Abstract
Elevated intracellular Ca2+ ([Ca2+]i) inhibition of NHE3 is reconstituted by NHERF2, but not NHERF1, by a mechanism involving the formation of multiprotein signaling complexes. To further evaluate the specificity of the NHERF family in calcium regulation of NHE3 activity, the current study determined whether NHERF3 reconstitutes elevated [Ca2+]i regulation of NHE3. In vitro, NHERF3 bound the NHE3 C terminus between amino acids 588 and 667. In vivo, NHE3 and NHERF3 associate under basal conditions as indicated by co-immunoprecipitation, confocal microscopy, and fluorescence resonance energy transfer. Treatment of PS120/NHE3/NHERF3 cells, but not PS120/NHE3 cells, with the Ca2+ ionophore, 4-bromo-A23187 (0.5 μm): 1) inhibited NHE3 Vmax activity; 2) decreased NHE3 surface amount; 3) dissociated NHE3 and NHERF3 at the plasma membrane by confocal immunofluorescence and fluorescence resonance energy transfer. Similarly, in Caco-2BBe cells, NHERF3 and NHE3 colocalized in the BB under basal conditions but after elevation of [Ca2+]i by carbachol, this overlap was abolished. NHERF3 short hairpin RNA knockdown (>50%) in Caco-2BBe cells significantly reduced basal NHE3 activity by decreasing BB NHE3 amount. Also, carbachol-mediated inhibition of NHE3 activity was abolished in Caco-2BBe cells in which NHERF3 protein expression was significantly reduced. In summary: 1) NHERF3 colocalizes and directly binds NHE3 at the plasma membrane under basal conditions; 2) NHERF3 reconstitutes [Ca2+]i inhibition of NHE3 activity and dissociates from NHE3 in fibroblasts and polarized intestinal epithelial cells with elevated [Ca2+]i; 3) NHERF3 short hairpin RNA significantly reduced NHE3 basal activity and brush border expression in Caco-2BBe cells. These results demonstrate that NHERF3 reconstitutes calcium inhibition of NHE3 activity by anchoring NHE3 basally and releasing it with elevated Ca2+.
In normal digestive physiology, the brush border (BB)2 Na+/H+ exchanger, NHE3, mediates the majority of the NaCl and NaHCO3 absorption in the ileum (1). Sequential inhibition and stimulation of NHE3 occur as part of digestive physiology. Short-term regulation of NHE3 activity is achieved through a variety of factors that affect NHE3 turnover number and/or surface expression and often involve a role for the cytoskeleton and accessory proteins, including the multi-PDZ domain containing proteins, NHERF1 and NHERF2 (1, 2). However, many details of this regulation are not understood.
The NHERF (Na+/H+ exchanger regulatory factor) family of multi-PDZ domain containing proteins consists of four evolutionarily related members, all of which are expressed in epithelial cells of the mammalian small intestine (2). NHERF1 and NHERF2 have been previously shown to contribute to acute NHE3 stimulation and inhibition (3–10). Recently, two additional PDZ domain containing proteins, termed NHERF3/PDZK1 and NHERF4/PDZK2/IKEPP, have been demonstrated to possess sequence homology with NHERF1 and NHERF2 (11–14). However, unlike NHERF1 and NHERF2, which are comprised of two tandem PDZ domains flanked by a C-terminal ezrin/radixin/moesin binding domain, NHERF3 and NHERF4 consist of four PDZ domains but no other protein-protein interacting domains (12).
NHERF3 was initially identified by a yeast two-hybrid screen from a human kidney cDNA library using the membrane-associated protein MAP17, as bait (12). NHERF3 is expressed in the brush border of epithelial cells of the kidney proximal tubule and the small intestine (12). NHERF3 associates with and, in a few cases, has been shown to regulate the activity of multiple apical membrane ion transporters including the cystic fibrosis transmembrane regulator (CFTR), urate anion exchanger 1 (URAT1), sodium-phosphate cotransporter type IIa (NaPiIIa), proton-coupled peptide transporter (PEPT2), and organic cation/carnitine cotransporter (OCTN2) (15–19). Furthermore, NHERF3 directly binds the C terminus of NHE3 (20). Recent studies have begun evaluating the effect of NHERF3 on mouse intestinal Na+ and Cl− transport. Basal electroneutral sodium absorption was decreased by >40% in the NHERF3 null mouse jejunum (21) and by >80% in the colon (22). In addition, Cinar et al. (22) demonstrated that cAMP and [Ca2+]i inhibition of NHE3 activity was abolished in the NHERF3 null mouse colon. However, the mechanism by which NHERF3 regulates NHE3 activity was not resolved.
Several physiological and pathophysiological agonists, acting through [Ca2+]i-induced second messenger systems, are known to inhibit electroneutral NaCl absorption in the small intestine (1, 23). Elevation of [Ca2+]i has previously been demonstrated to inhibit NHE3 activity in a NHERF2-, but not NHERF1-dependent manner (5). NHERF2 regulation of NHE3 involves the formation of multiprotein complexes at the plasma membrane that include NHE3, NHERF2, α-actinin-4, and PKCα, which induce endocytic removal of NHE3 from the plasma membrane by a PKC-dependent mechanism (5, 24). Because multiple PDZ proteins exist in the apical pole of epithelial cells (2), the current study was designed to determine whether NHERF3 could reconstitute Ca2+ regulation of NHE3 activity and to define how that occurred.
EXPERIMENTAL PROCEDURES
Reagents
4-Bromo-A23187, the non-fluorescent analog of the calcium ionophore, A23187, was from Biomol (25).
Antibodies
Affinity-purified rabbit polyclonal antibody to human NHERF3 was described previously (26). A separate rabbit polyclonal anti-NHERF3 antibody was used in Fig. 1D and was described previously (19). Mouse monoclonal anti-vesicular stomatitis virus (VSV)-G antibody P5D4 (hybridoma culture medium) was from Drs. T. Kreiss and D. Louvard. Mouse monoclonal anti-(VSV)-G Cy3-conjugated antibody was from Sigma.
FIGURE 1.
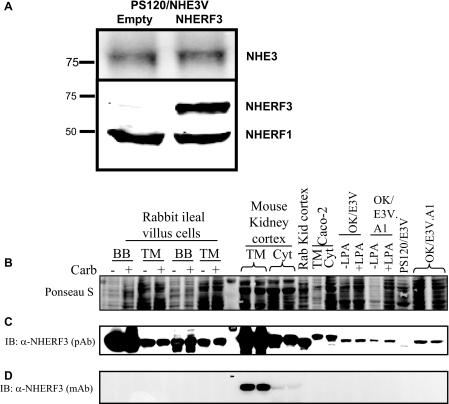
NHERF3 is expressed in renal and intestinal epithelial cells but not PS120 cells. A, PS120 cells express some endogenous NHERF1 but not NHERF2, NHERF3, or NHERF4. PS120 cells expressing NHE3 were stably transfected with either empty pcDNA3.1 vector or NHERF3 and used for the current studies. NHERF1 and NHERF3 proteins were detected by Western blot in cell lysates from PS120/NHE3V cells. B, membranes were stained with Ponceau S solution to estimate protein loading consistency among samples. C, NHERF3 protein is expressed in the brush border (BB), total membrane (TM), and cytosol (Cyt) made from the rabbit ileal sodium absorptive cells and mouse and rabbit kidney cortex. NHERF3 protein is also endogenously expressed in Caco2 TM and Cyt preparations as well as in OK cells but not in PS120 cells. D, a mouse-specific antibody (19) was used to detect NHERF3 in TM and Cyt preparations from the mouse kidney cortex. IB, immunoblot; pAb, polyclonal antibody; mAb, monoclonal antibody; LPA, lysophosphatidic acid; OK/E3V.A1 designates a clone.
Cell Lines
PS120 fibroblasts lack all endogenous plasma membrane NHEs, NHERF1 (minimal expression), NHERF2, NHERF3, and NHERF4/PDZK2/IKEPP (27). These cells, when stably expressing rabbit NHE3 with a C-terminal VSV-G protein epitope tag, are called PS120/NHE3 cells or E3V cells, as described (stable cell lines made using pcDNA 3.1; G418; Invitrogen) (5). All PS120 lines were grown in Dulbecco's modified Eagle's medium supplemented with 25 mm NaHCO3, 10 mm HEPES, 50 units/ml penicillin, 50 μg/ml streptomycin, 400 μg/ml G418, and 10% fetal bovine serum in a 5% CO2, 95% O2 incubator at 37 °C. PS120/NHE3 cells stably expressing NHERF3 were also generated (using pcDNA 3.1; hygromycin) and cultured in the above medium supplemented with 600 μg/ml hygromycin.
