Abstract
BdbD is a thiol:disulfide oxidoreductase (TDOR) from Bacillus subtilis that functions to introduce disulfide bonds in substrate proteins/peptides on the outside of the cytoplasmic membrane and, as such, plays a key role in disulfide bond management. Here we demonstrate that the protein is membrane-associated in B. subtilis and present the crystal structure of the soluble part of the protein lacking its membrane anchor. This reveals that BdbD is similar in structure to Escherichia coli DsbA, with a thioredoxin-like domain with an inserted helical domain. A major difference, however, is the presence in BdbD of a metal site, fully occupied by Ca2+, at an inter-domain position some 14 Å away from the CXXC active site. The midpoint reduction potential of soluble BdbD was determined as −75 mV versus normal hydrogen electrode, and the active site N-terminal cysteine thiol was shown to have a low pKa, consistent with BdbD being an oxidizing TDOR. Equilibrium unfolding studies revealed that the oxidizing power of the protein is based on the instability introduced by the disulfide bond in the oxidized form. The crystal structure of Ca2+-depleted BdbD showed that the protein remained folded, with only minor conformational changes. However, the reduced form of Ca2+-depleted BdbD was significantly less stable than reduced Ca2+-containing protein, and the midpoint reduction potential was shifted by approximately −20 mV, suggesting that Ca2+ functions to boost the oxidizing power of the protein. Finally, we demonstrate that electron exchange does not occur between BdbD and B. subtilis ResA, a low potential extra-cytoplasmic TDOR.
Disulfide bonds, formed upon oxidation of two cysteine residue side chain thiols, are key for the stability and/or function of many secreted and membrane-bound peptides and proteins in bacteria, and the failure to insert these correctly has wide ranging effects (1–4). To regulate the redox state of cysteine residues on the outside of the cytoplasmic membrane, intricate disulfide bond regulatory systems have evolved. These involve enzymes of the thiol:disulfide oxidoreductase (TDOR)3 family, which contain cysteine residues often arranged in a Cys-Xaa-Xaa-Cys thioredoxin motif (5, 6). These enzymes function in pathways that lead to the formation of disulfide bonds, rearrangement of incorrectly positioned disulfide bonds, or the removal of unwanted disulfide bonds, and the redox properties of the enzymes appear to correlate closely with function.
The paradigm system for disulfide bond formation is the DsbA-DsbB system of Escherichia coli, which has been characterized in great detail (7–11). DsbA is a soluble periplasmic TDOR, which has a thioredoxin-like fold with an additional helical domain (8). The protein oxidizes the di-thiol motifs of a range of substrates, generating in each a disulfide bond. Reduced DsbA is rapidly re-oxidized by DsbB, a membrane-bound TDOR (12) that channels the resulting electrons into the membrane quinol pool (13, 14). Dsb-like homologues appear to be extremely widespread in Gram-negative bacteria.
Gram-positive bacteria, unlike Gram-negatives, do not have an outer membrane and so have no spatially defined periplasmic compartment. This places different functional and structural demands on extra-cytoplasmic proteins, and one consequence of this is that extra-cytoplasmic TDORs are generally membrane-anchored in Gram-positive bacteria. Systems for the introduction of disulfide bonds appear to be variable in these organisms (15, 16). As an example, Mycobacterium tuberculosis does not contain close homologues of DsbA/B but contains another thioredoxin-like TDOR, DsbE, which has been shown to have redox properties similar to those of DsbA (17). Some Gram-positive bacteria, including Staphylococcus aureus, contain a DsbA homologue but no homologue of DsbB (18). The recent structural and biochemical characterization of S. aureus DsbA revealed major similarities with the E. coli protein but was also consistent with a distinct mechanism of re-oxidation (16).
Some Gram-positive bacteria, however, contain clear homologues of both DsbA and DsbB. In the model organism Bacillus subtilis, BdbD and BdbC/BdbB have been identified as homologues of DsbA and DsbB, respectively, and demonstrated to be involved in processes such as natural competence development, which requires the insertion of disulfide bonds (19). BdbD and BdbC are also involved in a number of other pathways that do not require the insertion of a disulfide bond; B. subtilis contains several extra-cytoplasmic TDORs, for example ResA and StoA (required for cytochrome c maturation and endospore biogenesis, respectively), which function to specifically reduce disulfide bonds introduced by BdbD (20, 21). To understand disulfide bond management systems in Gram-positive bacteria, detailed information on each of the different systems found is required.
Here we present the crystal structure of the catalytic domain of B. subtilis BdbD in both reduced and oxidized states. The structure is broadly similar to that of E. coli DsbA, but it also reveals the presence of a novel Ca2+-binding site remote from the CXXC active site. Using two-dimensional NMR methods and fluorescence kinetic studies of thiolate alkylation, we report the reduction potential and pKa properties of the soluble protein, which are entirely consistent with an oxidizing function for the protein in vivo. Conformational stability studies and NMR studies showed that the occupancy of the metal site by Ca2+ ion is not required for folding/stability but leads to a significant increase of the midpoint reduction potential. The possibility that the principal function of the metal site is to boost the oxidizing power of the protein is discussed.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
B. subtilis strains 1A1 (trpC2) and LUL10 (trpC2 bdbDΩTn10) (20) were used for preparation of extracts for immunoblot analysis. E. coli TOP10 (F− mcrA Δ[mrr-hsdRMS-mcrBC] F80lacZΔM15 ΔlacX74 recA1 araD139 Δ[ara-leu]7697 galU galK rpsL endA1 nupG) (Invitrogen) and B834(DE3) (F− ompT hsdSB(rB− mB−) gal dcm met (DE3)) (Novagen) containing pLLE34 (22) or pACN2 was used for production of sBdbD. pACN2 was constructed by PCR amplifying part of the bdbD gene using B. subtilis 1A1 genomic DNA as template and two primers (forward, 5′-A CAT ATG AAT AAC AAA ACG GAA CAA GGC 3′; reverse, 5′ CTC GAG CTT CCC TTT CAG CTC TTT TTC 3′; engineered NdeI and XhoI restriction sites are underlined). The resulting PCR product was blunt-end ligated into pUC18, and the NdeI and XhoI fragment was subsequently ligated into pET21a (Novagen) digested with the same enzymes and treated with shrimp alkaline phosphatase, and the construct was verified by DNA sequencing (MWG Biotec). Growth of B. subtilis strains and preparation of cell extracts for immunoblot analysis were done as recently described for StoA (23).
BdbD Antiserum and Immunoblot
sBdbD was produced in E. coli TOP10/pLLE34 and purified as described before (22). BdbD antiserum was obtained by immunizing New Zealand White rabbits with purified protein. For immunoblot, proteins were fractionated by SDS-PAGE (24) and transferred from the gel to a polyvinylidene fluoride blotting membrane (Immobilon P, Millipore) using a wet blot and 20 mm Tris, 150 mm glycine buffer containing 20% (v/v) methanol. Immunodetection was carried out by chemiluminescence using horseradish peroxidase-labeled secondary antibodies against rabbit (GE Healthcare) and Super Signal® West Pico chemiluminescent substrate (Pierce) and a Kodak image station 440 CF for signal recording.
