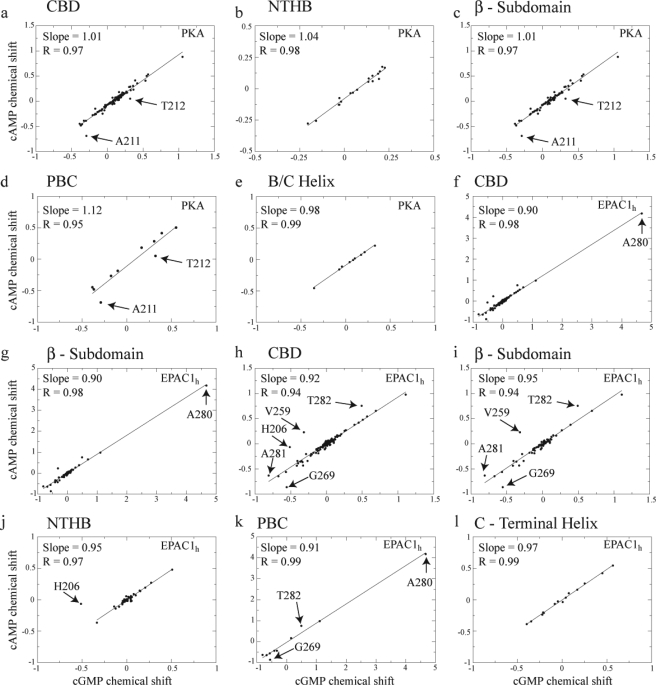
Correlations between cAMP- and cGMP-induced chemical shift changes in RIα–A-(119–244) (a–e) and EPAC1h-(149–318) (f–l). The horizontal axes report the differences in chemical shifts between the cGMP-bound and the apo states calculated as δω = 0.2Δδ15N + Δδ1H (28). The vertical axes report the differences in chemical shifts between the cAMP-bound and the apo states calculated using the same equation (28). a and f, all the assigned CBD residues were used in the linear correlations, whereas in the other panels the correlations were confined to subsets of residues to better identify possible outliers. Specifically, b and j are for the N-terminal helical bundle; c, g, and i are for the β-subdomain (including the PBC); d and k are for the PBC; and e and l are for the B/C helix. Because of the dominant chemical shift changes observed for Ala-280 of EPAC1h-(149–318), the correlations of f and g were recalculated in h and i, respectively, without Ala-280. Similarly, the correlation for the N-terminal helical-bundle region in j was calculated excluding His-206.
TABLE 3.
Summary of preferred syn/anti conformations in free and CBD-bound cNMPs
Comparative Analysis of the PKA CBD-A Structure in the cAMP- and cGMP-bound States
The magnitude of the structural differences between the cAMP- and cGMP-bound states of RIα-(119–244) was evaluated through the comparative analysis of the amide chemical shifts, which were assigned as explained in the supplemental material. Specifically, the cGMP-dependent amide chemical shift variations were correlated with the corresponding parts/million changes caused by cAMP binding, as shown in Fig. 3, a–e. The all-residue correlation (Fig. 3a) results in a slope close to 1 (i.e. 1.01) and an R value of 0.97. Similar results were obtained through subdomain specific linear regressions (Fig. 3, b–e), which preserve a high degree of correlation between the cAMP-bound and the cGMP-bound states. The strong correlation between these two states suggests that cGMP-bound RIα-(119–244) is structurally very similar to the cAMP-bound state. However, although the linearity of these correlations (Fig. 3, a–e) supports the absence of significant global conformational changes when cAMP is replaced by cGMP, a local perturbation is detected at the level of two adjacent residues located C-terminal to the phosphate binding cassette (PBC), i.e. Ala-211 and Thr-212, which consistently appear as regression outliers in Fig. 3, a, c, and d.
Our chemical shift-based assessment of the similarity between the cAMP- and cGMP-bound states of RIα is independently validated by the crystal structures of RIα in complex with cAMP and cGMP (PDB codes 1RGS and 1RL3, respectively) (46, 47), which reveal a high level of structural homology with a backbone-to-backbone root mean square deviation of only ∼0.4 Å. Furthermore, these crystal structures unveiled a slight cGMP-specific distortion at the C terminus of the PBC, which is in full agreement with the Ala-211 and Thr-212 outliers detected in the correlations of Fig. 3, a, c, and d, providing further confirmation of the validity of our chemical shift regressions for the purpose of assessing structural perturbations.
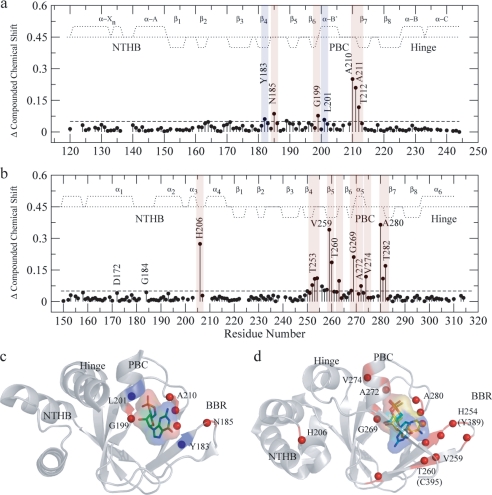
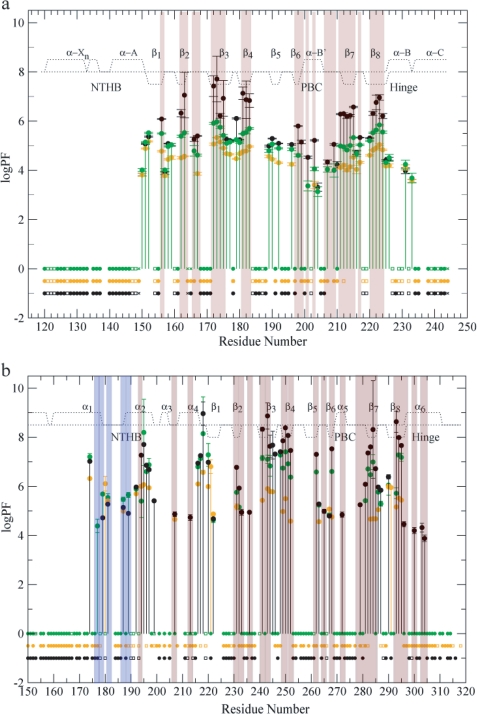
The binding of cGMP to RIα-(119–244) was also mapped through compounded chemical shifts (Fig. 4a). Unlike the chemical shift correlations discussed above (Fig. 3), the compounded chemical shift difference maps of Fig. 4 compare directly the cAMP-bound and cGMP-bound states without biases from the chemical shifts of the apo state and therefore provide a clearer picture of local perturbations linked to ligand changes. This is particularly important for residues that are unassigned in the apo state due, for instance, to line broadening beyond detection, as in the case of Ala-210 in RIα-(119–244). Such residues cannot be included in the apo versus cNMP correlations of Fig. 3, but are present in Fig. 4. Specifically, Fig. 4a reveals that cGMP induces only local chemical shift changes confined mainly to the β4 strand, in the base binding region (BBR), and to the PBC termini, i.e. Gly-199 at the N terminus of the PBC and Ala-210, Ala-211, and Thr-212 at the C terminus of the PBC, as also shown in Fig. 4c, which maps these cGMP-dependent perturbations into the structure of the RIα CBD-A.