Caco-2BBe cells express all four members of the NHERF gene family and small amounts of NHE3. Triple HA-tagged rabbit NHE3 in replication-deficient adenovirus was transiently infected into Caco-2BBe cells for subsequent biochemical analysis. Caco-2BBe cells were grown on Transwell filters (Corning) until 12 days post-confluence in Dulbecco's modified Eagle's medium supplemented with 25 mm NaHCO3, 10 mm HEPES, 0.1 mm nonessential amino acids, 50 units/ml penicillin, 50 μg/ml streptomycin, and 10% fetal bovine serum in a 5% CO2, 95% O2 incubator at 37 °C. Cells were serum-starved overnight and then treated with 6 mm EGTA for 2 h at 37 °C. Caco-2BBe cells were exposed to adenovirus 3HA-NHE3 for 6 h at 37 °C. Cells were allowed to recover in normal media over the next 40 h before study.
Immunofluorescence
PS120/NHE3/pcDNA3.1 and PS120/NHE3/NHERF3 cells were seeded on glass coverslips and grown to 70% confluence. Cells were serum starved for 3 h and then treated with vehicle or 4-bromo-A23187 (0.5 μm) for 15 min, washed three times in phosphate-buffered saline (PBS) and fixed for 15 min with 3% paraformaldehyde in PBS. The fixed cells were washed with PBS and treated with 20 mm l-glycine for 10 min. Cells were placed in blocking solution (PBS containing 15% fetal bovine serum, 2% bovine serum albumin, and 0.1% saponin) for 45 min at room temperature. Primary antibodies were incubated for 1 h at room temperature in blocking solution at the following dilutions: 1:100 for monoclonal anti-(VSV)-G Cy3-conjugated antibody (anti-NHE3 antibody) and 1:100 for polyclonal anti-NHERF3 antibody. Cells were then washed three times with PBS and incubated with anti-mouse Alexa Fluor 488-conjugated secondary antibodies (1:100) for 1 h at room temperature. Cells were washed three times with PBS and mounted with Gel Mount (Sigma) and then examined with a Zeiss LSM510 confocal fluorescence microscope. Results were then obtained from 6 to 8 individual experiments.
Fusion Proteins
His6 fusion proteins of full-length rabbit NHE3 C terminus (aa 475–832) and individual fragments of the C terminus of NHE3 (F1 (aa 475–588), F2 (aa 589–667), F3 (aa 668–747), and F4 (aa 748–832) were generated as described previously (28, 29).
Protein-Protein Interactions
Protein overlay (Far Western) assays were used to examine the interaction of recombinant NHE3 C terminus (3 μg of each full-length and fragments F1–4), on blots with recombinant NHERF3 (overlay) by subsequent incubation of blots with polyclonal anti-NHERF3 antibody, as described previously (29). Membranes were exposed to ECL (Amersham Biosciences) and x-ray film for ∼1 min. Results were obtained from 3 individual experiments.
NHERF3 shRNA
Sequence-verified shRNA lentiviral plasmids in hairpin-pLKO.1←-puromycin vector for NHERF3 gene silencing in Caco-2BBe cells were obtained through the Johns Hopkins HiT center from Open Biosystems (Huntsville, AL) and were used to generate lentiviral transduction particles. The shRNA NHERF3 constructs were: A, CCGGGCAAGGTTTGAGTGATAATATCTCGAGATATTATCACTCAAACCTTGCTTTTTG (TRCN0000059668); B, CCGGCCTATGATTATTTCCAAGCTACTCGAGTAGCTTGGAAATAATCATAGGTTTTTG (TRCN0000059669); and C, CCGGCTTAGGATCAATGGTGTCTTTCTCGAGAAAGACACCATTGATCCTAAGTTTTTG (TRCN0000059670).
For production of lentiviral particles, three components (pLKO.1 vector containing shRNA, a packaging vector pCMV-dR8.91 containing gag, pol, and rev genes, and envelop vector pCMV-VSV-G) were transfected into human embryonic kidney 293T cells. All plasmids were prepared by using an EndoFree Plasmid Maxi kit (Qiagen, Valencia, CA). Twenty-four hours before transfection, 20 × 106 human embryonic kidney 293T cells were plated on a 10-cm Petri dish. One hour before transfection the medium was changed to Opti-MEM serum-free media. The Lipofectamine 2000 method was used for transfection according to the manufacturer's protocol (Invitrogen). The transfection conditions were: 10 μg of packaging plasmid + 6 μg of envelope coding plasmid + 10 μg of shRNA coding plasmid in 500 μl of Opti-MEM. Lipofectamine 2000 solution was combined with plasmid solution and added to the cells. Production of lentiviruses was enhanced by replacement of the cell culture media at 16 h post-transfection with 5 ml of fresh media containing 10 mm sodium butyrate for 8 h (30, 31). After incubation, sodium butyrate was replaced with 5 ml of fresh media for another 16 h before virus harvesting. The lentivirus supernatants were passed through 0.45-μm pore PVDF Durapore filters (Millipore, Bedford, MA) and used immediately to transduce Caco-2BBe cells with shRNA of interest or stored at −80 °C for future use.
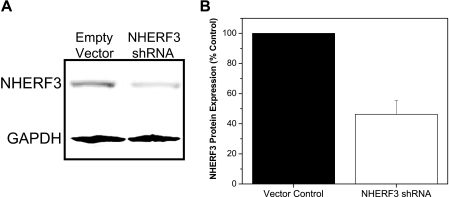
For lentiviral transduction, Caco-2BBe cells were plated on 6-well Transwell plates 24 h before to achieve 30–40% confluence. Harvested viral particles were mixed with equal volumes of complete Caco-2BBe cell media, incubated in the presence of 90 μg/ml Polybrene for 30 min at 37 °C, and added to the well. Twenty-four hours later, media was replaced with complete Caco-2BBe cell media containing 5 μg/ml puromycin. Cells transduced with either empty vector or NHERF3 shRNA constructs grew and achieved confluency in less than 7 days. Caco-2BBe cells containing empty vector or NHERF3 shRNA were studied at 14 days post-confluence. Efficiency of NHERF3 knockdown was assessed by Western blot (n = 4 for each shRNA construct) of total cell lysates prepared from Caco-2BBe cells transduced with each NHERF3 shRNA construct (A–C). NHERF3 shRNA construct A was ineffective in reducing endogenous NHERF3 expression in Caco-2BBe cells, whereas constructs B and C each knocked down endogenous NHERF3 in Caco-2BBe cells by ∼50%. In the current study, NHERF3 shRNA knockdown studies in Caco-2BBe cells used construct C.
Co-immunoprecipitation
Caco-2BBe cells were grown on 10-cm2 Transwell Petri dishes until 10 days post-confluence. On day 12, cells were serum starved for 4 h and then treated with 6 mm EGTA for 2.5 h. Cells were then incubated with the adenovirus 3HA-NHE3 construct for 6 h in serum-free media. After infection, cells were allowed to recover in normal media for 40 h prior to carbachol treatment. On day 14, cells were serum-starved again for 4 h and treated either with vehicle or 10 μm carbachol (Sigma) for 10 min at 37 °C. Adenovirus-infected Caco-2BBe cells were washed three times in ice-cold PBS containing 50 mm Tris. Cells were collected and lysed in 500 μl of ice-cold lysis buffer (10 mm HEPES, 50 mm NaCl, 5 mm EDTA, 1 mm benzamidine, 0.5% Triton X-100). Cell lysate was solubilized for 30 min at 4 °C with end-over-end rotation and subsequently homogenized 10 times using a 23-gauge needle. Cellular debris was cleared by centrifugation at 14,000 × g for 15 min. Supernatant was incubated with either anti-HA affinity matrix (Roche) or anti-VSV-G-agarose beads (Sigma) for 2 h with end-over-end rotation at 4 °C. Samples were washed five times with lysis buffer and immunoprecipitated proteins were eluted from beads with 2× sample buffer. Samples were resolved by 10% SDS-PAGE and proteins were detected with anti-HA and anti-NHERF3 antibodies and visualized on an Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE). Results were obtained from three individual experiments.