Production and Purification of sBdbD for Crystallization and NMR
A 23-kDa truncated form of the BdbD protein (sBdbD) consisting of residues 30–222 of the full-length BdbD protein (lacking the N-terminal transmembrane helix) augmented by an N-terminal methionine residue and a C-terminal His tag with the sequence LEHHHHHH was expressed from pACN2 in E. coli B834 (DE3). Cultures were grown at 37 °C on minimal glucose medium supplemented with either 60 μg liter−1 methionine or 60 μg liter−1 selenomethionine to an A600 of 0.6 and centrifuged at 6,000 × g for 10 min at 4 °C. Cells were resuspended in fresh minimal glucose medium, and expression of the bdbD gene was induced by the addition of 0.5 mm isopropyl β-d-thiogalactoside. Cells were grown for a further 12 h at 37 °C before harvesting at 6,000 × g for 10 min at 4 °C. Cells were broken by sonication, and the cell lysate was centrifuged to remove unbroken cells and debris. The supernatant was applied to a nickel affinity column equilibrated in 100 mm Tris, 500 mm NaCl, pH 8.0, and eluted with 100 mm Tris, pH 8.0, 500 mm NaCl, 250 mm imidazole, pH 8.0. Fractions containing sBdbD were then applied to a 5-ml anion exchange column (HiTrap Q, GE Healthcare) equilibrated in 100 mm Tris, 0.5 mm DTT, pH 8.2, and subsequently eluted with a 0–1 m gradient of NaCl in the same buffer. DTT was subsequently removed using a Sephadex G-25 column (PD10, GE Healthcare). MALDI-TOF mass spectrometry was used to confirm that the purified protein matched the predicted mass expected from the sequence. 15N-Labeled sBdbD was produced using the same method, but with 15NH4Cl (1 g/liter) as the sole nitrogen source.
The UV-visible absorption spectrum of sBdbD contained an intense band at 280 nm with an unusually pronounced peak at 290 nm, presumably due to tryptophan. ϵ280 nm was determined using the method of Pace et al. (25) as 21,033 m−1 cm−1, and this was used subsequently to determine the protein concentration. Metal ion content of the protein was determined by ICP-AE spectroscopy (Varian Vista); Ca2+ was removed from sBdbD by incubating the protein with 10 mm EDTA for 1 h at 25 °C. EDTA was subsequently removed using a PD10 desalting column. EDTA-treated samples were analyzed by ICP-AE and found to contain <0.1 Ca2+ per protein. Oxidized and reduced sBdbD samples were prepared by incubation in the dark for 2 h in the presence of either 1 mm diamide or 1 mm DTT before removing redox buffers using a PD10 desalting column.
Crystallization and Structure Determinations
Purified sBdbD (native or selenomethionine-labeled) was concentrated to 12 mg/ml in 10 mm Mops, pH 7.0, and crystallized using the sitting drop vapor diffusion method. Equal volumes (1 μl) of protein and crystallization reagent (25–32.5% PEG 2000 (w/v), 0.1 m ammonium acetate, 0.1 m Mes, pH 6.5) were mixed and equilibrated over an 800-μl reservoir of the crystallization reagent alone. Low quality crystals grown at high concentrations of PEG (e.g. 30–32.5% (w/v)) were broken using a pipette tip and used for streak-seeding of drops containing lower concentrations of PEG (25–27% (w/v)). Crystals deemed suitable for diffraction were then cryo-protected in a solution composed of 30% (w/v) PEG 2000, 20% (v/v) ethylene glycol, 0.1 m ammonium acetate, 0.1 m Mops, pH 7.0 (with or without 1 mm DTT as required), before being flash-frozen in preparation for data collection.
X-ray data sets for both native and selenomethionine-labeled protein crystals were collected on beam line ID23-1 of the European synchrotron radiation facility. Diffraction data were indexed and integrated with MOSFLM (26) and then scaled with SCALA (27). Phases were determined using BP3 of the CCP4 suite (28), having already identified the selenium sites by manual inspection of Patterson maps. Further phase improvement was accomplished by solvent flattening and histogram matching with DM, and the vast majority of the structure was built automatically using ARP/WARP (29). Further model building and refinement (including refinement of individual anisotropic b-factors) utilized COOT (30) and REFMAC (31) as part of the CCP4 suite (28). Structure validation was aided by PROCHECK (32), and the final models were submitted to the Protein Data Bank with accession codes 3EU3 (reduced), 3EU4 (oxidized), 3GH9 (EDTA-treated/oxidized), and 3GHA (EDTA-treated/reduced).
Analysis of the metal site was conducted by systematically refining different metals and examining electron density difference maps (i.e. |Fo| − |Fc| maps) for indications of the quality of fit. To quantify the degree of difference density associated with each candidate metal, MAPMAN (33, 34) from the RAVE suite was used to integrate the difference density within a 3.5-Å radius of the metal center. The anomalous scattering characteristics of the metal site were also compared with those of the selenium atoms in our selenomethionine structure. FFT (CCP4 suite) was used to generate an anomalous difference map using data obtained from the “peak” data set of the selenomethionine MAD experiment (with phases from the refined model), and MAPMAN was used to integrate the anomalous density around both the unknown metal site and the selenium atom of SeMet187. The experimentally determined f″ value for selenium at this wavelength was 5.55 electrons, and the integrated anomalous density associated with the metal site was 24% that of the ordered selenium of SeMet187, suggesting that the f″ of the unknown metal is ∼1.33 electrons. The f″ values of Ca2+, K+, and Fe2+ were all roughly consistent with the anomalous scattering at the unknown metal site, although other metals such as Na+, Mg2+, Ni2+ Cu2+, and Zn2+ were not.
Assignment of the metal site geometry as “capped octahedral” was based on a consideration of all possibilities; angles between each ligand and the central metal were measured from the refined structure and compared with ideal angles calculated for each potential geometry. The root mean square deviation from the ideal angles (root mean square angle) was then used to find the geometry that best represents the site. The root mean square angle values for each possible 7-ligand geometry were as follows: pentagonal bipyramid (18.57°); square face-capped trigonal prism (20.23°); triangle face-capped trigonal prism (32.21°), and capped octahedron (12.15°).
Reduction Potential Determinations
15N-Labeled sBdbD (100 μm final protein concentration) in 50 mm potassium phosphate, pH 7, was treated with 10 mm GSSG or with 1 mm diamide (reacted overnight in the dark) to generate the fully oxidized protein or with 10 mm GSH or 5 mm DTT to generate the fully reduced protein. 1H 15N-HSQC spectra of oxidized and reduced sBdbD showed that the exchange between oxidized and reduced proteins was slow on the NMR time scale, and so spectra of mixtures corresponded to a superposition of spectra arising from the individual oxidized and reduced proteins. Several resonances that were dependent on the redox state of the protein were selected for analysis. sBdbD was oxidized at 4 °C overnight with 1 mm diamide, and excess oxidant was removed using a PD10 column and the protein equilibrated in 50 mm potassium phosphate buffer, pH 7. Fully oxidized sBdbD (100 μm final concentration) in the same buffer was then prepared as above, and a final concentration of 10 mm GSSG was added. Increasing concentrations of GSH (from a ratio of GSH:GSSG of 1:50 to 1:1.5) were added, and the samples were incubated at 25 °C. Redox equilibrium was established after 1 h, and 1H 15N-HSQC spectra were recorded for each, and the intensity changes at the selected chemical shift values were monitored. The resonance at 1H/15N = 9.516/115.587 was used as an intensity standard to minimize variation between samples. Spectra of fully reduced and oxidized sBdbD, which provided the end points of the titration, were recorded following the removal of reductant/oxidant by gel filtration. GSH and GSSG concentrations were calibrated, respectively, by reaction with 5,5′-dithiobis-(2-nitrobenzoic acid), and by absorbance measurements at 248 nm using ϵ248 nm = 382 m−1 cm−1 (35). The fraction of sBdbD reduced, fr, at each point in the titration and for each selected resonance in the NMR spectrum was calculated, and the midpoint reduction potential of sBdbD was determined as described previously (23, 36); further details are given in the supplemental material.