FIGURE 4.
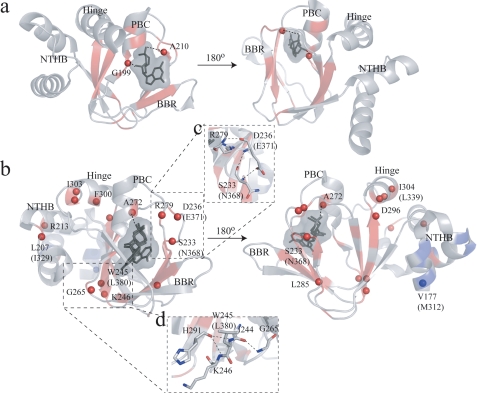
Compounded chemical shift changes between the cAMP- and cGMP-bound states of RIα-A-(119–244) (a) and of EPAC1h-(149–318) (b). The chemical shifts observed for Ala-210 in a and for A280 in b are downscaled by a factor of 2 to obtain a better visual dynamic range. The average chemical shift is indicated by the dashed horizontal lines. The secondary structure for RIα-A (according to PDB 1RGS) and EPAC1h (according to PDB 1O7F) is indicated by dotted lines. The positive and negative values indicate α-helical and β-sheet probabilities, respectively. The residues with compounded chemical shift changes above the average chemical shift are highlighted in red, and the residues with compounded chemical shift changes comparable with the average are highlighted in blue. The compounded chemical shifts between the cAMP- and cGMP-bound states are mapped into the ribbon diagrams of RIα-A (c) (PDB 1RL3) and EPAC (PDB- 3CF6) (d). d, cAMP is shown in syn and cGMP is shown in anti conformations (as discussed in the text) along with their van der Waals surfaces. The anti conformation of cGMP was created based on the structure of anti-cAMP in cAMP-bound HCN (PDB code 1VP6), and the cAMP syn conformation was obtained from (Sp)-cAMPS-bound EPAC2m (PDB code 3CF6). The backbone NHs showing chemical shift changes > average are indicated by red spheres. The amino acid residues are labeled according to the EPAC1h sequence, and the nonconserved EPAC2m residues are shown within parentheses.
Comparative Analysis of the EPAC1 CBD Structure in the cAMP- and cGMP-bound States
The linear correlation analysis approach was also used to evaluate cAMP versus cGMP structural differences for EPAC1h-(149–318) (Fig. 3, f–l), for which no cGMP-bound structure has yet been solved. Again, the strong correlations observed in the plots of Fig. 3, f–l, as evident from the slope and R values close to unity, suggest that the binding of cGMP induces similar global structural rearrangements as that of cAMP. However, a close inspection of the chemical shift variations in EPAC1h-(149–318) (Fig. 3, h–k) reveals several outliers indicative of local structural differences between the cGMP- and cAMP-bound structures. For instance, His-206 in α3 within the N-terminal helical bundle (Fig. 3, h and j), Val-259 in the BBR, and the PBC residues Gly-269, Ala-280, Ala-281, and Thr-282 display significant chemical shift variations as cAMP is replaced by cGMP (Fig. 3, h, i, and k). Despite the presence of these multiple local perturbations involving α3 and the cAMP-binding site, interestingly the α6 helix and the ionic latch region (i.e. 168–197) in the N-terminal helical bundle did not exhibit any significant structural change upon substitution of cAMP with cGMP (Fig. 3, h–l), as also independently supported by the 2° structure analysis (supplemental Fig. S2) based on the secondary chemical shifts. Specifically, the secondary structure comparison between the apo-, the cAMP-, and the cGMP-bound states of EPAC1h-(149–318) (supplemental Fig. S2) indicates that, similarly to cAMP, binding of cGMP significantly destabilizes the C-terminal region of the α6 hinge helix (supplemental Fig. S2, b and c).
As for our analysis of the PKA system, the effect of cGMP on the structure of EPAC1h-(149–318) was mapped also by compounded chemical shift changes (Fig. 4, b and d). Fig. 4, b and d, consistently shows that, unlike in PKA, in EPAC the substitution of cAMP with cGMP causes perturbations that propagate well beyond the PBC termini and the BBR β4. Specifically, b and d of Fig. 4 indicate that the binding of cGMP also affects the other strand of the BBR (i.e. β5) as well as His-206 in α3, within the N-terminal helical bundle, and residues in the α5 region of the PBC (i.e. Ala-272 and Val-274).
Comparative Analysis of the PKA CBD-A Solvent Protection in the cAMP- and cGMP-bound States
Chemical shifts probe mainly the most stable structures accessible to proteins and may not sense ligand-dependent perturbations at the level of partially unfolded excited states, which are detected, however, through variations in H/D exchange rates and in the corresponding protection factors (PFs), as shown in Fig. 5a for cGMP-bound RIα-(119–244). The resulting cGMP-dependent changes in PFs are mapped into the structure of the RIα CBD-A in Fig. 6a. The H/D exchange data in Fig. 5a and Fig. 6a show that cGMP binding not only dramatically increases the solvent exposure of the PBC amides of Gly-199 and Ala-210, but it also causes an overall reduction, relative to the cAMP-bound state, in the PFs measured for highly protected residues. For instance, the PFs observed for the deeply buried amides of the inner strands of the β-barrel (i.e. β3, -4, -7, and -8), which typically exchange through transient global unfolding pathways (20), display in the cGMP-bound state intermediate values between those measured for the apo- and cAMP-bound states. Interestingly, the ΔΔGunfolding (1.65 ± 0.70 kcal/mol) calculated based on the average PFmax for the β-subdomain of the cGMP-bound state (i.e. 〈PFmax〉β = 5.47 ± 0.06; ΔGunfolding = 7.64 ± 0.06 kcal/mol) and of the cAMP-bound state (i.e. 〈PFmax〉β = 6.66 ± 0.60; ΔGunfolding = 9.29 ± 0.70 kcal/mol) agrees well with the ΔΔGunfolding of 1.5 kcal/mol calculated based on the dissociation constants of cGMP and cAMP for this domain (50), as explained in the supplemental material. This observation indicates that the observed reduction in the maximal PFs occurring as cAMP is replaced by cGMP is mainly due to the reduced binding ability of cGMP compared with cAMP. However, replacement of the adenine of cAMP with the guanine of cGMP does not significantly affect regions outside the β-subdomain, which are subject mainly to local rather than global transient unfolding events. For instance, the PFs measured for the N- and C-terminal helices of RIα-(119–244) remain largely unperturbed going from cAMP to cGMP (Fig. 5a).