Measurement of Na+/H+ Exchange
Cellular Na+/H+ exchange activity in PS120 cells grown to ∼70% confluence on glass coverslips and confluent Caco-2BBe cells grown on Transwell filters was determined fluorometrically using the intracellular pH-sensitive dye, 2′,7′-bis(carboxyethyl)5–6-carboxyfluorescein-acetoxymethyl ester (BCECF-AM, 5 μm; Molecular Probes, Eugene, OR), as described previously (32). PS120 cells were exposed to 40 mm NH4Cl alone or with 4-bromo-A23187 (0.5 μm) during a 15-min dye loading, as described previously (32). Caco-2BBe cells were exposed to 50 mm NH4Cl alone or with carbamylcholine chloride (carbachol; 10 μm) for the final 10 min of the 50-min dye loading period. For Caco-2BBe cells, endogenous NHE1 and NHE2 activity were inhibited by 50 μm HOE694 (kindly provided by Dr. H. Lang, Hoechst, Germany) present in all solutions. At the end of each experiment, the fluorescence ratio was converted to pHi using the high potassium/nigericin method (33). Na+/H+ exchange activity data in PS120 cells was calculated as the product of Na+-dependent changes in pHi × the buffering capacity at each pHi, and individual points shown in figures are rates of Na+/H+ exchange calculated at multiple pHi values using at least three coverslips per condition in a single experiment. Kinetics of Na+/H+ exchange were analyzed by Hill plot using Origin (Microcal Software) to estimate Vmax and K′(H+)i in individual experiments (33). For Caco-2BBe cells, the initial rate (about the first minute) of NHE3 activity was measured and expressed as change in pHi/min (34, 35). Mean ± S.E. were determined from nine separate experiments (31).
Measurement of Surface NHE3
To measure surface NHE3 amount, PS120 cells were treated with either 4-bromo-A23187 (15 min) or vehicle at 37 °C and then surface-labeled with biotin at 4 °C as described previously (29, 36). The total, intracellular, and surface fractions were resolved by SDS-PAGE and transferred to NC membrane, and NHE3 was quantified by labeling with anti-VSV-G antibody. Protein amounts were analyzed on the Odyssey Infrared Imaging System (Li-Cor). The efficiency of cell surface biotinylation of NHE is estimated to be at least 89% (36). Mean ± S.E. were determined from at least three separate experiments.
Acceptor Photobleaching Fluorescence Resonance Energy Transfer (FRET)
Fluorescence microscopy was performed as described previously on PS120 cells fixed in 3% paraformaldehyde (5, 34), with minor revisions. NHE3 (VSV-G-tagged) expression was visualized with anti-VSV-G Cy3-conjugated primary antibody. NHERF3 expression was detected with anti-NHERF3 antibody and goat anti-rabbit Cy5 secondary antibody (Jackson ImmunoResearch). Cells were imaged on a Zeiss LSM 410 confocal microscope (Carl Zeiss, Thornwood, NY) using a 1.3 NA ×100 Plan-neofluor objective. Cy3 and Cy5 fluorescence was excited using lasers at 534 and 633 nm and detected using appropriate filter sets (Cy3 filter cube: excitation 515–560 nm, 565-nm long pass dichroic, emission 573–648 nm; Cy5 filter cube: excitation 590–650 nm, 660-nm long pass dichroic, emission 663–738 nm) (Chroma Technology, Brattleboro, VT). No fluorescence was observed from a Cy3-labeled specimen using the Cy5 filters, nor was Cy5 fluorescence detected using the Cy3 filter set.
In the present study, the acceptor bleaching FRET method was used, as described in detail (29, 37, 38). Images were collected from 50 to 100 different regions of interest from a single monolayer, stored on a computer disk, and fluorescence intensity of donor before and after acceptor photobleaching was analyzed for 50–100 identical 10 × 10 pixel regions of interest in each individual image using Metamorph (Universal Imaging Corp.) with macros. FRET efficiency was calculated as,
where ID1 and ID2 are the donor fluorescence intensities before and after acceptor photobleaching, respectively (30). FRET efficiency E was analyzed and plotted as a function of acceptor fluorescence intensity. Mean ± S.E. were determined from at least three separate experiments.
Statistics
Results were expressed as mean ± S.E. Statistical evaluation was by analysis of variance or Student's t test.
RESULTS
Expression of NHERF3
PS120 cells lack endogenous NHERF3 as well as NHERF2 and NHERF4 but do express small amounts of NHERF1 (Fig. 1A) (27). OK proximal tubule cells and polarized intestinal Caco-2BBe cells express NHERF3 protein (Fig. 1, B and C). Using a polyclonal antibody, NHERF3 protein is also expressed in the BB, total membrane, and cytoplasm (Cyto) preparations from rabbit ileal mucosa and mouse and rabbit kidney cortex (Fig. 1C). A rodent-specific monoclonal antibody (19) was also used to probe for NHERF3 expression and detected NHERF3 protein expression in mouse kidney cortex (Fig. 1D). As determined by Western blot, NHERF3 is highly enriched in the ileal BB as well as the mouse and rabbit kidney proximal tubule membranes (Fig. 1C).
Although NHERF3 has been shown to regulate NHE3 activity in the mouse intestine, the mechanism responsible for this regulation remains unclear (21, 22). Therefore, the current study examined the role of NHERF3 in the PS120 fibroblast cell model, which lacks NHERF2 and NHERF4 as well as the polarized intestinal epithelial cell model, Caco-2BBe, which expresses all four members of the NHERF family. NHERF3 or empty pcDNA3.1 vector control were transfected into PS120/NHE3V cells and protein expression of NHE3 and NHERF3 was determined by Western blot (Fig. 1A). NHERF3 has been suggested to play a significant role in basal NHE3 activity in mouse intestine (21, 22). We have previously demonstrated that NHERF1 and NHERF2 directly bind NHE3 between aa 589 and 667 (28). Therefore the next study was designed to determine whether NHERF3 also bound the same region of the NHE3 C terminus.
NHERF3 Directly Interacts with NHE3 by in Vitro Overlay Assay
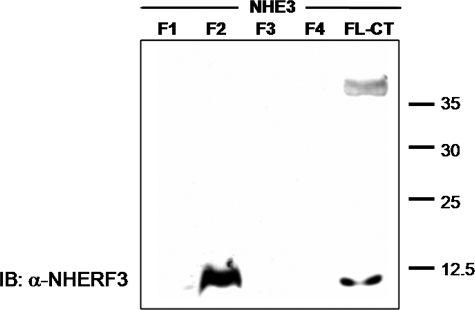
To assess direct physical interaction of NHE3 and NHERF3/PDKZ1, protein overlay assays were employed using the full-length C terminus of NHE3 (aa 475–832) as well as the C terminus divided into four contiguous fragments. NHERF3 bound to NHE3 directly. This interaction occurred in the full-length C terminus of NHE3 as well as in the F2 C-terminal fragment (aa 589–667) of NHE3 but not F1, F3, or F4 (Fig. 2). This is the same region previously shown to bind NHERF1 and NHERF2 (28). Thus, NHERF3 can directly bind NHE3 in vitro between aa 588 and 667.
FIGURE 2.
NHERF3 directly binds to the C terminus of NHE3. Purified His6 fusion proteins making up the entire NHE3 C terminus (CT) were generated and divided into four parts (F1–4), separated on SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were incubated with purified recombinant human NHERF3 protein and visualized by primary antibody to NHERF3 and exposed to enhanced chemiluminescence. Protein overlay assays demonstrate that purified recombinant human NHERF3 binds to the full-length C-terminal fragment of NHE3 (aa 475–832) and the F2 region of NHE3 (aa 589–667). Lower band in the full-length NHE3 C-terminal fragment is assumed to be a breakdown product containing the F2 domain. Results were obtained from three individual experiments. IB, immunoblot.