NMR spectra were acquired using a Bruker Avance III 800 MHZ spectrometer equipped with a triple resonance, pulsed field gradient probe, operating at 1H frequency of 800.23 MHz and 15N frequencies of 81.09 MHz, using pulse sequences incorporated into the Bruker Topspin 2.1 software. Two-dimensional 1H 15N-HSQC spectra were recorded at 25 °C. The 1H carrier frequency was positioned at the resonance of the water during the experiments. The 15N carrier frequency was at 115 ppm. Spectra were processed using NMRPipe (37) and peak intensities analyzed in NMRDraw (37). Before Fourier transformation, a cosine-bell window function was applied to each dimension for apodization. The indirect dimensions were first linearly predicted to double the number of data points and then zero-filled to round up the number of data points to the nearest power of 2.
Determination of pH Stability Range and Cysteine Thiol pKa Values
For pKa determination experiments, a mixed buffer system (PCTC buffer: K2HPO4, sodium citrate, Tris, and Ches, all at 50 mm) was used to maintain constant pH. To determine the range of pH over which sBdbD was stable (and therefore the range within which pKa values could be measured), changes in the intrinsic tryptophan fluorescence of sBdbD (0.25 μm final concentration) as a function of pH were followed upon excitation at 280 nm. For pKa measurements, pre-reduced sBdbD was reacted with 6-bromoacetyl-2-dimethylaminonaphthalene (badan) under pseudo-first order conditions, and analyzed as described previously (38). Further details are given in the supplemental material.
Protein Conformational Stability Measurements
sBdbD (both with and without Ca2+) was pre-reduced or oxidized by incubation in the dark for 2.5 h at 4 °C in 2 mm DTT or diamide (39), respectively. Excess reductant/oxidant was removed using a PD10 column equilibrated in 50 mm potassium phosphate, pH 7.0. Equilibrium unfolding experiments were conducted by incubating sBdbD in various quantities of urea in 50 mm phosphate buffer, pH 7.0 (reduced samples included 1 mm DTT to prevent re-oxidation), and monitoring intrinsic tryptophan fluorescence as a probe of the degree of unfolding. Refolding experiments were performed in a similar manner starting with protein that was incubated overnight in 6 m urea. In all cases, sBdbD (0.25 μm final concentration) was allowed to equilibrate at 25 °C for 4 h before measurements were made. Tryptophan fluorescence was measured at 25 °C using an LS55 luminescence spectrophotometer (PerkinElmer Life Sciences) with an excitation wavelength of 290 nm. Fluorescence intensity at 353 nm was used to determine Gibbs free energies of stabilization (ΔGstab) and midpoints of unfolding, as described previously (36); further details are given in the supplemental material.
Other Methods
For experiments involving substrate analogue peptides, pre-oxidized 15N-labeled sBdbD (300 μm in 50 mm potassium phosphate buffer, pH 7) was mixed with 600 μm reduced peptide and incubated at 25 °C for 1 h before measuring 1H 15N-HSQC spectra. Peptides were purchased from Severn Biotech, Kidderminster, UK) and contained the sequences IYKANCIACHGENYE (referred to as CXXCH peptide) and IYKANSIASHGENYE (SXXSH peptide), which correspond to the wild-type sequence and a double Cys to Ser variant of residues 55–69 of cytochrome c550 (CccA) from B. subtilis. For MALDI-TOF experiments, 10 mg/ml sinapinic acid prepared in 50% acetonitrile and 0.05% trifluoroacetic acid was used as the matrix. Spectra were recorded using a Shimadzu Biotech AXIMA-CFR instrument operating in linear mode, calibrated using cytochrome c (equine) 12,361.96 Da, apo-myoglobin (equine) 16,952.27 Da, and aldolase (rabbit muscle) 39,212.28 Da (Sigma). The soluble domain of ResA (sResA) was produced in E. coli and purified as described before (21).
RESULTS AND DISCUSSION
BdbD Is Membrane-localized
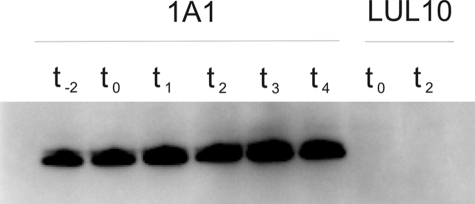
The B. subtilis bdbD gene encodes a protein of 222 amino acid residues with a putative N-terminal signal peptide for export to the outside of the cytoplasmic membrane. Although BdbD was predicted to have a type I signal peptidase cleavage site between residue 36 and 37 (40), such predictions are unreliable. To establish if BdbD is a membrane-bound protein, B. subtilis strain 1A1 was grown in NSMP medium (nutrient sporulation medium with phosphate). Because BdbD plays roles in both growing and stationary phase cells, samples were taken from the culture at various points in the growth curve. Soluble and membrane fractions of cell lysates were analyzed for BdbD by using immunoblot with polyclonal antiserum directed against BdbD without the putative membrane anchor. BdbD was not detected in the soluble fraction but was present in the membrane fraction, at a level that was largely independent of the growth phase (Fig. 1). Thus, we conclude that the TDOR domain of BdbD is attached to the membrane by the N-terminal transmembrane segment.
FIGURE 1.
Immunoblot analysis of the membrane fraction of B. subtilis strains 1A1 and LUL10 for BdbD. Strain 1A1 is the wild-type strain, and LUL10 has the bdbD gene inactivated. Cells were harvested at different time points during growth in NSMP. Time points t−2 and t0 are 2 h before and at entry to stationary growth phase, respectively. Subsequent numbers indicate hours into stationary phase. Approximately 10 μg of membrane protein was loaded in each lane for 1A1 and 20 μg for LUL10. BdbD antigen was not detected in the soluble cell fraction.
High Resolution Crystal Structures of BdbD in Reduced and Oxidized Forms
The soluble domain of BdbD (residues 37–222, sBdbD) was produced in E. coli, purified, and crystallized. The crystal structure of sBdbD was solved using selenomethionine MAD with subsequent refinement against high resolution native data sets obtained for crystals of both oxidized and reduced forms of the protein. Crystals of sBdbD belonged to space group P21 with a single molecule in the asymmetric unit. Full data collection and refinement statistics for the oxidized and reduced structures are given in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses represent the highest resolution shell. r.m.s. indicates root mean square. Multiplicity and completeness are “anomalous” for seleno-methionine data but not for the native datasets. Rmerge refers to cross-dataset scaling using the reduced native dataset as reference. Free c.c. is the correlation coefficient calculated with the free reflection set.