FIGURE 5.
PFs of RIα-A-(119–244) (a) and EPAC1h-(149–318) (b). a, PFs measured with 10-fold excess cAMP (black), cGMP (green) and without 10-fold excess cAMP (orange) (15) are plotted against the residue number. b, EPAC1h-(149–318) PFs are measured with cAMP (black), cGMP (green) and apo states (orange) are plotted against the residue number. The filled circles at the base line refer to the fast exchanging residues within the dead time (20 min) of the experiment or by the first or second HSQCs. The residues affected by overlap are denoted as squares, and the residues for which no data are reported are either ambiguous or proline. The backbone amides that exchange in the cGMP-bound state faster than the cAMP-bound state are highlighted with a pink background, and the residues exchanging more slowly relative to the cAMP-bound state are highlighted in blue. The secondary structures are denoted by the dotted line as explained in Fig. 4.
FIGURE 6.
Changes in the PFs between the cAMP- and cGMP-bound states mapped into the ribbon diagrams of RIα-A (PDB 1RL3) (a) and EPAC (PDB- 3CF6) (b). a, cGMP is shown as stick bonds with its van der Waals surface. The backbone ribbon is highlighted according to the color coding of Fig. 5. Residues exchanging fast (within the dead time of the H/D experiment or by the second HSQC) are shown as spheres and the dotted line represents hydrogen bonds between the ligand and the protein backbone. b, cyclic nucleotide conformations are as described in Fig. 4, and the backbone ribbon is highlighted according to the color coding of Fig. 5. The amino acid residues are labeled according to the EPAC1h sequence, and the nonconserved EPAC2m residues are shown within parentheses. Selected hydrogen bond networks destabilized by the replacement of cAMP with cGMP are shown in c and d.
Comparative Analysis of the EPAC1 CBD Solvent Protection in the cAMP- and cGMP-bound States
The H/D PFs for EPAC1h-(149–318) are reported in Figs. 5b and 6b. These data indicate that in the cGMP-bound state the protection of the Ala-272 NH in the PBC is significantly lower than that observed for the cAMP-bound state (Fig. 5b and Fig. 6b). Another striking feature of the EPAC1h-(149–318) PFs shown in Fig. 5b and Fig. 6b is that several residues in α1 and α2 are more protected in the cGMP-bound state than in the cAMP-bound state, suggesting that these helices are at least partially stabilized when cAMP is replaced by cGMP. Unlike α1 and α2, binding of cGMP causes a distinct reduction in several PFs measured for the helices adjacent to the β-barrel (i.e. α4 and α6) and for the β-subdomain. For instance, Leu-207 and Arg-213 in α4 and Phe-300, Ile-303, and Ile-304 in α6 all become fast exchanging upon cGMP binding (i.e. they exchange within the dead time of the H/D experiment or by the second HSQC after exposure to D2O) similarly to the apo state (Fig. 5b and Fig. 6b). Similar transitions from protected amides in the cAMP-bound state to fast exchanging residues in the cGMP-bound state are also observed for two clusters of adjacent residues in the β-subdomain (Fig. 6, c and d). One cluster is formed by Trp-245 and Lys-246 in β3 and Gly-265 in the neighboring β6 strand (Fig. 6d), whereas another cluster includes Arg-279 at the C terminus of the PBC as well as Ser-233 and Asp-236 in the adjacent β2–3 loop (Fig. 6c). The remaining cGMP-dependent changes in the maximal protection factors (Fig. 6b) observed for the highly buried β-strands are consistent with the KD values previously reported for cGMP binding to EPAC (9).
Overall, our H/D exchange data indicate, consistently with our compounded chemical shift results, that in EPAC, unlike in PKA, replacement of cAMP with cGMP induces long range effects that propagate well beyond the PBC into the α- and β-subdomains. To further explore the nature of these long range perturbations, we have extended our comparative NMR investigations to HSQC line width analyses and to 15N relaxation measurements, designed to probe dynamics in the picosecond to nanosecond and millisecond to microsecond time scales.
Comparative Analysis of the PKA CBD-A Dynamics in the cAMP- and cGMP-bound States
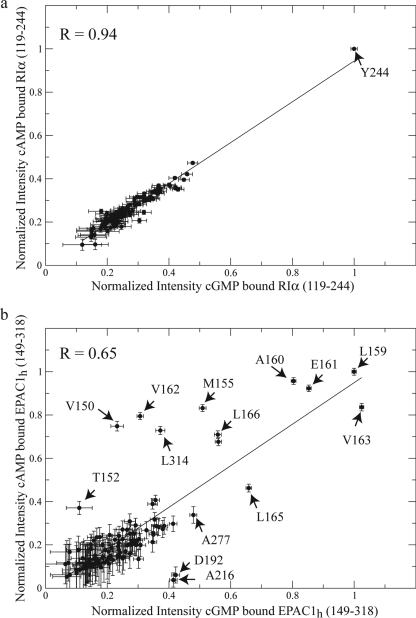
The changes in dynamics in RIα-(119–244) occurring as cAMP is replaced by cGMP were probed through line width analyses. The differential line broadening of HSQC cross-peaks between the two bound states is sensitive to picosecond to nanosecond and millisecond to microsecond backbone dynamics (14, 22). For instance, millisecond to microsecond chemical exchange significantly broadens the line width resulting in a decreased HSQC cross-peak intensity, whereas the presence of fast (picosecond to nanosecond) motions will sharpen the peaks leading to an increased HSQC cross-peak intensity. We therefore analyzed the correlation between the normalized HSQC cross-peak intensities in the cAMP- and in the cGMP-bound states of RIα-(119–244) (Fig. 7a). The intensities of the HSQC spectra of RIα-(119–244) are normalized against Tyr-244 (Fig. 7a), because this C-terminal residue has the highest intensity in both ligand-bound forms and its intensity is independent of ligand binding. The relatively high correlation coefficient (0.94) observed between the cGMP- and cAMP-bound states of RIα-(119–244) (Fig. 7a) suggests that the binding of cGMP does not induce any significant line broadening or change in dynamics for this CBD of PKA. Because of the similarity between the cAMP-bound and cGMP-bound dynamic patterns observed for PKA, we did not investigate further the dynamics of cGMP-bound PKA.