Elevation of [Ca2+]i Inhibits NHE3 Activity in PS120 Cells Co-expressing NHERF3 by Decreasing Surface NHE3
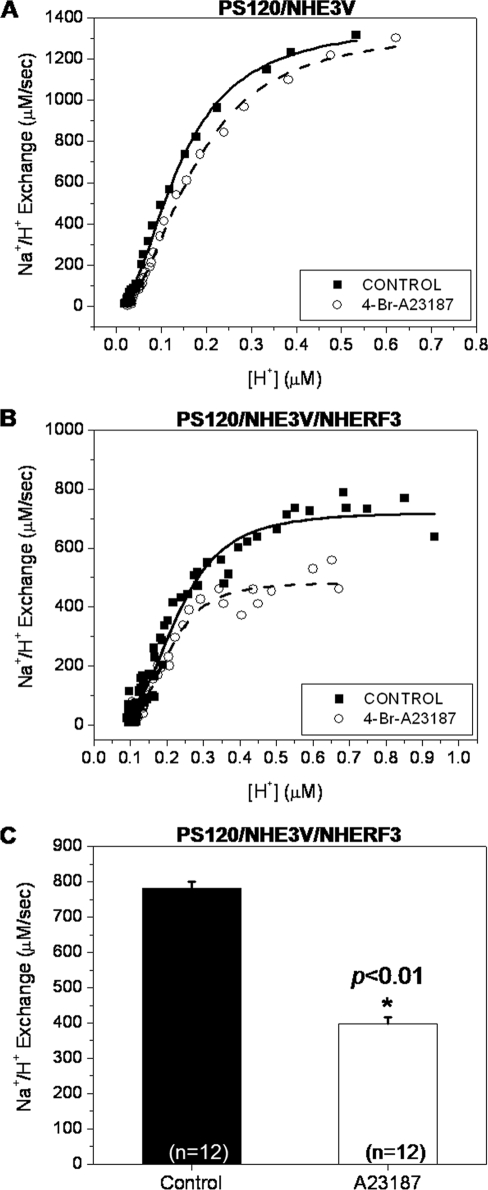
Although PS120 cells express a small amount of NHERF1, which does not reconstitute Ca2+ regulation of NHE3 activity, the current study examined the role of NHERF3 in calcium regulation of NHE3 activity (5). Na+/H+ exchange was measured in PS120 cells using the pH-sensitive dye, BCECF. In PS120 cells expressing NHE3 but not NHERF3, elevation of [Ca2+]i using 4-bromo-A23187 had no effect on NHE3 activity (Fig. 3A; n = 12). However, in PS120 cells stably expressing NHE3 and NHERF3, A23187 treatment significantly decreased NHE3 activity, by more than 40% (Fig. 3, B and C; n = 12). These experiments demonstrate that NHERF3 can reconstitute Ca2+ regulation of NHE3 activity, a result similar to that demonstrated previously with NHERF2 (5).
FIGURE 3.
NHERF3 inhibits NHE3 activity in response to elevated [Ca2+]i. Na+/H+ exchange was measured in PS120/NHE3V cells expressing NHERF3 or empty vector control, using the pH-sensitive dye, BCECF, which was loaded into cells during a 15-min incubation with 40 mm NH4Cl. A, in PS120 cells not expressing NHERF3, elevation of [Ca2+]i by the calcium ionophore, 4-bromo-A23187 (0.5 μm), had no effect on NHE3 activity. B, A23187 treatment in PS120 cells co-expressing NHE3 and NHERF3, resulted in decreased NHE3 activity by ∼40%. Panels A and B represent measurements taken from single experiments. C, bar graph summarizes the mean ± S.E. of 12 experiments similar to that in B. p value (*) is in comparison with control (paired t test).
In PS120/NHE3V/NHERF3 cells, under basal conditions, NHE3 surface expression was 11.9 ± 2.3% and decreased by ∼39% (p < 0.05) to 7.3 ± 1.4% in cells treated with A23187 (supplemental Fig. S1, A and B; n = 4). Based on these data, NHERF3-mediated inhibition of NHE3 activity is associated with decreased surface expression, similar to results previously shown for NHERF2. Previous co-immunoprecipitation studies have demonstrated that the association of NHERF2 and NHE3 did not change under elevated calcium conditions (5). We thus hypothesized it was likely that calcium regulation of NHE3 activity involving NHERF2 and NHERF3 occurred through a similar mechanism. Therefore, the next study asked whether there were changes in NHERF3 association with NHE3 under elevated calcium conditions.
NHERF3 and NHE3 Interactions Are Dynamic and Decrease in Response to Elevated Calcium in PS120/E3V/NHERF3 Cells
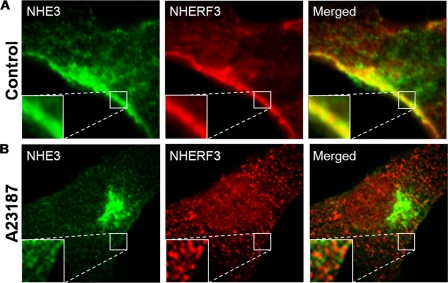
In PS120/E3V/NHERF3 cells, NHE3 and NHERF3 colocalize in distinct areas within the plasma membrane (Fig. 4A, as shown in the insets) and some intracellular vesicles under basal conditions. However, after elevation of [Ca2+]i using A23187, NHERF3 and NHE3 no longer colocalize as determined by immunofluorescence (Fig. 4B). NHE3 and NHERF3 dissociation is evident throughout the cell and at the plasma membrane (shown in the insets). Furthermore, the amount of NHE3 in the intracellular compartment is increased after elevation of [Ca2+]i (Fig. 4B), which is consistent with decreased surface expression of NHE3 (supplemental Fig. S1B).
FIGURE 4.
NHE3 and NHERF3 dissociate after elevation of intracellular calcium in PS120 cells. A, PS120/NHE3V/NHERF3 cells were seeded onto glass coverslips and grown to 70–80% confluence. Cells were serum starved for 4 h and treated with either vehicle or 4-bromo-A23187 (0.5 μm) for 15 min at 37 °C. Cells were then fixed with 3% paraformaldehyde, permeabilized, and stained with antibodies to NHE3 and NHERF3 and localization determined by confocal microscopy. In PS120/NHE3V/NHERF3 cells, NHE3 and NHERF3 colocalize in distinct areas within the plasma membrane as well as within intracellular vesicles. B, after elevation of intracellular calcium, NHE3 and NHERF3 no longer colocalize either at the plasma membrane or in intracellular compartments (×100 oil objective lens; 0.5-μm confocal sections). Insets magnify (×3 zoom) areas along the plasma membrane. Results were obtained from six to eight individual experiments.
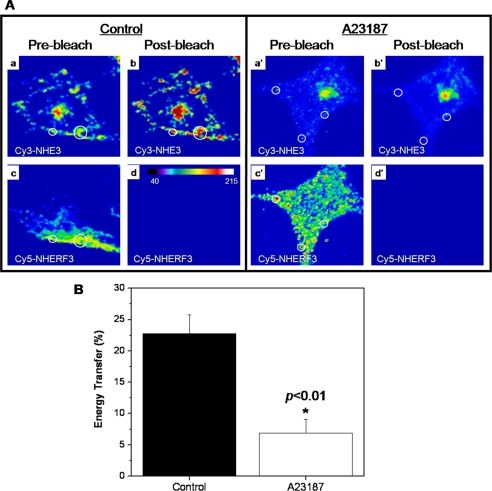
Cy3/Cy5 acceptor photobleaching FRET was used to confirm direct protein-protein interactions of NHE3 and NHERF3/PDKZ1 in PS120 cells under basal conditions and to further examine whether that interaction was dynamic with elevated [Ca2+]i. NHE3 (Cy3) and NHERF3/PDKZ1 (Cy5) directly bound to one another at the plasma membrane in PS120 cells under basal conditions (Fig. 5, A and B). Using confocal microscopy, initial images of Cy3-labeled NHE3 and Cy5-labeled NHERF3 were obtained by focusing on the plasma membrane localized at the edge of the cell (Fig. 5A). Cy3 (NHE3) fluorescence (Fig. 5A, b and b′) was measured once Cy5 fluorescence (Fig. 5A, d and d′) was no longer detectable after photobleaching. Quantification of fluorescence energy transfer was performed under basal and elevated [Ca2+]i examining NHE3 and NHERF3 localization at the plasma membrane by using 0.5-μm xy sections. Analysis of PS120 cells revealed that NHE3 and NHERF3 exhibit FRET under basal conditions (Fig. 5A). This was significantly reduced 15 min after elevating [Ca2+]i with A23187. Moreover, the total percent energy transfer between NHERF3 and NHE3 decreased in response to elevated [Ca2+]i (Fig. 5B). These data suggest that NHE3 and NHERF3 physically interact under basal [Ca2+]i conditions and there is decreased association after [Ca2+]i was elevated for 15 min.