| Selenomethionine sBdbD | Reduced native | Oxidized native | |||
|---|---|---|---|---|---|
| Space group | P21 | ||||
| Unit cell | 39.9, 43.5, and 54.7 Å (β = 107.5°) | ||||
| Unique reflections | 28,058 | 27,977 | 27,961 | 27,450 | 7,198 |
| Energy | 12,654 eV | 12,653 eV | 12,661 eV | 12,654 eV | 13,288 eV |
| f′ | −9.18 | −10.41 | −5.52 | ||
| f″ | 5.55 | 3.61 | 4.53 | ||
| Resolution | 27.51 to 1.50 Å (1.58 to 1.50 Å) | 28.21 to 1.50 Å (1.58 to 1.50 Å) | 52.2 to 2.3 Å (2.42 to 2.3 Å) | ||
| Rsym | 0.072 (0.444) | 0.060 (0.393) | 0.030 (0.308) | 0.052 (0.267) | 0.046 (0.119) |
| I/σ(I) | 11.1 (1.8) | 14.3 (3.8) | 15.8 (4.1) | 15.0 (3.4) | 15.7 (7.3) |
| Completeness | 97.7% (98.7%) | 97.2% (98.6%) | 92.1% (90.0%) | 97.8% (89.5%) | 94.8% (81.2%) |
| Multiplicity | 1.8 (1.8) | 1.8 (1.8) | 1.9 (1.9) | 3.5 (2.9) | 2.3 (2.2) |
| Rmerge | 0.078 | 0.068 | 0.077 | 0 | |
| Rwork | 0.2051 | 0.1937 | |||
| Rfree | 0.2273 | 0.2480 | |||
| r.m.s. angle | 1.225° | 1.262° | |||
| r.m.s. bond | 0.0098 Å | 0.0090 Å | |||
| Free c.c. | 0.9425 | 0.8918 | |||
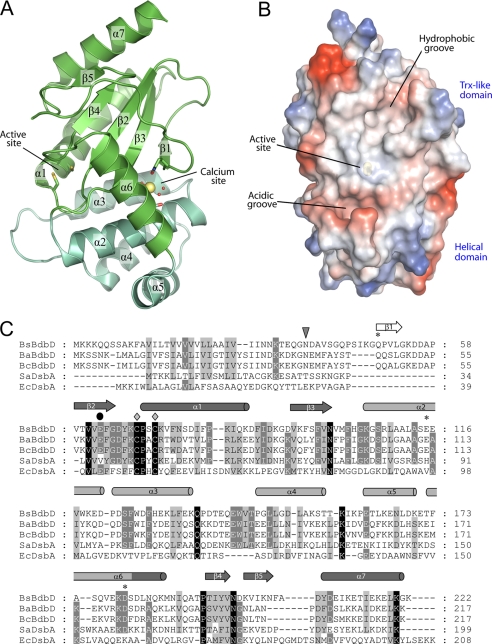
The overall crystal structure of BdbD is shown in Fig. 2A. The fold of BdbD is clearly related to the DsbA family of TDORs, for which a number of proteins have been structurally characterized. Like all DsbA-like proteins, BdbD is composed of two domains, a classical thioredoxin-like domain and an α-helical domain (Fig. 2A, green and teal, respectively). The thioredoxin-like domain consists of residues 59–103 (forming the βαβ component of the fold) and 182–222 (forming the αββα component of the fold). The helical domain consists of residues 104–181 that form five α-helices, the fifth of which is as an extension of the first helix of the thioredoxin αββα fold, therefore providing a structurally defined connection between the two domains. Preceding the βαβ component of the thioredoxin fold at the N terminus is an additional β-strand, which forms a β-sheet with the other β-strands of the thioredoxin fold. The arrangement of secondary structure elements is shown in Fig. 2C.
FIGURE 2.
Structure of the soluble domain of B. subtilis BdbD. A, three-dimensional structure of sBdbD showing that the protein exhibits a thioredoxin-like fold with an inserted helical domain. Secondary structure elements are labeled from the N terminus with the predicted transmembrane helix of the full-length protein (covered by residues 1–36 that were not present in the protein used for crystallization and therefore not indicated on the figure) being 0. The active site and Ca2+-binding sites are indicated. The thioredoxin-like part of BdbD is colored green, and the all-helical domain is colored teal. Note that the same color scheme is used in Figs. 3 and 4. B, surface representation of sBdbD. Regions colored red indicate areas of high negative electrostatic potential, and blue areas indicate areas of high positive potential. Neutral regions are in white. Electrostatic potentials of surfaces were calculated using PyMOL (73). All structural figures were prepared with PyMOL and annotated with GIMP. C, amino acid residue sequence comparison of B. subtilis BdbD (BsBdbD) with other DsbA-like proteins from Bacillus anthracis (BaBdbD), Bacillus cereus (BcBdbD), S. aureus (SaDsbA), and E. coli (EcDsbA). Invariant residues are marked in black and highly conserved residues in gray. Diamonds indicate the active site residues Cys69 and Cys72, and asterisks indicate the residues acting as ligands to the Ca2+ ion. A black circle indicates the near-active site glutamate residue (Glu63), and an inverted triangle indicates the beginning of the soluble domain of B. subtilis BdbD studied here. Secondary structural elements, derived from the structure of BdbD, are indicated above the sequence. Elements belonging to the thioredoxin-fold are indicated in dark gray; those belonging to the helical domain are indicated in light gray, and those not belonging to either are indicated in white. Residue numbers are indicated on the right; these refer to the non-processed predicted translated sequence. The alignment was obtained using ClustalW (74) and annotated in Genedoc (75).
BdbD is most similar to S. aureus DsbA (SaDsbA), sharing a number of structural features with this protein that differ from E. coli DsbA (EcDsbA) (16). For example, BdbD possesses a loop between helices α3 and α4 of the all-helical domain that is considerably longer than the equivalent loop observed in EcDsbA and also lacks the hydrophobic groove observed on the surface of EcDsbA, which was recently shown to be important for interaction of DsbA with DsbB (41). BdbD can be superposed with SaDsbA and EcDsbA with root mean square deviation values of 2.5 and 3.1 Å, respectively. Despite the overall similarities, two features clearly distinguish BdbD from all previously characterized DsbA proteins. First, and most strikingly, BdbD contains a unique metal site located between the helical domain and the central β-sheet of the thioredoxin-like domain. Second, the β-sheet at the center of the thioredoxin-like domain has a different topology to that of previously characterized DsbA proteins. In BdbD, four of the five β-strands that make up the central β-sheet are structurally equivalent to those of the DsbA family of proteins (i.e. strands β2, β3, β4, and β5), but the location of the fifth strand in sBdbD is topologically distinct from its location in DsbA, despite the fact that the strand is located at the same position in the primary sequence of both proteins (i.e. the strand is β1 in both DsbA and BdbD). In BdbD, strand β1 runs anti-parallel to strand β3 (the strand preceding the helical domain), whereas in DsbA, strand β1 runs anti-parallel to strand β5 (the strand that precedes the final helix). Thus, in BdbD, strand β1 is the closest β-strand to the all-helical domain, although in previously characterized DsbA proteins strand β1 is the most distant from the helical domain. The topological differences between DsbA and BdbD have two important structural consequences. First, the repositioning of strand β1 means that the packing between the all-helical and thioredoxin domains is much denser in BdbD than in other DsbA proteins. This means that domain motions of the type previously observed for EcDsbA (42) are unlikely to occur in BdbD because strand β1 occupies the void that permits hinge movements between the domains in DsbA. Second, the close approach of strand β1 and the helical domain forms an important structural backbone to the metal-binding site that is observed exclusively in BdbD (see below).
Structural Features of the BdbD Active Site
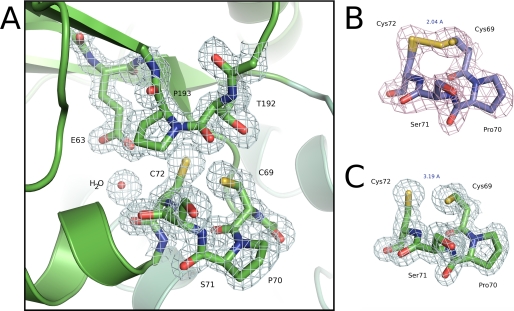
The active site thioredoxin-like CXXC motif of BdbD is located at the N terminus of a long helix (α1) and has the sequence Cys69–Pro70–Ser71–Cys72. The active site structure of the reduced protein is shown in Fig. 3A, with an accompanying electron density map (Fig. 3C). In addition to the CPSC motif itself, we also note the conserved cis-proline loop directly opposite the active site cysteines and an unusual buried glutamate residue (Glu63) in close proximity to Cys72.
FIGURE 3.
Active site region of BdbD. A, detailed view of the N terminus of helix α1 of sBdbD showing the Cys-Pro-Ser-Cys active site of sBdbD and the closely lying cis-proline (Pro193), which is invariant in all thioredoxin-like proteins. B and C, electron density (contoured at 1.0σ) of the active site region of sBdbD in oxidized and reduced states, respectively.