FIGURE 7.
Correlation plot of the normalized HSQC intensities for the cAMP-bound and cGMP-bound states of RIα-A-(119–244) (a) and EPAC1h-(149–318) (b). a, in RIα-A-(119–244) the HSQC intensities were normalized against residue Tyr-244. The high correlation (R = 0.94) between the cAMP- and cGMP-bound states indicates that no significant change in dynamics occurs between the two states. b, intensities were normalized against that of the Leu-159 peak in both cAMP- and cGMP-bound states. The linear fit between the cAMP and cGMP intensities is indicated by a solid straight line. The amino acid residues showing significant changes in intensity between the cAMP-bound and cGMP-bound states are labeled. The low correlation (R = 0.65) observed for EPAC1h-(149–318) points to significant changes in dynamics upon substitution of cAMP with cGMP.
Comparative Analysis of the EPAC1 CBD Dynamics in the cAMP- and cGMP-bound States
Similarly to PKA, the changes in dynamics in EPAC1 CBD occurring as cAMP is replaced by cGMP were initially assessed through the correlation between the normalized HSQC cross-peak intensities in the cAMP- and in the cGMP-bound states of EPAC1h-(149–318) (Fig. 7b). In Fig. 7b, the HSQC intensities are normalized against Leu-159, because Leu-159 has the highest intensity in both ligand-bound forms, and its intensity is independent of ligand binding. Fig. 7b shows that, unlike PKA, for EPAC1h-(149–318) a poor correlation (R = 0.65) is observed between the differential line broadenings of the cGMP- and cAMP-bound states suggesting that the binding of cGMP significantly perturbs the dynamics of EPAC. This conclusion is also supported by the presence of multiple outliers in the correlation shown in Fig. 7b.
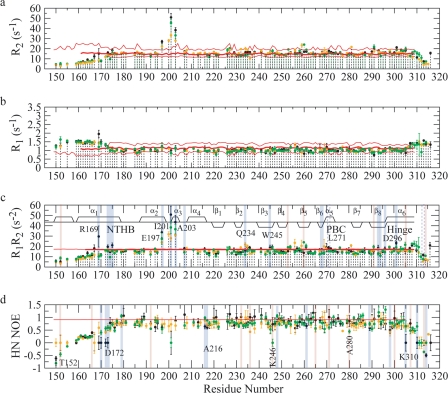
To understand how cGMP binding affects the dynamic patterns of the different subdomains of EPAC, we further probed the picosecond to nanosecond and millisecond to microsecond dynamics of cGMP-bound EPAC1h-(149–318) through the measurement of 15N longitudinal (R1) and transverse (R2) relaxation rates and of heteronuclear {1H}-15N NOEs (Fig. 8, a, b, and d), and we compared our results with those previously reported for the apo- and cAMP-bound states of the same EPAC domain (14). The relaxation data were also analyzed through reduced spectral density mapping (Fig. 9, a–c). The cGMP-dependent changes in dynamics relative to the cAMP-bound state revealed by our 15N relaxation experiments (Fig. 8 and Fig. 9) were then mapped into the three-dimensional structures of EPAC in Fig. 10, a and b, and c and d, for the picosecond to nanosecond and millisecond to microsecond time scales, respectively. The maps of Fig. 10 reveal that cGMP modulates the dynamics of the EPAC CBD at multiple sites affecting both the β- and α-subdomains.
FIGURE 8.
Backbone 15N relaxation rates for EPAC1h-(149–318) in its apo (yellow) (14), cAMP-bound (black) (14), and cGMP-bound (green) states are plotted against residue numbers. The experimental relaxation rates were measured at a field of 700 MHz. a, spin-spin relaxation rate R2; b, spin-lattice relaxation rate R1; c, product of the R1 and R2 relaxation rates; d, 15N{1H} NOE calculated as Isat/Inonsat. c and d, residues experiencing quenching or enhancement of dynamics upon cGMP binding relative to the cAMP-bound state of EPAC1h-(149–318) are highlighted with blue and red, respectively. In all panels, the rates calculated based on hydrodynamic bead models of EPAC1h-(149–309), -(161–309), and -(173–309), respectively, are represented by pink horizontal lines, as described previously (14). These values provide an assessment of the contribution to each rate from overall tumbling. The secondary structure is also shown in c similarly to Fig. 4. The residues for which an apparent quenching in millisecond to microsecond relaxation dynamics may also be due to concurrently enhanced picosecond to nanosecond relaxation rates are highlighted in blue dashed lines. Residues for which no relaxation data are available are prolines or are overlapped and/or broadened beyond detection.
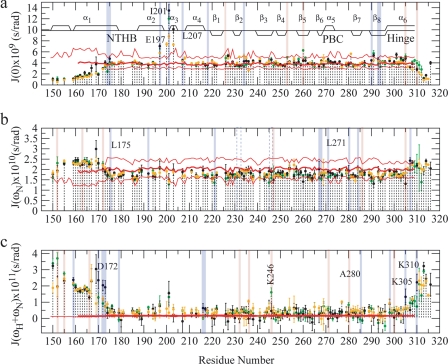
FIGURE 9.
Reduced spectral densities for EPAC1h-(149–318) in its apo (orange) (14), cAMP-bound (black) (14), and cGMP-bound (green) states are plotted against residue numbers. The spectral density values, J(0) (a), J(ωN) (b), and J(ωH +ωN) were computed based on the relaxation rates reported in Fig. 8. a, J(0) value includes contributions from chemical exchange effects. Secondary structure elements are as described in Fig. 4. The J(ωN) (b) and J(ωH +ωN) (c) values were computed using the frequencies ωN = −γN Bo and ωH = −γH Bo with Bo = 16.44 tesla at 700 MHz. The red lines in all three panels indicate the reduced spectral densities computed based on the hydrodynamic bead models, as shown in Fig. 8 (14). In all the panels the residues showing enhanced or reduced dynamics upon cGMP binding relative to the cAMP-bound state are highlighted in red and blue, respectively.
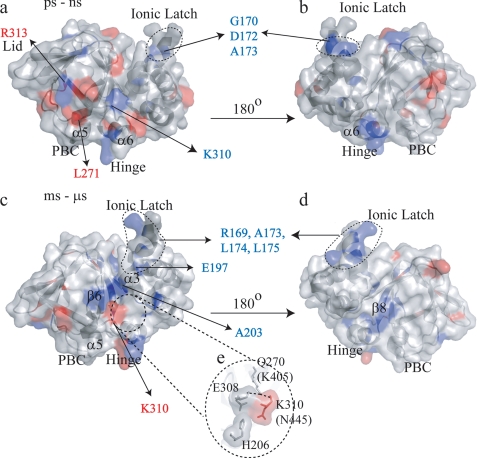
FIGURE 10.