FIGURE 5.
NHE3 and NHERF3 directly associate as determined by fluorescence resonance energy transfer and this association decreases in response to elevated calcium. PS120 cells were prepared as described above and NHE3 and NHERF3 protein expression detected using secondary antibodies conjugated to Cy3 (NHE3) or Cy5 (NHERF3). A, NHE3 and NHERF3 directly interact in PS120/NE3V/NHERF3 cells as determined by acceptor photobleaching FRET. Using confocal microscopy, initial fluorescent images of NHE3 and NHERF3 localization were captured before photobleaching (a, a′, c, and c′). Cy5-labeled NHERF3 signal was bleached with continuous laser excitation until Cy5 fluorescence was completely eliminated (d and d′). NHE3 Cy3 (donor) images (b and b′) were captured after Cy5 photobleaching and analyzed for changes in fluorescent intensity using the MetaMorph image analysis software. B, the results of the current study demonstrated that NHE3 and NHERF3 exhibit FRET under basal conditions and the total percent of energy transfer was significantly (p < 0.01) decreased in PS120/NHE3V/NHERF3 cells treated for 15 min with 4-bromo-A23187. 50–100 regions of interest were analyzed and data summarized in B. *, p in comparison to control.
NHERF3 and NHE3 Overlap in the Apical Membrane of Caco-2BBe Cells under Basal Conditions, and This Interaction Decreases in Response to Elevated Calcium
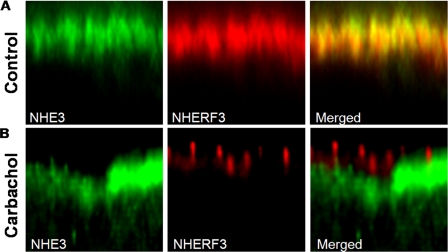
Overlap under basal conditions and dissociation of NHE3 from NHERF3 with elevated [Ca2+]i was also observed in the polarized intestinal Na+ absorptive epithelial cell line, Caco-2BBe. Serial confocal sections of polarized (filter grown) confluent Caco-2BBe monolayers infected with 3HA-NHE3 adenovirus demonstrated that NHE3 and NHERF3 overlap exclusively in the BB under basal conditions (Fig. 6A). However, after 10 μm carbachol treatment for 10 min, NHERF3 appeared to form clusters in the BB and no longer colocalized with NHE3 (Fig. 6B). NHE3 distribution appeared to be lower in the BB or in a subapical region in Caco-2BBe cells (Fig. 6B).
FIGURE 6.
NHERF3 and NHE3 colocalize in BB of Caco-2BBe cells under basal conditions and dissociate after 10 μm carbachol treatment. Caco-2BBe cells were grown on Anapore filters until they reached 12 days post-confluence. Cells were then infected with adenovirus containing the 3HA-tagged NHE3 construct. On day 14, cells were fixed with 3% paraformaldehyde, permeabilized, and stained with antibodies to NHE3 and NHERF3 and localization determined by confocal microscopy. A, in the BB of Caco-2BBe cells, NHE3 and NHERF3 colocalize under basal conditions. B, after treatment with carbachol (10 min), NHERF3 expression becomes clustered within the BB and no longer colocalizes with NHE3. NHE3 also appears to move to a subapical region after carbachol treatment. Similar results were obtained from three individual experiments.
NHE3 and NHERF3 Dissociate under Elevated Ca2+ Conditions in Vivo
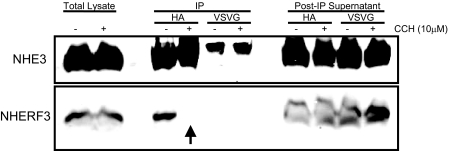
The decreased association of NHE3 and NHERF3 by immunofluorescence was also observed in co-immunoprecipitation studies (Fig. 7). To confirm that the in vivo interaction of NHE3 and NHERF3 was reduced in Caco-2BBe cells treated with carbachol, we immunoprecipitated NHE3 from Caco-2BBe cell lysates. Under basal conditions, NHE3 co-precipitated NHERF3 (Fig. 7). However, after carbachol treatment the association of NHE3 and NHERF3 could not be demonstrated. The results of these studies demonstrate that NHE3 and NHERF3 interact under basal conditions and this interaction is reduced after carbachol treatment (i.e. elevated [Ca2+]i conditions). To determine whether [Ca2+]i was sufficient to mediate this dissociation, we performed Far Western blot analysis of purified NHERF3 and NHE3 C-terminal fragments. In the presence or absence of free Ca2+, direct binding of NHERF3 to the F2 fragment of NHE3 was not altered (supplemental Fig. S2). This suggests that carbachol-initiated signaling effectors separate from elevated [Ca2+]i are required for the dissociation of NHERF3 and NHE3. Given that the dissociation of NHE3 and NHERF3 was associated with decreased NHE3 activity and reduced NHE3 surface expression, we hypothesized that the role of NHERF3 in intestinal epithelial cells was to retain NHE3 in the BB. To test this hypothesis, we knocked down NHERF3 in Caco-2BBe cells by shRNA and determined the effect on basal and calcium regulation of NHE3 activity.
FIGURE 7.
NHE3 and NHERF3 dissociate under elevated Ca2+ conditions in vivo. NHE3 was immunoprecipitated (IP) using an anti-HA antibody from Caco-2BBe cell lysates treated with either vehicle or 10 μm carbachol. NHE3 and NHERF3 expression were detected by Western blot, which determined that NHE3 and NHERF3 co-precipitated under basal conditions. However, this interaction was completely lost in Caco-2BBe cells treated with carbachol. Anti-VSV-G antibody was used as a negative control, showing nonspecific IgG bands of larger size than immunoprecipitated NHE3. Please note that after 10 min of carbachol treatment, NHE3 no longer co-precipitated NHERF3 (as demonstrated by arrow). Results were obtained from three individual experiments.
NHERF3 Contributes to Basal NHE3 Activity and Is Necessary for Calcium Inhibition of NHE3 Activity
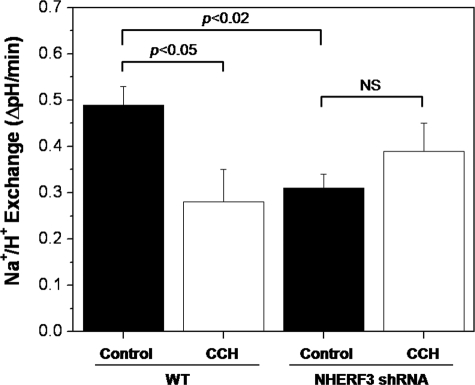
Using the pH-sensitive dye, BCECF, NHE3 activity was measured in Caco-2BBe cells. 10 μm carbachol treatment significantly reduced the rate of sodium-dependent pH recovery by more than 40% (Fig. 8). These data suggest that carbachol elevation of [Ca2+]i significantly decreased NHE3 activity and was associated with dissociation of NHE3 and NHERF3 as demonstrated by immunofluorescence (Fig. 6A) and co-immunoprecipitation studies (Fig. 7).
FIGURE 8.
NHERF3 anchors NHE3 to the BB of Caco-2BBe cells and regulates basal and calcium-mediated inhibition of NHE3 activity. Using BCECF to measure initial rates of sodium/hydrogen exchange, basal NHE3 activity was significantly reduced in Caco-2BBe cells with stably knocked down NHERF3 compared with empty vector control. In addition, carbachol-mediated inhibition of NHE3 activity was abolished in NHERF3 shRNA Caco-2BBe cells. Bar graph summarizes the mean ± S.E. of nine experiments. p value is in comparison with control (analysis of variance).
To further understand the role of NHERF3 in NHE3 regulation, a lentivirus-mediated delivery system was used to stably knock down NHERF3 in Caco-2BBe cells (Fig. 9, A and B). In Caco-2BBe cells infected with NHERF3 shRNA lentivirus, endogenous NHERF3 protein levels were reduced by >50% (Fig. 9B). Furthermore, this reduction was maintained in culture media containing puromycin for at least five passages (data not shown). NHERF3 shRNA reduced basal NHE3 activity by ∼50% and abolished carbachol-mediated inhibition of NHE3 activity (Fig. 8; n = 9).