In the structure of reduced sBdbD, the cysteine sulfur atoms are separated by a distance of 3.19 Å (Fig. 3C), with clear indications from the electron density that, despite their close proximity, the cysteine sulfurs are in the free thiol/thiolate forms (“unbiased” omit maps for the oxidized and reduced structures are shown in supplemental Fig. S1A). In contrast, the oxidized structure showed continuous electron density between the sulfur atoms of Cys69 and Cys72 demonstrating the formation of a disulfide bond (Fig. 3B and supplemental Fig. S1B). Although crystallized as a fully oxidized sample, refinement of a disulfide bond in the oxidized structure did show some residual electron density indicating a small fraction of sBdbD was in the reduced state, probably due to a degree of photo-reduction in the x-ray beam. We thus refined the “oxidized” structure as a mixture of oxidized (70%) and reduced states (30%), with the active site dominated by the oxidized form (a more completely oxidized disulfide structure was obtained for a sample treated with EDTA, see below and supplemental Fig. S1C). The structural data show that there are very few redox-linked conformational changes between the oxidized and reduced forms of the protein, such that superposition of oxidized and reduced BdbD gave a root mean square deviation of 0.268 Å (calculated for all atoms of residues 37–221).
Thiolate Form of Cys69 Is Significantly Stabilized Relative to the Thiol Form
The reduction potentials of thioredoxin-like TDORs are intimately linked with the pKa values of their active site cysteines: in general, the more extensive the stabilization of the thiolate form of the active site cysteines, the lower their corresponding pKa value and the higher the active site reduction potential (43–45). Furthermore, within the thioredoxin-like family of TDORs, it is the N-terminal active site cysteine that exhibits the greatest variation and is thus likely to be the key determinant of reduction potential. In the structure of reduced sBdbD, the Cys69 sulfur atom is hydrogen-bonded to the backbone amide groups of both Ser71 (3.4 Å) and Cys72 (3.2 Å) as well as to the side chain hydroxyl group of Thr192. The extensive hydrogen bonding network around the Cys69 sulfur is clearly an important factor in maintaining a low cysteine pKa for Cys69 and is therefore likely to be a key structural determinant of the reduction potential of the protein.
Structural Implications for the Interaction of BdbD with Substrates
The loop region directly opposite the active site (composed of Thr192 and cis-Pro193) is very likely to be involved in the interaction of BdbD with its substrates (46). This cis-proline residue is conserved among all known thioredoxin-like proteins and is likely to be important in presenting the carboxyl oxygen of the preceding residue for hydrogen bond interactions with substrate cysteine residues as observed in various TDOR-substrate complexes (47–49). Substitution of the DsbA equivalent of BdbD Pro193 has been shown previously to lead to an accumulation of covalent DsbA complexes in vivo (50). Furthermore, the crystal structure of EcDsbA in a covalent complex with DsbB has shown that this loop is important in mediating contacts between DsbA and DsbB (41). We propose that similar interactions are likely to be important for the interaction of BdbD with its substrates. However, we note that although BdbC is clearly a homologue of DsbB, the manner by which BdbD is re-oxidized by BdbC cannot be the same as for DsbA/DsbB, because BdbC lacks the external loop domain with which DsbA interacts (20). Furthermore, as noted above, the peptide-binding groove identified in EcDsbA (which appears to interact with a well defined portion of DsbB) does not exist in the structure of BdbD (Fig. 2B). Indeed, the electrostatic surface view of BdbD (Fig. 2B) shows that little of the surface shape or the electrostatic properties are conserved between BdbD and DsbA-like proteins (8, 16).
Role of Glu63
Our structures show that Glu63 is buried in a position very close to the Cys72 sulfur atom (Fig. 3A). Buried polar or charged residues, which destabilize folding, are usually functionally important, and in support of this, a negative charge at the equivalent position of Glu63 is highly conserved among BdbD and DsbA homologues although, interestingly, it is not present in SaDsbA (Fig. 2C). The role of Glu63 is not yet clear, but one possibility is that Glu63 acts as a general acid/base during the mechanism of disulfide exchange. Asp26 in E. coli thioredoxin, which is located spatially in a similar position to Glu63 of BdbD, was proposed to act in just such a way during interaction with its substrates (51). We note that buried glutamate residues also occur in ResA (Glu80) and StoA (Glu71) which, like BdbD, are extra-cytoplasmic membrane-bound TDORs from B. subtilis (23, 52). The buried glutamates of StoA and ResA are not in equivalent positions to Glu63 in terms of primary sequence, but the location of the negative charge close to the C-terminal active site cysteine is similar in all of these TDORs. Furthermore, these glutamate residues of ResA and StoA have both been confirmed as mechanistically important through the investigation of Glu-to-Gln mutants (23, 38).
BdbD Contains a Ca2+ Ion in a Capped Octahedral Geometry
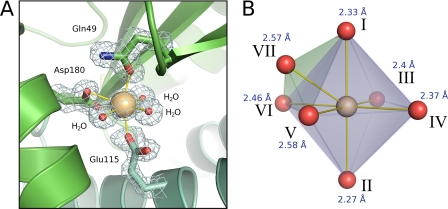
The metal ion site located between the thioredoxin and helical domains is 14 Å from the nearest active site cysteine sulfur and is coordinated by three protein residues as follows: Gln49, Glu115, and Asp180. In addition to these three residues, the central ion also forms coordinate bonds with three low b-factor solvent molecules (Fig. 4A). The metal therefore has a total of seven ligands (the carboxylate group of Asp180 is bidentate) arranged with a capped octahedral geometry (Fig. 4B). We identified the metal as Ca2+ on the basis of several lines of evidence. First, ICP-AE spectroscopy of purified sBdbD demonstrated the presence of 0.9 ± 0.1 Ca2+ (and no other metal ions) per protein. Second, only Ca2+ and K+ refined well against the x-ray data. Smaller candidate ions such as Na+ or Mg2+ refined to unrealistically low b-factors and gave positive peaks in difference maps (|Fo| − |Fc|), suggesting they are not electron dense enough to fit the observed data. Larger ions such as Zn2+ or Fe2+ also refined poorly, with strong negative density in difference maps suggestive of a smaller metal or a partial occupancy. The latter possibility can be ruled out because the anomalous scattering of the metal site (at a wavelength of 0.9798 Å) was very small; a low occupancy of Zn2+ (or another transition metal) would be expected to give much stronger anomalous scattering. Third, the metal-to-water bond lengths derived from the high resolution structure of sBdbD (2.37, 2.40, and 2.56 Å) are consistent with those of Ca2+ (typically 2.40 Å) but not of K+ (typically 2.82 Å) (53).
FIGURE 4.
Ca2+-binding site of BdbD. A, detailed view (contoured at 1.2 σ) of the Ca2+-binding site of sBdbD, showing monodentate ligands Gln49 and Glu115, bidentate Asp180, and three crystallographically ordered water molecules. B, an ideal capped octahedral site for comparison with that observed in BdbD. Positions of the idealized capped octahedron correspond to the following: I, Gln49O-ϵ; II, Glu115O-ϵ; III–V, waters; VI and VII, Asp180O-δ1 and Asp180O-δ2. Ca2+-ligand distances are indicated.
A coordination number of seven is extremely common among protein-bound Ca2+ sites, although a search of the Protein Data Bank did not reveal any that exactly matched the site in BdbD in terms of the ligand set. Furthermore, many of these Ca2+ sites have pentagonal bipyramidal geometry, with the capped octahedral geometry observed here being much less common.