Changes in picosecond to nanosecond (a and b) and millisecond to microsecond (c and d) dynamics for cGMP-bound EPAC1h-(149–318) relative to the cAMP-bound state are mapped onto the holo-EPAC2m structure (PDB 3CF6) (11). b and d were obtained from a and c, respectively, through a 180° rotation. The residues experiencing quenching or enhancement in dynamics upon replacement of cAMP with cGMP are colored according to the color coding of Fig. 8. The amino acid residues are labeled according to the sequence of EPAC1h. The residues reported within parentheses correspond to EPAC2m, and they are not conserved between EPAC1h and EPAC2m. e shows the putative interactions between Gln-270 in the PBC, the 308, 310 residues, and the N-terminal helical-bundle (i.e. α3).
One of the most striking features revealed by the relaxation data recorded for cGMP-bound EPAC1h-(149–318) (Figs. 8–10) is a quenching of dynamics at several critical locations in the N-terminal helical bundle. For instance, cGMP binding results in quenching of millisecond to microsecond and/or picosecond to nanosecond dynamics for several residues in the ionic latch region (i.e. Arg-169, Gly-170, Asp-172, Ala-173, Leu-174, Leu-175, and Glu-197), as evident from the R1R2, J(0), NOE, and J(ωH + ωN) values. This result is confirmed also by the statistical analysis of the relaxation rates shown in Table 4, which reveals that for the ionic latch residues the average R1R2 value observed in the cGMP-bound state is significantly closer to that of the apo-form than to that of the cAMP-bound state. A similar pattern was also observed for the cGMP dependence of the NOE and J(ωH + ωN) averages over the ionic latch region (Table 4). The statistical analysis of Table 4 therefore confirms that cGMP consistently quenches the dynamics of the ionic latch region in both the picosecond to nanosecond and the millisecond to microsecond time scales, bringing the relaxation rates and corresponding spectral densities to values comparable with those previously reported for the inactive apo state (14).
TABLE 4.
Relaxation data for the ionic latch, the α3 and the α6 helices
| Ionic latch, residues 168–173a |
α3 helix, residues 201–205a |
α6 helix, residues 297–318a |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Apo | cGMP | cAMP | Apo | cGMP | cAMP | Apo | cGMP | cAMP | |
| J(0) × 109 (s/radians) | 2.85 ± 0.11 | 3.18 ± 0.18 | 3.62 ± 0.51 | 8.07 ± 0.29 | 10.3 ± 0.86 | 11.8 ± 0.81 | 3.28 ± 0.05 | 3.83 ± 0.20 | 3.85 ± 0.17 |
| J(ωN) × 1010 (s/radians) | 2.22 ± 0.11 | 2.19 ± 0.14 | 2.29 ± 0.21 | 1.65 ± 0.10 | 1.49 ± 0.23 | 1.75 ± 0.28 | 1.87 ± 0.07 | 1.91 ± 0.14 | 1.88 ± 0.13 |
| J(ωH+ ωN) × 1011 (s/radians) | 0.95 ± 0.31 | 0.83 ± 0.18 | 2.33 ± 0.53 | 0.45 ± 0.32 | 1.51 ± 0.35 | 0.88 ± 0.59 | 0.98 ± 0.18 | 0.88 ± 0.22 | 1.13 ± 0.31 |
| R1R2 (s−2) | 14.9 ± 0.82 | 13.9 ± 1.56 | 21.6 ± 4.35 | 29.3 ± 1.79 | 37.4 ± 5.24 | 47.1 ± 7.32 | 14.4 ± 0.51 | 16.3 ± 1.11 | 16.4 ± 1.23 |
| NOE | 0.54 ± 0.14 | 0.57 ± 0.09 | 0.0 ± 0.20 | 0.70 ± 0.21 | 0.0 ± 0.19 | 0.43 ± 0.31 | 0.50 ± 0.10 | 0.59 ± 0.09 | 0.44 ± 0.15 |
a To avoid introduing biases in the comparisons between different states of EPAC1h-(149–318), only the residues were considered for which data are available in all three states.
The consistent quenching of both picosecond to nanosecond and millisecond to microsecond dynamics to values closer to those of the inactive state as cAMP is replaced by cGMP is a feature that is quite unique of the ionic latch site. Indeed other regions, such as the α3 helix, for instance, display a mixed pattern in which cGMP quenches the millisecond to microsecond dynamics but enhances the picosecond to nanosecond flexibility relative to the cAMP-bound state (Table 4). In addition, cGMP binding quenches relatively to cAMP the millisecond to microsecond dynamics of Gln-234 at the β2-β3 loop, β6 (Asp-267 and Phe-268) and β8 (Leu-293, Arg-294, and Asp-296). On the other hand, the NOE data and J(ωH + ωN) values indicate that upon substituting cAMP with cGMP an increase in picosecond to nanosecond dynamics occurs for Phe-232 and Asp-236 at the β2-β3 region, Lys-246 at the β4, Gly-265 at the N terminus of β6, and Leu-271 at the N terminus of PBC (Figs. 8–10). A mixed pattern of cGMP-dependent dynamic changes is observed also for α6 (Figs. 8–10). However, for this C-terminal region the average R1R2 and J(0) values (Table 4) reveal that there is still a net enhancement in millisecond to microsecond dynamics in the cGMP-bound form relative to the apo state, as also observed previously for the cAMP-bound state (14).
DISCUSSION
Binding of cAMP to PKA Is Coupled with an anti to syn Transition
The CBDs of the R subunit of PKA bind cAMP in a syn conformation (46), although our data indicate a predominantly anti orientation for cAMP free in solution (Table 3). These observations suggest that the binding of cAMP to PKA is coupled with an anti to syn transition, most likely driven by several hydrophobic and stacking interactions between the adenine base of cAMP and the PKA CBDs (supplemental Table S1).