FIGURE 9.
NHERF3 shRNA knockdown in Caco-2BBe cells. A, Western blot analysis of total cell lysates prepared from Caco-2BBe cells infected with lentivirus empty vector control or NHERF3 shRNA. Glyceraldehyde-3-phosphate dehydrogeanse (GAPDH) was used as internal control. B, NHERF3 protein expression was significantly reduced in Caco-2BBe cells stably knocked down for NHERF3 in complete media containing puromycin as selection marker. Bar graph summarizes the mean ± S.E. of four experiments. p value is in comparison with control (paired t test).
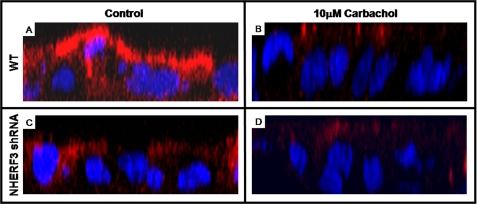
Using stably knocked down NHERF3 shRNA Caco-2BBe cells, we tested whether decreased basal NHE3 activity was due to decreased BB expression of NHE3. Confocal microscopy demonstrated that BB NHE3 expression was significantly decreased in Caco-2BBe cells in which NHERF3 was knocked down compared with empty vector controls (Fig. 10, A versus C). In addition, NHE3 expression was decreased after carbachol exposure in control Caco-2BBe cells (Fig. 10, A versus B), which correlated with the changes in NHE3 transport activity (Fig. 8). In Caco-2BBe cells infected with NHERF3 shRNA, carbachol treatment (Fig. 10D) did not seem to alter NHE3 expression at the BB when compared with empty vector Caco-2BBe cells treated with carbachol (Fig. 10B). The results of these experiments suggest that NHERF3 regulates basal NHE3 activity by anchoring NHE3 to the BB of intestinal epithelial cells and dissociates from NHE3 containing complexes to possibly allow NHE3 internalization.
FIGURE 10.
BB NHE3 expression was decreased in NHERF3 knockdown Caco-2BBe cells. Caco-2BBe cells grown on filters were infected with 3HA-NHE3 adenovirus, fixed, and stained for NHE3 using anti-HA primary antibody. Confocal microscopy of serial sections (0.5 μm) of Caco-2BBe cells infected with empty vector control (A and B) or NHERF3 shRNA (C and D) stained with NHE3 and Hoescht 33342 (nuclear counterstain). Z-series images were reconstructed using MetaMorph Image Analysis software. BB NHE3 expression was significantly reduced in NHERF3 shRNA Caco-2BBe cells (C) as well as in cells (empty vector and NHERF3 shRNA) treated with 10 μm carbachol for 10 min (B and D). Results were obtained from three individual experiments.
DISCUSSION
Acute regulation by ligands and second messengers (i.e. cAMP, cGMP, and [Ca2+]i) of intestinal neutral NaCl absorption, which includes NHE3, is known to depend on the NHERF family of multi-PDZ domain containing proteins (1, 2). The involvement of the NHERF family has added another level of complexity to the understanding of the post-prandial changes in Na+ absorption and the abnormalities of this process that occur in diarrheal diseases.
Several approaches have been taken to understand the contribution of individual NHERF proteins to NHE3 regulation. Cell lines lacking endogenous NHERF family members have been transfected with individual NHERF proteins and knock-out mouse models of individual NHERFs have been created. Studies performed in NHERF1 KO mice not only confirmed previous cell line studies suggesting a role for NHERF1 in cAMP-mediated inhibition of NHE3 activity, but also demonstrated that cAMP regulation via NHERF1 was tissue dependent (i.e. NHERF1 was necessary for cAMP inhibition of NHE3 activity in renal proximal tubule but not in distal ileum) (10, 39). Moreover, studies of the NHERF3 KO mouse demonstrated that whereas these mice appeared healthy, intestinal NHE3 activity was significantly reduced and these mice failed to respond to both cAMP and elevated [Ca2+]i with NHE3 inhibition (21, 22). Although these studies strongly suggest a role for NHERF proteins in NHE3 regulation, the cellular and molecular mechanisms responsible for NHERF-dependent regulation remain unclear. In addition, studies of NHERF1 (6, 40) and NHERF2 KO (40) mice have shown changes in expression of other NHERF protein family members, making it challenging to decide which NHERF protein accounts for changes in function. Intestinal studies of NHERF3 KO mice have not yet addressed whether changes in NHE3 regulation could also be due to compensatory changes of the other NHERF proteins (21, 22).
The current study began a reductionist evaluation of the contribution of NHERF3 to NHE3 regulation by examining a fibroblast cell model (PS120), which lacks most members of the NHERF family (except for a small amount of NHERF1) as well as a Na+ absorptive intestinal cell line (Caco-2BBe), which endogenously expresses all four NHERF family members. These models allowed us to examine the role of NHERF3 alone (in PS120 cells) in NHE3 regulation and when it was the only NHERF protein knocked down (in Caco-2BBe cells). This Caco-2BBe NHERF3 shRNA model had the additional advantage that NHE3 regulation was not affected by other NHERF-containing, non-epithelial cells that affect NHE3 function when studying intact mouse models. Our current study demonstrated that NHERF3 reconstituted Ca2+ inhibition of NHE3 in both the fibroblast and Caco-2BBe cell models, by what appears to be a similar process. The mechanism appears to involve NHERF3 anchoring NHE3 to the plasma membrane under basal conditions and release of NHE3 anchoring with elevated [Ca2+]i. Our study confirmed a previous observation that NHERF3 directly binds NHE3 (20). However, our results indicated that this interaction involved an internal NHE3 binding site, which was unlike a previous report that indicated that the binding of NHERF3 to NHE3 involved the C-terminal three amino acids of NHE3 (only a C-terminal fusion protein was used in these studies, precluding the ability to identify an internal binding site) (20). The involvement of an internal NHE3 binding site for NHERF3 has been recently suggested by Gisler et al. (42) using a modified split-ubiquitin membrane yeast two-hybrid system, which demonstrated that the same internal NHE3 binding domain also interacts with NHERF1 and NHERF4, although the role for binding was not defined further.
In this study, we demonstrate that the NHE3-NHERF3 interaction is dynamic, decreasing with elevated [Ca2+]i. This NHE3-NHERF3 dissociation was associated with decreased NHE3 membrane expression and activity. To further determine whether NHERF3 played an anchoring role in NHE3 expression on the plasma membrane, shRNA was used to knockdown NHERF3 in Caco-2BBe cells. These studies revealed that apical NHE3 expression was significantly reduced in the absence of NHERF3. We interpret these results to indicate that NHERF3 anchors NHE3 to the plasma membrane under basal conditions. Interrupting the NHE3-NHERF3 interaction either by knocking down NHERF3 or by elevated Ca2+ leads to increased internalization of NHE3, presumably by ongoing endocytosis in the first case and via stimulated endocytosis of NHE3 shown to occur with elevated Ca2+ in the second case (5). This interpretation is supported by mutation studies of the CFTR C-terminal PDZ binding motif. Deletion of this motif did not alter apical sorting of CFTR but rather decreased the membrane half-life of CFTR suggesting that the CFTR-PDZ motif served as a membrane retention signal (43). Whether the internal binding site of NHE3 for NHERF3 is also a membrane retention signal, requires further study.
The results of the current study along with our preliminary evidence that NHERF2 and NHE3 associate dynamically (44) represent the first evidence that binding between NHE3 and members of the NHERF family occurs dynamically. However, the Ca2+-dependent association between NHE3 and NHERF3 is not the first time NHERF proteins have been shown to bind dynamically to their substrates. Dynamic association of PDZ proteins during signal transduction has been previously demonstrated for NaPiIIa, CFTR, β2-adrenergic receptor, and the platelet-derived growth factor receptor (45–47). For example, following parathyroid hormone receptor activation, NaPiIIa dissociates from NHERF1 (47). In the case of CFTR, PKC activation causes CFTR to be released from the PDZ2 domain of NHERF1 thereby removing one of the two CFTR/NHERF1 binding sites (47). The mechanisms demonstrated to be responsible for dynamic NHERF/substrate interactions include, presence of substrate, substrate phosphorylation, NHERF phosphorylation, and/or NHERF dimerization (6, 47–49). In addition, multiple other aspects of signaling (e.g. endocrine) seem to influence the relationship of NHERF proteins with their substrates. Phosphorylation of NHERF3 at Ser509 by PKA and glucagon was associated with increased expression of the scavenger receptor class B type I and PDZK1, which resulted in decreased plasma high density lipoprotein levels (50). Although the details responsible for NHERF3-dependent regulation of NHE3 activity remain to be determined, the results of the current study appear to represent another example of the highly regulated association of the NHERFs with their ligands.