This is the first example of a TDOR that contains a metal ion binding site remote from the CXXC active site, raising the question of what the functional significance of the metal site is. Multiple sequence alignments of BdbD/DsbA proteins show that the three residues involved in coordinating the metal site are strictly conserved among all Bacillus species, but not in Gram-negative bacteria nor in other Gram-positive organisms such as S. aureus (Fig. 2C). This suggests that the site is likely to be relevant to the function of BdbD. A second question raised by the structure is whether or not Ca2+ is the native metal ion in B. subtilis. The protein was heterologously produced in the cytoplasm of E. coli cells, and it is possible that Ca2+ was the only ion available in sufficient quantities to fill the site in the absence of an alternative. However, Ca2+ concentrations in bacterial cells, as in eukaryotic cells, are tightly regulated and are estimated to be between 100 and 300 nm (54). Therefore, Ca2+ is not normally found at sites usually occupied by other metal ions. Furthermore, we can rule out that Ca2+ was added during or after purification as it was not used at any stage during the purification or as a component in the crystallization buffer. Therefore, the data indicate that Ca2+ is the natural metal ion in BdbD.
Ca2+ is extremely important for a range of processes in eukaryotic cells. Its role in bacterial cells is much less clear, but there are a number of examples of bacterial Ca2+-binding proteins, which are proposed to be involved in a range of processes, including the maintenance of cell structure and motility, and in cell differentiation processes such as sporulation (55). Many of these are related to eukaryotic EF-hand proteins that contain a conserved helix-loop-helix structural motif in which Ca2+ is coordinated by the side chains of three residues from the loop, a backbone atom from another loop residue, a water molecule, and the side chain of an acidic residue in the second helix (56, 57). The structures of several bacterial EF-hand-like proteins have been determined, revealing that although they share the conserved Ca2+-binding motif DX(D/N)XDG within the central loop of the EF-hand domain, they exhibit significant diversity in the structural elements surrounding the motif. Ca2+ is also found in other types of bacterial proteins, including those belonging to the RTX family of bacterial cytolysins (58) that are normally secreted, which includes toxins (59) and lipases (60), as well as a lectin (61) and a collagenase (62), which both contain unusual binuclear Ca2+ sites. In all of these structures, Ca2+ is bound in a loop that provides the majority of the ligands, and therefore all are distinct from the site described here in sBdbD, in which the three residue side chains acting as ligands are derived from three different parts of the primary sequence (Fig. 2C). A greater degree of similarity is observed for the Ca2+ sites of peroxidases and, in particular, the di-heme cytochrome c peroxidase, in which Ca2+ is coordinated by three residues and four water molecules and lies at the interface between the two domains of the protein (63).
Ca2+ Site Is Not Required for Folding of BdbD
To investigate the importance of the Ca2+ site for the structure of sBdbD, the crystal structure of a metal chelator-treated sample was solved in both oxidized and reduced states (data collection and refinement statistics are given in supplemental Table S1). The sample used for crystallization contained <10% Ca2+ according to metal analysis, and the chelator, EDTA, was maintained at 100 μm in all crystallization conditions and cryo-protectants used during preparation of the crystals. Despite this, some residual electron density was present within the metal site of both the oxidized and reduced EDTA-treated sBdbD structures, although this was substantially less than in the as-isolated sBdbD structures. Overall structures of reduced and oxidized chelator-treated sBdbD were very similar to those of the as-isolated protein (see supplemental Fig. S2). There were several minor changes to the local structure of residues at the site, including Glu115, which adopted multiple distinct side chain conformations consistent with heterogeneity at the site. Furthermore, there was essentially no anomalous scattering at the position of the metal. Best fits to the data were obtained using either a low occupancy Ca2+ ion or a full occupancy Na+ ion. Given that EDTA-treated sBdbD contained some Ca2+, at least some of the observed intensity must arise from Ca2+ occupation, but it is possible that Na+ replaces Ca2+ in some BdbD molecules. We note that other metals (such as K+) can be ruled out on the basis of the metal-to-water coordination distances that are consistent with occupation by Na+ or Ca2+. Although it has not yet been possible to completely remove all traces of metal(s) from the observed site, it is nonetheless clear that occupation by Ca2+ is drastically reduced in the EDTA-treated structures. This indicates that Ca2+ is not required to maintain the native three-dimensional fold of the protein.
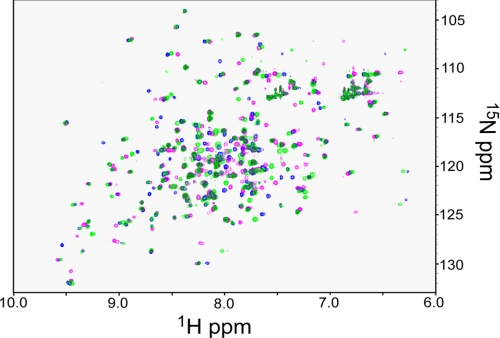
Two-dimensional NMR methods were also used to investigate conformational changes upon removal of Ca2+. The 1H 15N-HSQC spectrum of reduced 15N-labeled sBdbD was recorded before and after treatment with EDTA (Fig. 5). A significant proportion of resonances was observed to have shifted upon depletion of Ca2+, indicating that the presence of the ion does have structural consequences in solution. Similar results were obtained for the oxidized form of the protein (data not shown). In light of the crystal structure of Ca2+-depleted sBdbD, we conclude that these changes must be subtle (for example, similar to those caused by an oxidation state change, which do not cause major structural rearrangement but result in shifts in many resonance positions (see below)). Titration of EDTA-treated sBdbD with Ca2+ ions resulted in a 1H 15N-HSQC spectrum identical to that recorded for the native protein (data not shown), whereas titration with Mg2+ ions resulted in an HSQC spectrum distinct from those of Ca2+-bound and Ca2+-depleted sBdbD (Fig. 5). These data indicate that the structural effects of Ca2+ are specific to this metal ion.
FIGURE 5.
Structural effects of metal binding to BdbD monitored by NMR. 1H 15N-HSQC spectra of reduced sBdbD (200 μm) in 50 mm potassium phosphate, pH 7.0. The spectrum in green corresponds to the as-isolated Ca2+-bound form, and the spectrum in magenta resulted from Ca2+-depleted sBdbD. The spectrum in blue was obtained following the addition of an excess of Mg2+ ions to metal-depleted sBdbD.
Reduction Potential Determination for BdbD
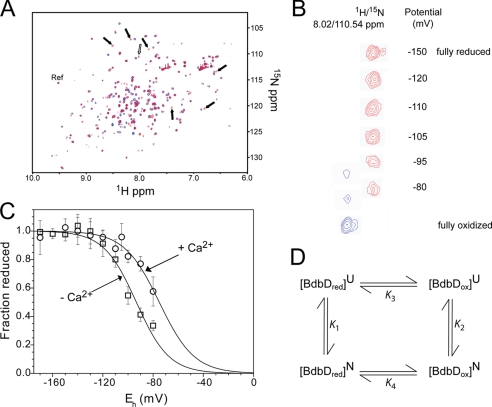
The sensitivity of tryptophan fluorescence intensity to the redox state of TDOR proteins has provided a convenient means to measure their redox potential (23, 36). BdbD lacks the near-active site tryptophan, and so fluorescence methods could not be used to monitor redox state. Therefore, an alternative method was sought. Two-dimensional NMR methods have been used previously to determine the reduction potential of human thioredoxin 1 (64). NMR has the potential to provide much more detailed information about changes occurring in the protein during a redox transition. For example, the effect of redox buffers can readily be monitored through the 1H 15N-HSQC spectrum of a 15N-labeled protein, and the formation of adducts (e.g. mixed disulfides between the protein and glutathione) can be detected. 1H 15N-HSQC spectra of fully oxidized and reduced sBdbD (Fig. 6, A and B) revealed differences in a number of resonances that, although not assigned to specific residues, enabled the redox composition of sBdbD samples to be followed as a function of the potential generated by the GSSG/GSH couple (Fig. 6C). Under the conditions of concentration required for the NMR experiments, the highest potential that could be reliably generated by the GSSG/GSH couple was limited to approximately −80 mV. Normally, this would be more than sufficient to cover the entire transition for a thioredoxin-like protein. In this case, the protein was not fully oxidized at −80 mV, and so a complete redox titration data set was not obtained. Nevertheless, the data could be fitted to the Nernst equation, giving a midpoint reduction potential (Em) of −75 ± 5 mV versus NHE at pH 7 and 25 °C with n, the number of electrons involved in the reduction, at 1.85 ± 0.26. This value of Em is similar to those reported for DsbA from E. coli and S. aureus and is consistent with an oxidative function for BdbD on the outside of the cytoplasmic membrane in B. subtilis.