Binding of cGMP to PKA Does Not Require an anti to syn Transition but Introduces Short Range Perturbations Confined to the Cyclic Nucleotide-binding Site
Unlike cAMP, cGMP free in solution prefers a syn conformation, and it preserves this base orientation even after binding PKA (47). The syn structure of PKA-bound cGMP ensures that the guanine base can interact with the same hydrophobic and aromatic residues that drive the binding of syn-cAMP (supplemental Table S1) (46). However, the binding of syn-cGMP to PKA introduces steric clashes between the N2H2 of cGMP and the C-β of Ala-210, which result in a slight distortion of the C terminus of the PBC relative to the cAMP-bound structure, as shown before by crystallography (47). This structural perturbation is also confirmed by our chemical shift (Fig. 3, a, c, and d, and Fig. 4, a and c) and H/D (Fig. 5a and Fig. 6a) maps on RIα-(119–244), showing that the hydrogen bond between the Ala-210 NH and the axial exocyclic phosphate oxygen is significantly weakened as cAMP is replaced by cGMP. In addition, our NMR data also show that the distortion of the PBC C terminus caused by cGMP propagates to the N terminus of the PBC and to the BBR (Fig. 4, a and c, Fig. 5a, and Fig. 6a). Specifically, the hydrogen bond between the Gly-199 NH and the 2′-OH of the ribose becomes weaker as cAMP is replaced by cGMP and the local environment of Asn-185 in the BBR is perturbed.
cAMP versus cGMP Selectivity Determinants in PKA
The local perturbations in RIα-(119–244) caused by the replacement of cAMP with cGMP and confined to the PBC termini and the BBR are reminiscent of the effects reported previously (16) for the phosphorothioate (Sp)-cAMPS analog. The (Sp)-cAMPS ligand is a cAMP agonist where the axial exocyclic phosphate oxygen hydrogen-bonded to Ala-210 is isolobally replaced by a bulky sulfur atom. For both cGMP and (Sp)-cAMPS the steric hindrance at the C terminus of the PBC results in reduced binding affinities compared with cAMP (9). Furthermore, for both cGMP and (Sp)-cAMPS, the distortion of Ala-210 is propagated through the adjacent Gln-165 to Asn-185 (16). Asn-185 is located in the hyper-variable β4-β5 loop, which is known to be subject to high sequence variability among different cAMP binding domains (51), and it is not considered critical for the allosteric activation of PKA. Furthermore, no significant cGMP-dependent perturbations relative to the cAMP-bound form are observed for regions known to be essential “hot spots” of the allosteric network of PKA (i.e. the α-B′ helix in the PBC, the hinge α-B/C helix, Gly-169 and Asp-170 in the β2–3 loop) (15, 16, 52, 53). In addition, the dynamic profile of cGMP-bound RIα-(119–244) does not differ significantly from that of cAMP-bound RIα-(119–244) (Fig. 7a), suggesting that also the entropic determinants of the allosteric control of PKA are preserved by cGMP.
Overall, it therefore appears that, similarly to (Sp)-cAMPS, the perturbations on RIα caused by cGMP relative to the cAMP-bound state are limited to recognition rather than allosteric effects, explaining why both (Sp)-cAMPS and cGMP act as weak agonists of cAMP with respect to the activation of PKA. In other words, our data explain why the cAMP versus cGMP second messenger selectivity of PKA is controlled primarily at the level of binding affinity rather than allostery.
Binding of cAMP to EPAC Is Coupled with an anti to syn Transition
Similarly to PKA, the essential CBD of EPAC recognizes cAMP in a syn conformation as shown by our NMR data for EPAC1h-(149–318) bound to cAMP and also by crystallography for the longer EPAC2mΔ305 construct bound to the agonist (Sp)-cAMPS and Rap1B (11). These observations indicate that cAMP binding is coupled to an anti to syn transition for both PKA and EPAC. However, the nature of the CBD-base interactions is quite different in these two systems. In PKA the adenine of cAMP interacts primarily with hydrophobic and aromatic residues in both CBDs without requiring the N6H2 group of the adenine (supplemental Table S1) (46). Unlike PKA, in EPAC the adenine base is involved in both hydrophobic and hydrophilic interactions (supplemental Table S1), including an adenine-specific hydrogen bond with the N6H2 of cAMP (11).
Binding of cGMP to EPAC Is Coupled with a syn to anti Transition
As a result of the adenine-specific interactions in EPAC mentioned above, when cAMP is substituted with cGMP the hydrogen bond involving the N6H2 is lost and the base is now free to change orientation from syn to anti to release the steric hindrance between the N2H2 of the guanine and the Cβ of the conserved alanine at the C terminus of the PBC (i.e. Ala-280 in EPAC1h and Ala-415 in EPAC2m) (Fig. 1c). Indeed, our NMR data support an anti conformation for EPAC-bound cGMP (Fig. 2d and Tables 1 and 3), confirming that binding of cGMP to EPAC is coupled with a syn to anti transition, i.e. the reverse of the base re-orientation observed for cAMP binding to both PKA and EPAC (Table 3).
The anti conformation adopted by cGMP when docked into the binding site of EPAC is also confirmed by the significant chemical shift changes observed for the BBR (i.e. β4 and β5) when cAMP-bound and cGMP-bound EPAC1h-(149–318) are compared (Fig. 3, h and i, and Fig. 4, b and d). These changes reflect an overall decreased protection of the BBR in the cGMP-bound state relative to the cAMP-bound form, as indicated by our comparative H/D analysis (Fig. 5b and Fig. 6b). Our chemical shift and H/D perturbation maps (Fig. 3, f–i and k, Fig. 4, b and d, and Fig. 5b, and Fig. 6b) also show that the effects of the replacement of cAMP with cGMP in EPAC1h-(149–318) are not limited to the BBR, but they extend to most of the remaining parts of the cNMP-binding site, i.e. the N and C termini of the PBC as well as its short α5 helix, suggesting that the docking of the cyclic phosphate and ribose moieties of cGMP into the EPAC CBD may differ at least slightly from that of cAMP.
cAMP Versus cGMP Selectivity Determinants in EPAC1
Another major difference between the PKA and EPAC CBDs is that in the latter the perturbations caused by cGMP propagate well beyond the cNMP-binding site (supplemental material). However, our cAMP versus cGMP comparative NMR analyses reveal that the cGMP-dependent perturbations at the cNMP-binding site and beyond do not significantly compromise the overall structural integrity of cAMP-bound EPAC1h-(149–318) and most remarkably preserve the conformational features of this CBD that are known to be pivotal for allosteric activation (i.e. α5 stabilization, α6 C terminus destabilization, and local environment of Gly-238) (14, 20, 54).
Despite the structural similarities between the cAMP-bound and cGMP-bound states of the EPAC CBD, a critical difference is observed between cAMP and cGMP at the level of dynamic profiles measured for their complexes with EPAC1h-(149–318). Specifically, the most striking effect of the replacement of cAMP with cGMP is a quenching of the picosecond to nanosecond and millisecond to microsecond dynamics for several residues in the N-terminal helical bundle α1–2 ionic latch region (Fig. 10; Table 4), which is involved in the inhibitory RR-CR interactions stabilizing the inactive state. Consistently with the decreased ionic latch flexibility of the cGMP-bound state, multiple sites within helices α1–2 become more protected when cGMP replaces cAMP, in sharp contrast with the concurrent destabilization observed for the rest of the domain (Fig. 5b and Fig. 6b).