Previous studies examining acute NHE3 regulation by cAMP, cGMP, and elevated [Ca2+]i identified a role for members of the NHERF family in forming NHE3-containing, multiprotein complexes (2, 51). Of these identified regulatory processes, the most complicated appears to be calcium inhibition. Studies of murine KO models, although still in progress, have demonstrated that NHERF3 is necessary for basal as well as calcium and cAMP inhibition of intestinal neutral NaCl absorption and NHE3 inhibition (best illustrated in the colon) and that NHERF2 is necessary for basal activity and calcium inhibition of NHE3 activity in the ileum (21, 22, 41). Supporting these findings, NHERF2 and, as shown here, NHERF3 can reconstitute calcium inhibition in fibroblasts that lack all endogenous NHERFs other than a small amount of NHERF1 expression. That both NHERF2 and NHERF3 are involved in NHE3 inhibition with elevated Ca2+ indicates the complexity of this regulatory process. In a parallel study, we generated stable NHERF2 knockdown in Caco-2BBe cells to determine whether NHERF2, which is also present in the BB of Caco-2BBe cells (2), and contributes to basal and/or Ca2+ regulation of NHE3 activity (data not shown and in preparation for submission elsewhere). In contrast to NHERF3 KD, basal NHE3 activity was not altered in the NHERF2 KD Caco-2BBe cells. However, NHERF2 KD prevented carbachol-mediated inhibition of NHE3 activity. This result supports previous studies demonstrating that Ca2+ inhibition of NHE3 activity requires NHERF2 (5). Although both NHERF2 and NHERF3 are required for Ca2+ inhibition, only NHERF3 contributes to basal NHE3 activity suggesting the NHERF2- and NHERF3-regulated NHE3 activity by different mechanisms that may involve their differential subcellular localization. Although mechanistic studies have suggested the role of NHERF2 in Ca2+ regulation of NHE3 involves formation of NHE3 containing complexes that include α-actinin-4 and PKCα (5) and dynamic fixation of NHE3 to the cytoskeleton (44), the role of NHERF3 was not understood until the current studies. We reasoned that to understand how a complex regulatory process functions, studies of simple cells as well as studies in epithelial cell culture models would be useful.
In the current studies, it was shown that elevation of [Ca2+]i inhibits NHE3 in cells containing NHERF3 by decreasing the amount of NHE3 on the plasma membrane of fibroblasts and the BB of Caco-2BBe cells. A similar decrease of BB NHE3 was demonstrated in the BB of the rabbit ileum treated with carbachol, supporting that these cell culture models might provide insights in the mechanism of calcium inhibition of NHE3 (52). The mechanistic insights provided from both cell models was that although NHE3 associates with NHERF3 under basal conditions, acute elevation of intracellular [Ca2+]i dissociates NHE3 from NHERF3. Moreover, confocal microscopy and acceptor photobleaching FRET showed that NHERF3 does not traffic with NHE3 from the plasma membrane to the recycling compartment of fibroblasts. Also, in Caco-2BBe cells, NHERF3 remained in the BB, whereas NHE3 moved to the lower part of the BB, compatible with movement to the intervillus clefts, from which increased endocytosis occurs. Because NHE3 association with NHERF proteins limits NHE3 mobility in the BB, as assessed by FRAP (44), we suggest that the dissociation from NHERF3 is a necessary part of the mechanism that frees NHE3 to move from its microvillus or plasma membrane location where it associates with the cytoskeleton to the microvillus clefts from which it can be endocytosed. The similarity in the apparent role of NHERF3 to fix NHE3 in the membrane under basal conditions and to release NHE3 with elevated Ca2+ further indicates that some NHE3 regulatory steps are similar in polarized epithelial cells and fibroblasts despite the great differences in structure of the membrane domains involved.
In conclusion, an important implication from the current studies is that multiple NHERF family proteins appear to be involved in a single NHE3 regulatory cascade at the level of interactions with NHE3 itself. The results of the current study demonstrate that NHERF3 directly binds NHE3 and anchors NHE3 to the BB of intestinal epithelial cells. Moreover, this interaction occurs under basal conditions but is dynamic in response to elevated calcium. Although NHE3 and NHERF3 dissociate after elevated [Ca2+]i, the coordinated role of other members of the NHERF family in previous or subsequent steps requires future study. Moreover, the effects of NHERF3 knockdown on NHE3 activity in our studies were similar to effects on NHE3 activity of knocking out NHERF3 in mouse colon, both lowering basal and preventing Ca2+ dependent inhibition of NHE3 activity. However, the current studies provide some potential mechanistic understanding of the phenomenon described in the intact intestine. Our results suggest that the role of NHERF3 is to anchor NHE3 to BB under basal conditions and to release NHE3 to allow subsequent endocytosis with elevated Ca2+, which is a mechanism different from NHE3 complex formation previously described as the mechanism by which NHERF2 contributes to Ca2+ inhibition of NHE3 (5, 24).
Supplementary Material
Acknowledgment
We acknowledge the expert editorial assistance of H. McCann.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK26523, R01-DK61765, PO1-DK72084, K01-DK080930, and R24-DK64388 (The Hopkins Basic Research Digestive Diseases Development Core Center) from the NIDDK. This work was also supported by Grant T32-DK07632 from The Hopkins Center for Epithelial Disorders.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- BB
- brush border
- CFTR
- cystic fibrosis transmembrane regulator
- FRET
- fluorescence resonance energy transfer
- PKC
- protein kinase C
- VSV
- vesicular stomatitis virus
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- aa
- amino acid
- shRNA
- short hairpin RNA
- KO
- knockout
- BCECF
- 2′,7′-bis(carboxyethyl)5–6-carboxyfluorescein.