FIGURE 6.
Reduction potential determination for BdbD. A, 1H 15N-HSQC spectra of sBdbD (100 μm) in 50 mm potassium phosphate, pH 7.0, in reduced (red) and oxidized (blue) states. Resonances that were used to follow oxidation state changes are indicated by arrows. The open arrow indicates the resonance shown in more detail in B. The selected reference resonance, which is insensitive to oxidation state, is also indicated (as Ref). B, plot showing potentially dependent changes for one of the selected resonances (reduced resonance at 1H/15N = 8.02/110.54 ppm) at the indicated potential (the reduced resonance is in red and in the oxidized resonance is in blue). As the potential increases the reduced resonance intensity decreases, and intensity corresponding to the oxidized protein (1H/15N = 8.077/110.81 ppm) is observed. C, plot of fraction of reduced sBdbD as a function of the cell potential. The standard deviation for each titration point is indicated on the plot. The solid line shows a fit to supplemental Equation S1. D, thermodynamic cycle connecting the oxidized and reduced forms of native sBdbD with those of unfolded sBdbD (10). K1–K4 are equilibrium constants calculated from ΔG values measured here or, in the case of K3, from literature values for unfolded TDORs/model peptides.
Oxidized BdbD Contains a Destabilizing Disulfide Bond
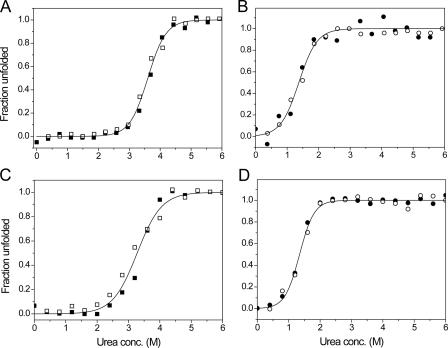
The conformational stabilities of sBdbD in both reduced and oxidized states were investigated through equilibrium unfolding (and refolding) experiments using urea, in which the intrinsic tryptophan fluorescence of sBdbD was used to monitor the state of folding (Fig. 7, A and B, respectively). Urea-induced unfolding of sBdbD was found to be completely reversible and was analyzed in terms of a two-state model, yielding a free energy of stabilization of −30.4 ± 2.5 kJ mol−1 for the reduced protein and −11.8 ± 1.6 kJ mol−1 for the oxidized protein. The greater stability of the reduced state compared with the oxidized state is a characteristic of oxidizing TDORs. For sBdbD, ΔΔGox/red = 18.6 ± 4 kJ mol−1, a value similar to that measured for EcDsbA (14.8 ± 4 kJ mol−1) (65). The associated m values for the folding/unfolding transition were similar for both redox states (Table 2), indicating that the protein undergoes a similar degree of unfolding in each state. This is consistent with there being few structural changes between the reduced and oxidized states and that the disulfide bond is between two cysteines residues located close to one another and, therefore has little effect on the degree of unfolding.
FIGURE 7.
Unfolding profiles of native and EDTA-treated sBdbD. 1 μm pre-reduced and pre-oxidized sBdbD protein was incubated in various concentrations of urea at 25 °C. Fraction unfolded was calculated from the fluorescence intensity. A and B show data from reduced (squares) and oxidized (circles) as-isolated protein, respectively, in 0.1 m Tris-HCl, pH 8.0. The folding (open symbols) and unfolding (filled symbols) data were analyzed using supplemental Equation S3, and the resulting fits are drawn as solid lines. C and D show folding and unfolding data from equivalent experiments performed with Ca2+-depleted sBdbD.
TABLE 2.
Free energies of stabilization of oxidized and reduced sBdbD in the presence and absence of Ca2+ ions at 25 °C
| sBdbD | Midpoint of transition (D0.5, mGdn-HCl) | Cooperativity | ΔGstab | ΔΔGox/reda |
|---|---|---|---|---|
| m, kJ mol−1m−1 | kJ mol−1 | kJ mol−1 | ||
| With Ca2+/reduced | 3.60 | 8.5 ± 0.7 | −30.4 ± 2.5 | 18.6 |
| With Ca2+/oxidized | 1.37 | 8.6 ± 1.1 | −11.8 ± 3.5 | |
| Without Ca2+/reduced | 3.28 | 6.4 ± 0.6 | −20.9 ± 2.1 | 6.5 |
| Without Ca2+/oxidized | 1.35 | 10.7 ± 0.8 | −14.4 ± 1.1 |
aΔΔGox/red indicates the difference between the conformational stabilities of the oxidized and reduced proteins, i.e. ΔGox − ΔGred.
With information about the folding stabilities of the reduced and oxidized protein, the thermodynamic cycle (10) (illustrated in Fig. 6D) can be used to estimate the reduction potential of the folded protein. From the cycle, the difference in folding stabilities between the oxidized and reduced states (ΔΔGox/red) is equivalent to the difference in ΔG between oxidized and reduced states in folded and unfolded states. In other words ΔΔGox/red is equal to the difference in reduction potentials of the protein in the folded and unfolded states. Using the expression K2/K1 = K4/K3, in which K1 and K2 are equilibrium constants for the unfolding of reduced and oxidized sBdbD, respectively, and K3 and K4 are redox equilibrium constants for the unfolded and native proteins, respectively, the value of K4 (which gives the reduction potential of the folded protein) can be obtained. To do this, the reduction potential of the unfolded protein is required to calculate K3. It has been shown previously that reduction potentials of a range of unfolded thioredoxin-like proteins, including E. coli glutaredoxin 1 (Grx1) and glutaredoxin 3 (Grx3), DsbA, and DsbC, exhibit remarkably little variation, with values of approximately −215 mV (10, 44, 66, 67). Furthermore, the reduction potential of the CXXC motifs of a range of peptides that contain the active site motifs of TDOR proteins also exhibits little variation, with values of approximately −205 mV (68). Therefore, it is reasonable to assume that the reduction potential of unfolded sBdbD is approximately ∼ −210 mV. Using this, a reduction potential of approximately −110 mV (versus NHE at pH 7) was calculated for folded sBdbD. This is somewhat lower than that determined by redox titration (Fig. 6) but is entirely consistent with an oxidative role of BdbD.