The decreased solvent exposure and the suppression of picosecond to nanosecond and millisecond to microsecond dynamics observed in the α1-α2 ionic latch region when cAMP is substituted with cGMP suggest that one of the determinants of the cGMP antagonism in EPAC is the reduction of the entropic penalty, which is otherwise used by cAMP to weaken and release the inhibitory ionic latch interactions (14). Such a dynamically driven allosteric mechanism promotes the selective response of EPAC systems to cAMP as opposed to cGMP, and it may play a key role for the in vivo reduction of the cross-talk between cAMP- and cGMP-dependent signaling cascades. Although it is clear that in vivo the selective activation of cAMP- and/or cGMP-dependent pathways is also controlled through modulations in cNMP synthesis, degradation, and compartmentalization (5), our results indicate that dynamically driven allostery (14, 55–57) provides an additional level of control for the cNMP second messenger selectivity.
Conclusions
The CBDs of both PKA and EPAC selectively sense the signal of cAMP rather than that of other second messengers such as cGMP. However, the mechanisms underlying these selective CBD responses to cAMP are remarkably different in these two systems. In PKA the cAMP versus cGMP selectivity has evolved mainly at the level of binding affinity, whereas in EPAC the cNMP selectivity is mostly defined at the level of allosteric propagations. Our comparative NMR analyses of the structural and dynamic features of the PKA and EPAC CBDs in their cGMP-bound states have provided the molecular basis for the different selectivity mechanisms of these two critical eukaryotic signaling cNMP sensors. Specifically, the PKA CBD responds to cGMP similarly to a previously characterized cAMP agonist, i.e. (Sp)-cAMPS, whereby the same syn base orientation of cAMP is preserved at the price of steric clashes between the NH2 in position 2 of the guanine and the C terminus of the PBC. This hindrance causes a structural distortion in the C-terminal region of the PBC, which accounts for the reduced affinity for PKA. However, the allosteric network of this CBD is not compromised by cGMP, as the only significant cGMP-specific effect that goes beyond the PBC is confined to the hyper-variable β4–5 loop, which has not been shown to be critical for the activation of PKA.
In contrast to PKA, cGMP in EPAC is recognized in an anti conformation and causes multiple perturbations in the cNMP-binding site and beyond relative to the cAMP-bound form. Although these cGMP-dependent effects do not significantly modify the structural features of the cAMP-bound EPAC1 CBD investigated, a remarkable difference is observed between the cAMP- and cGMP-bound states of the EPAC1 CBD at the level of the dynamic profile of the inhibitory ionic latch region. These observations suggest that in EPAC, unlike PKA, a key determinant of the selective response to cAMP is the differential dynamics at critical functional residues. Our study therefore highlights the evolution of allosteric mechanisms for determining signaling selectivity as a function of dynamic changes, even in the absence of significant differences in structure and in ligand affinities. Furthermore, the models of cNMP selectivity outlined here are anticipated to facilitate the design of future drug leads targeting cAMP-dependent pathways.
Supplementary Material
Acknowledgments
We are grateful to Dr. Xiaodong Cheng for generously providing the EPAC1h cDNA and together with Dr. Susan S. Taylor and E. Tyler McNicholl for helpful discussions.
This work was supported by the Canadian Institutes of Health Research.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S3, and Figs. S1 and S2.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S3, and Figs. S1 and S2.
- PKA
- cAMP-dependent protein kinase A
- BBR
- base binding region
- CBD
- cAMP binding domain
- cAMPS
- phosphorothioate analog of cAMP
- CR
- catalytic region
- EPAC
- exchange protein directly activated by cAMP
- GEF
- guanine-nucleotide exchange factor
- HSQC
- heteronuclear single-quantum coherence
- PBC
- phosphate binding cassette
- PF
- protection factor
- R
- regulatory subunit of PKA
- RIα
- isoform Iα of the R subunit of PKA
- RR
- regulatory region
- PDB
- Protein Data Bank
- ROESY
- rotating frame nuclear Overhauser enhancement spectroscopy
- NOE
- nuclear Overhauser effect
- MES
- 4-morpholineethanesulfonic acid
- H/D
- hydrogen/deuterium exchange
- CPMG
- Carr-Purcell-Meiboom-Gill pulse program.
REFERENCES
- 1.Kim C., Vigil D., Anand G., Taylor S. S. (2006) Eur. J. Cell Biol. 85,651–654 [DOI] [PubMed] [Google Scholar]
- 2.Rehmann H., Wittinghofer A., Bos J. L. (2007) Nat. Rev. Mol. Cell Biol. 8,63–73 [DOI] [PubMed] [Google Scholar]
- 3.Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. (2007) Cell 130,1032–1043 [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Dillon T. J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P. J. (2006) Mol. Cell. Biol. 26,2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelligrino D. A., Wang Q. (1998) Prog. Neurobiol. 56,1–18 [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Robinson P. J. (1997) J. Neurochem. 68,443–456 [DOI] [PubMed] [Google Scholar]
- 7.Shabb J. B., Buzzeo B. D., Ng L., Corbin J. D. (1991) J. Biol. Chem. 266,24320–24326 [PubMed] [Google Scholar]
- 8.Shabb J. B., Ng L., Corbin J. D. (1990) J. Biol. Chem. 265,16031–16034 [PubMed] [Google Scholar]
- 9.Rehmann H., Schwede F., Døskeland S. O., Wittinghofer A., Bos J. L. (2003) J. Biol. Chem. 278,38548–38556 [DOI] [PubMed] [Google Scholar]
- 10.Christensen A. E., Selheim F., de Rooij J., Dremier S., Schwede F., Dao K. K., Martinez A., Maenhaut C., Bos J. L., Genieser H. G., Døskeland S. O. (2003) J. Biol. Chem. 278,35394–35402 [DOI] [PubMed] [Google Scholar]