REFERENCES
- 1.Zachos N. C., Tse M., Donowitz M. (2005) Annu. Rev. Physiol. 67,411–443 [DOI] [PubMed] [Google Scholar]
- 2.Donowitz M., Cha B., Zachos N. C., Brett C. L., Sharma A., Tse C. M., Li X. (2005) J. Physiol. 567,3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha B., Kim J. H., Hut H., Hogema B. M., Nadarja J., Zizak M., Cavet M., Lee-Kwon W., Lohmann S. M., Smolenski A., Tse C. M., Yun C., de Jonge H. R., Donowitz M. (2005) J. Biol. Chem. 280,16642–16650 [DOI] [PubMed] [Google Scholar]
- 4.Cunningham R., Steplock D., Wang F., Huang H., Xiaofei E., Shenolikar S., Weinman E. J. (2004) J. Biol. Chem. 279,37815–37821 [DOI] [PubMed] [Google Scholar]
- 5.Kim J. H., Lee-Kwon W., Park J. B., Ryu S. H., Yun C. H., Donowitz M. (2002) J. Biol. Chem. 277,23714–23724 [DOI] [PubMed] [Google Scholar]
- 6.Morales F. C., Takahashi Y., Kreimann E. L., Georgescu M. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101,17705–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shenolikar S., Voltz J. W., Minkoff C. M., Wade J. B., Weinman E. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,11470–11475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinman E. J., Cunningham R., Wade J. B., Shenolikar S. (2005) J. Physiol. 567,27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinman E. J., Steplock D., Shenolikar S. (2003) FEBS Lett. 536,141–144 [DOI] [PubMed] [Google Scholar]
- 10.Yun C. H., Oh S., Zizak M., Steplock D., Tsao S., Tse C. M., Weinman E. J., Donowitz M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94,3010–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernando N., Gisler S. M., Pribanic S., Déliot N., Capuano P., Wagner C. A., Moe O. W., Biber J., Murer H. (2005) J. Physiol. 567,21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kocher O., Comella N., Tognazzi K., Brown L. F. (1998) Lab. Invest. 78,117–125 [PubMed] [Google Scholar]
- 13.Scott R. O., Thelin W. R., Milgram S. L. (2002) J. Biol. Chem. 277,22934–22941 [DOI] [PubMed] [Google Scholar]
- 14.Thelin W. R., Hodson C. A., Milgram S. L. (2005) J. Physiol. 567,13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anzai N., Miyazaki H., Noshiro R., Khamdang S., Chairoungdua A., Shin H. J., Enomoto A., Sakamoto S., Hirata T., Tomita K., Kanai Y., Endou H. (2004) J. Biol. Chem. 279,45942–45950 [DOI] [PubMed] [Google Scholar]
- 16.Capuano P., Bacic D., Stange G., Hernando N., Kaissling B., Pal R., Kocher O., Biber J., Wagner C. A., Murer H. (2005) Pflugers Arch. 449,392–402 [DOI] [PubMed] [Google Scholar]
- 17.Kato Y., Sugiura M., Sugiura T., Wakayama T., Kubo Y., Kobayashi D., Sai Y., Tamai I., Iseki S., Tsuji A. (2006) Mol. Pharmacol. 70,829–837 [DOI] [PubMed] [Google Scholar]
- 18.Noshiro R., Anzai N., Sakata T., Miyazaki H., Terada T., Shin H. J., He X., Miura D., Inui K., Kanai Y., Endou H. (2006) Kidney Int. 70,275–282 [DOI] [PubMed] [Google Scholar]
- 19.Wang S., Yue H., Derin R. B., Guggino W. B., Li M. (2000) Cell 103,169–179 [DOI] [PubMed] [Google Scholar]
- 20.Thomson R. B., Wang T., Thomson B. R., Tarrats L., Girardi A., Mentone S., Soleimani M., Kocher O., Aronson P. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102,13331–13336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillesheim J., Riederer B., Tuo B., Chen M., Manns M., Biber J., Yun C., Kocher O., Seidler U. (2007) Pflugers Arch. 454,575–586 [DOI] [PubMed] [Google Scholar]
- 22.Cinar A., Chen M., Riederer B., Bachmann O., Wiemann M., Manns M., Kocher O., Seidler U. (2007) J. Physiol. 581,1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen M. E., Wesolek J., McCullen J., Rys-Sikora K., Pandol S., Rood R. P., Sharp G. W., Donowitz M. (1991) J. Clin. Invest. 88,855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee-Kwon W., Kawano K., Choi J. W., Kim J. H., Donowitz M. (2003) J. Biol. Chem. 278,16494–16501 [DOI] [PubMed] [Google Scholar]
- 25.Deber C. M., Hsu L. C. (1986) Biochem. Biophys. Res. Commun. 134,731–735 [DOI] [PubMed] [Google Scholar]
- 26.Malmberg E. K., Pelaseyed T., Petersson A. C., Seidler U. E., De Jonge H., Riordan J. R., Hansson G. C. (2008) Biochem. J. 410,283–289 [DOI] [PubMed] [Google Scholar]
- 27.Ahn W., Kim K. H., Lee J. A., Kim J. Y., Choi J. Y., Moe O. W., Milgram S. L., Muallem S., Lee M. G. (2001) J. Biol. Chem. 276,17236–17243 [DOI] [PubMed] [Google Scholar]
- 28.Yun C. H., Lamprecht G., Forster D. V., Sidor A. (1998) J. Biol. Chem. 273,25856–25863 [DOI] [PubMed] [Google Scholar]
- 29.Zachos N. C., Hodson C., Kovbasnjuk O., Li X., Thelin W. R., Cha B., Milgram S., Donowitz M. (2008) Cell Physiol. Biochem. 22,693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasmi M., Glynn J., Jin M. J., Jolly D. J., Yee J. K., Chen S. T. (1999) J. Virol. 73,1828–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakoda T., Kasahara N., Hamamori Y., Kedes L. (1999) J. Mol. Cell. Cardiol. 31,2037–2047 [DOI] [PubMed] [Google Scholar]
- 32.Levine S. A., Nath S. K., Yun C. H., Yip J. W., Montrose M., Donowitz M., Tse C. M. (1995) J. Biol. Chem. 270,13716–13725 [DOI] [PubMed] [Google Scholar]
- 33.Levine S. A., Montrose M. H., Tse C. M., Donowitz M. (1993) J. Biol. Chem. 268,25527–25535 [PubMed] [Google Scholar]
- 34.Janecki A. J., Montrose M. H., Zimniak P., Zweibaum A., Tse C. M., Khurana S., Donowitz M. (1998) J. Biol. Chem. 273,8790–8798 [DOI] [PubMed] [Google Scholar]
- 35.Watson A. J., Levine S., Donowitz M., Montrose M. H. (1991) Am. J. Physiol. 261,G229–G238 [DOI] [PubMed] [Google Scholar]
- 36.Cavet M. E., Akhter S., de Medina F. S., Donowitz M., Tse C. M. (1999) Am. J. Physiol. 277,C1111–C1121 [DOI] [PubMed] [Google Scholar]
- 37.Kenworthy A. K., Edidin M. (1998) J. Cell Biol. 142,69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovbasnjuk O., Edidin M., Donowitz M. (2001) J. Cell Sci. 114,4025–4031 [DOI] [PubMed] [Google Scholar]
- 39.Murtazina R., Kovbasnjuk O., Zachos N. C., Li X., Chen Y., Hubbard A., Hogema B. M., Steplock D., Seidler U., Hoque K. M., Tse C. M., De Jonge H. R., Weinman E. J., Donowitz M. (2007) J. Biol. Chem. 282,25141–25151 [DOI] [PubMed] [Google Scholar]
- 40.Donowitz M., Singh S., Salahuddin F. F., Hogema B. M., Chen Y., Gucek M., Cole R. N., Ham A., Zachos N. C., Kovbasnjuk O., Lapierre L. A., Broere N., Goldenring J., deJonge H., Li X. (2007) J. Proteome Res. 6,4068–4079 [DOI] [PubMed] [Google Scholar]
- 41.Murtazina R., Kovbasnjuk O., Chen Y., Hogema B. M., Seidler U., Weinman E. J., De Jonge H., Donowitz M. (2008) Gastroenterology 134,A749 [Google Scholar]
- 42.Gisler S. M., Kittanakom S., Fuster D., Wong V., Bertic M., Radanovic T., Hall R. A., Murer H., Biber J., Markovich D., Moe O. W., Stagljar I. (2008) Mol. Cell. Proteomics 7,1362–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swiatecka-Urban A., Duhaime M., Coutermarsh B., Karlson K. H., Collawn J., Milewski M., Cutting G. R., Guggino W. B., Langford G., Stanton B. A. (2002) J. Biol. Chem. 277,40099–40105 [DOI] [PubMed] [Google Scholar]
- 44.Cha B., Kenworthy A., Murtazina R., Donowitz M. (2004) J. Cell Sci. 117,3353–3365 [DOI] [PubMed] [Google Scholar]
- 45.Déliot N., Hernando N., Horst-Liu Z., Gisler S. M., Capuano P., Wagner C. A., Bacic D., O'Brien S., Biber J., Murer H. (2005) Am. J. Physiol. Cell Physiol. 289,C159–C167 [DOI] [PubMed] [Google Scholar]
- 46.Raghuram V., Hormuth H., Foskett J. K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,9620–9625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voltz J. W., Brush M., Sikes S., Steplock D., Weinman E. J., Shenolikar S. (2007) J. Biol. Chem. 282,33879–33887 [DOI] [PubMed] [Google Scholar]
- 48.Hegedüs T., Sessler T., Scott R., Thelin W., Bakos E., Váradi A., Szabó K., Homolya L., Milgram S. L., Sarkadi B. (2003) Biochem. Biophys. Res. Commun. 302,454–461 [DOI] [PubMed] [Google Scholar]
- 49.Shenolikar S., Minkoff C. M., Steplock D. A., Evangelista C., Liu M., Weinman E. J. (2001) FEBS Lett. 489,233–236 [DOI] [PubMed] [Google Scholar]
- 50.Nakamura T., Shibata N., Nishimoto-Shibata T., Feng D., Ikemoto M., Motojima K., Iso-O N., Tsukamoto K., Tsujimoto M., Arai H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102,13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donowitz M., Li X. (2007) Physiol. Rev. 87,825–872 [DOI] [PubMed] [Google Scholar]
- 52.Li X., Zhang H., Cheong A., Leu S., Chen Y., Elowsky C. G., Donowitz M. (2004) J. Physiol. 556,791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.