- 11.Rehmann H., Arias-Palomo E., Hadders M. A., Schwede F., Llorca O., Bos J. L. (2008) Nature 455,124–127 [DOI] [PubMed] [Google Scholar]
- 12.Zagotta W. N., Olivier N. B., Black K. D., Young E. C., Olson R., Gouaux E. (2003) Nature 425,200–205 [DOI] [PubMed] [Google Scholar]
- 13.Rehmann H., Das J., Knipscheer P., Wittinghofer A., Bos J. L. (2006) Nature 439,625–628 [DOI] [PubMed] [Google Scholar]
- 14.Das R., Mazhab-Jafari M. T., Chowdhury S., SilDas S., Selvaratnam R., Melacini G. (2008) J. Biol. Chem. 283,19691–19703 [DOI] [PubMed] [Google Scholar]
- 15.Das R., Esposito V., Abu-Abed M., Anand G. S., Taylor S. S., Melacini G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104,93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das R., Melacini G. (2007) J. Biol. Chem. 282,581–593 [DOI] [PubMed] [Google Scholar]
- 17.Das R., Abu-Abed M., Melacini G. (2006) J. Am. Chem. Soc. 128,8406–8407 [DOI] [PubMed] [Google Scholar]
- 18.Abu-Abed M., Das R., Wang L., Melacini G. (2007) Proteins 69,112–124 [DOI] [PubMed] [Google Scholar]
- 19.Esposito V., Sjoberg T., Das R., Brown S., Taylor S. S., Melacini G. (2006) J. Biomol. NMR 36, Suppl. 1, 64. [DOI] [PubMed] [Google Scholar]
- 20.Mazhab-Jafari M. T., Das R., Fotheringham S. A., SilDas S., Chowdhury S., Melacini G. (2007) J. Am. Chem. Soc. 129,14482–14492 [DOI] [PubMed] [Google Scholar]
- 21.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6,277–293 [DOI] [PubMed] [Google Scholar]
- 22.Cavanagh J., Fairbrother W. J., Palmer A. G., 3rd, Skelton N. J. (1996) Protein NMR Spectroscopy: Principles and Practice, pp. 100–102, Academic Press, New York [Google Scholar]
- 23.Goddard T. D., Kneller D. G. (2006) SPARKY Version 3, University of California, San Francisco [Google Scholar]
- 24.Farrow N. A., Zhang O., Forman-Kay J. D., Kay L. E. (1994) J. Biomol. NMR 4,727–734 [DOI] [PubMed] [Google Scholar]
- 25.Vialle-Printems C., van Heijenoort C., Guittet E. (2000) J. Magn. Reson. 142,276–279 [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez J. C., Jennings P. A., Melacini G. (2004) J. Biomol. NMR 30,155–161 [DOI] [PubMed] [Google Scholar]
- 27.Mulder F. A., Bouakaz L., Lundell A., Venkataramana M., Liljas A., Akke M., Sanyal S. (2004) Biochemistry 43,5930–5936 [DOI] [PubMed] [Google Scholar]
- 28.Adén J., Wolf-Watz M. (2007) J. Am. Chem. Soc. 129,14003–14012 [DOI] [PubMed] [Google Scholar]
- 29.Sattler M., Schleucher J., Griesinger C. (1999) Prog. Nucleic Magn. Reson. Spectrosc. 34,93–158 [Google Scholar]
- 30.Eghbalnia H. R., Bahrami A., Wang L., Assadi A., Markley J. L. (2005) J. Biomol. NMR 32,219–233 [DOI] [PubMed] [Google Scholar]
- 31.Gronenborn A. M., Clore G. M., Blazy B., Baudras A. (1981) FEBS Lett. 136,160–164 [DOI] [PubMed] [Google Scholar]
- 32.Desvaux H., Berthault P. (1999) Progr. NMR Spectr. 35,295–340 [Google Scholar]
- 33.Ni F. (1994) Prog. Nucleic Magn. Reson. Spectrosc. 26,517–606 [Google Scholar]
- 34.Otting G., Wuthrich K. (1989) J. Magn. Res. 85,586–594 [Google Scholar]
- 35.de Ven F. J. M. v. (1995) Multidimensional NMR in Liquids: Basic Principles and Experimental Methods, pp. 99–105, Wiley-VCH, New York [Google Scholar]
- 36.Palmer A. G. (1998) Curvefit, Columbia University, New York [Google Scholar]
- 37.Bai Y., Milne J. S., Mayne L., Englander S. W. (1993) Proteins 17,75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33,5984–6003 [DOI] [PubMed] [Google Scholar]
- 39.Fischer M. W., Majumdar A., Zuiderweg E. R. (1998) Prog. Nucleic Magn. Reson. Spectrosc. 33,207–272 [Google Scholar]
- 40.Yip G. N., Zuiderweg E. R. (2004) J. Magn. Reson. 171,25–36 [DOI] [PubMed] [Google Scholar]
- 41.Renner C., Schleicher M., Moroder L., Holak T. A. (2002) J. Biomol. NMR 23,23–33 [DOI] [PubMed] [Google Scholar]
- 42.Bernadó P., García de la Torre J., Pons M. (2002) J. Biomol. NMR 23,139–150 [DOI] [PubMed] [Google Scholar]
- 43.García de la Torre J. G. (2001) Biophys. Chem. 93,159–170 [DOI] [PubMed] [Google Scholar]
- 44.Atkinson R. A., Kieffer B. (2004) Prog. Nucleic Magn. Reson. Spectrosc. 44,141–187 [Google Scholar]
- 45.Houben K., Wasielewski E., Dominguez C., Kellenberger E., Atkinson R. A., Timmers H. T., Kieffer B., Boelens R. (2005) J. Mol. Biol. 349,621–637 [DOI] [PubMed] [Google Scholar]
- 46.Su Y., Dostmann W. R., Herberg F. W., Durick K., Xuong N. H., Ten Eyck L., Taylor S. S., Varughese K. I. (1995) Science 269,807–813 [DOI] [PubMed] [Google Scholar]
- 47.Wu J., Brown S., Xuong N. H., Taylor S. S. (2004) Structure 12,1057–1065 [DOI] [PubMed] [Google Scholar]
- 48.Clayton G. M., Silverman W. R., Heginbotham L., Morais-Cabral J. H. (2004) Cell 119,615–627 [DOI] [PubMed] [Google Scholar]
- 49.Passner J. M., Steitz T. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94,2843–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seguí-Lines G., Gavina J. M., D'Amaral J. C., Britz-McKibbin P. (2007) Analyst 132,741–744 [DOI] [PubMed] [Google Scholar]
- 51.Johnson D. A., Akamine P., Radzio-Andzelm E., Madhusudan M., Taylor S. S. (2001) Chem. Rev. 101,2243–2270 [DOI] [PubMed] [Google Scholar]
- 52.Kannan N., Wu J., Anand G. S., Yooseph S., Neuwald A. F., Venter J. C., Taylor S. S. (2007) Genome Biol. 8,R264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kornev A. P., Taylor S. S., Ten Eyck L. F. (2008) PLoS Comput. Biol. 4,e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harper S. M., Wienk H., Wechselberger R. W., Bos J. L., Boelens R., Rehmann H. (2008) J. Biol. Chem. 283,6501–6508 [DOI] [PubMed] [Google Scholar]
- 55.Cooper A., Dryden D. T. (1984) Eur. Biophys. J. 11,103–109 [DOI] [PubMed] [Google Scholar]
- 56.Stevens S. Y., Sanker S., Kent C., Zuiderweg E. R. (2001) Nat. Struct. Biol. 8,947–952 [DOI] [PubMed] [Google Scholar]
- 57.Popovych N., Sun S., Ebright R. H., Kalodimos C. G. (2006) Nat. Struct. Biol. 13,831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.