The removal of Ca2+ from sBdbD resulted in some conformational changes (see above); therefore, it was of interest to determine the relative stabilities of reduced and oxidized sBdbD in the absence of Ca2+. Equilibrium unfolding experiments were carried out as above (Fig. 7, C and D, and Table 2). Reversible unfolding was observed in each case, and the data could be fitted to a two-state model. For both reduced and oxidized states, the data demonstrated that Ca2+ is not essential for stability; folded sBdbD is stable in both oxidation states, consistent with the structural data above. However, reduced sBdbD had a folding stability of −20.9 ± 2.0 kJ mol−1, ∼10 kJ mol−1 lower than in the presence of Ca2+, and so the metal ion significantly stabilizes the reduced protein. The oxidized protein, on the other hand, in the absence of Ca2+ exhibited a folding stability similar to and even somewhat greater than that of Ca2+-replete sBdbD (−14.4 ± 1.0 kJ mol−1 compared with −11.8 ± 1.6 kJ mol−1 for the as-isolated protein). Therefore, Ca2+ does not contribute to the stability of the oxidized state and may even destabilize it relative to the Ca2+-free form. It is also of note that the m values for Ca2+-depleted sBdbD are significantly different for the reduced and oxidized states, implying that the extent of unfolding is different, consistent with a difference in structure between the oxidized and reduced states in their native and/or denatured forms.
The data indicate clearly that the loss of Ca2+ leads to a drop in the value of ΔΔGox/red and therefore a decrease in the difference between reduction potentials in folded and unfolded states. From the cycle in Fig. 7D, the reduction potential of sBdbD in the absence of Ca2+ was calculated to be ∼60 mV lower than when Ca2+ is present, suggesting that a major role of Ca2+ is to increase the oxidizing power of BdbD.
Ca2+ Site of BdbD Serves to Increase the Active Site Reduction Potential
To determine directly the effect of the presence of Ca2+ on the redox properties of sBdbD, the mid-point reduction potential of the Ca2+-free protein was determined by NMR methods, as described above for the as-isolated Ca2+-containing protein (Fig. 6C). At the highest potential that could be generated by the GSSG/GSH couple (−80 mV), the extent of reduction was much more significant than in the Ca2+-containing protein, and fitting of the data to the Nernst equation, giving a midpoint reduction potential (Em) of −95 ± 5 mV versus NHE at pH 7 and 25 °C, with n, the number of electrons involved in the reduction, at 1.98 ± 0.2. This value of Em is significantly lower than that determined for the Ca2+-containing protein, demonstrating that the Ca2+ site, although 14 Å away from the nearest active site cysteine, exerts an important effect on the redox properties of the protein, such that in its presence the potential is ∼20 mV more positive. This difference, although less than that predicted from unfolding studies, is qualitatively in good agreement.
pKa Values of BdbD Active Site Cysteines
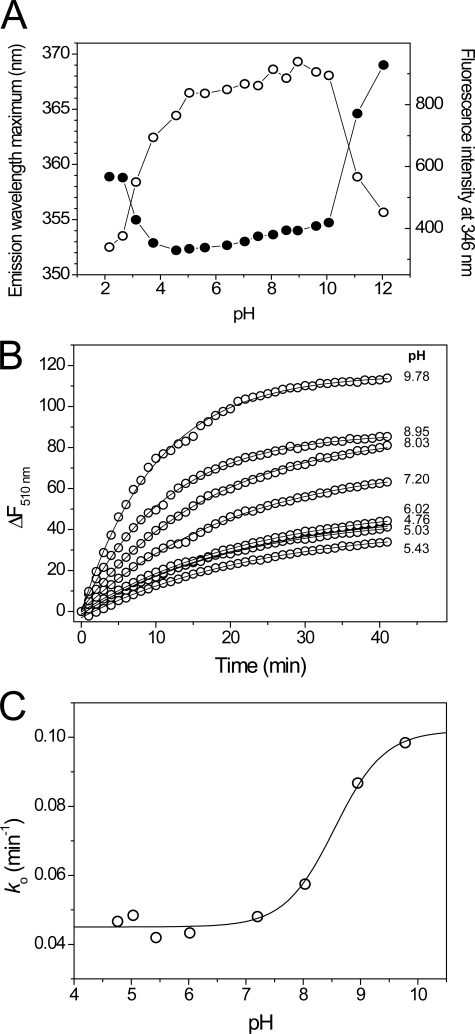
To define the range over which the pKa values of the sBdbD active site cysteines can be measured, the pH stability of folded sBdbD was investigated using the intrinsic fluorescence of tryptophan. A plot of emission wavelength maximum versus pH (Fig. 8A) showed that pH-induced unfolding of sBdbD led to a sizeable shift in emission wavelength from around 353 nm at pH 7 to much longer wavelengths at extremes of pH. The apparent stability range of BdbD lies between pH 4.5 and pH 10 and is similar to both ResA and StoA from B. subtilis (23, 36). Within the stable range itself, there was little evidence for titration of other residues affecting the fluorescence of tryptophan residues. The data indicate that pKa values lower than 4.5 cannot be measured. This is particularly important for sBdbD because, as a DsbA-like protein, it is likely to have a highly nucleophilic N-terminal cysteine in the CXXC motif (11, 44).
FIGURE 8.
pH stability and pKa determination for BdbD. A, plots of tryptophan fluorescence emission maxima and emission intensity at 346 nm as a function of pH for solutions of sBdbD (0.15 μm in PCTC buffer), as a function of pH. B, time-dependent increases in fluorescence at 510 nm upon reaction with wild-type sBdbD (1 μm) with badan (12 μm) in a mixed buffer system at pH values from 4.76 to 9.78, as indicated, at 25 °C. Plots were fitted (solid lines) to obtain an observed, pseudo-first order rate constant ko. C, plots of ko as a function of pH. The solid line show a fit to supplemental Equation S2.
The acid-base properties of the active site cysteines of sBdbD were investigated by measuring rates of reaction with the fluorescent probe badan, as described under “Experimental Procedures” (Fig. 8B). During the badan modification experiment, a single fluorescence peak with a maximum emission at 510 nm was observed for badan-treated sBdbD, corresponding to covalent badan-adduct(s) of protein cysteine residue(s). A plot of the pseudo-first order rate constant versus pH revealed a significant rate constant at low pH and an apparent protonation/deprotonation event at pH 8.6 (Fig. 8C). The rate constant at low pH is roughly constant (∼0.045 min−1), whereas that measured after the transition is estimated to be ∼0.102 min−1 on the basis of the fitted Henderson-Hasselbalch curve. sBdbD contains only two cysteines (Cys69 and Cys72) and, although only a single titration event is detected, the high rate of badan modification at low pH strongly suggests that a cysteine residue must be reacting with badan at acidic pH values. We therefore conclude that the apparent pKa event is likely to be due to titration of Cys72, whereas the high rate at low pH is because of the thiolate form of Cys69, which is likely to have a pKa value below the minimum pH at which sBdbD is stable. This is consistent with pKa values reported for other DsbA-like proteins where the N-terminal cysteine usually has a very low pKa value (11, 16, 44).
The pH stability studies reported here highlight that the determination of accurate cysteine pKa values requires knowledge of the pH-induced unfolding, because the pKa values of cysteines in the unfolded protein are likely to be significantly higher than in the folded state. This raises the question of how the effects of protein unfolding at low pH have been taken into account in previous determinations of cysteine pKa values in EcDsbA and other DsbA-like proteins. We note that for SaDsbA, a pKa of 3.37 was determined (16), but the pH stability of the protein was not reported, although Charbonnier et al. (69) concluded that the pKa value of wild-type EcDsbA could not be accurately determined because of protein unfolding at low pH. Nelson and Creighton (11) directly addressed this issue by monitoring thiolate formation in EcDsbA using the absorbance change at 240 nm to report on thiol/thiolate status, while simultaneously measuring the 288 nm signal as indicative of folding/unfolding transitions. From these data, a pKa of ∼3.5 for the N-terminal cysteine was determined, but it was also shown that the protein unfolded below pH 3. Thus, deprotonation and unfolding events overlap, raising the question of whether the pKa values reported are really because of protonation/deprotonation of the active site cysteine in the native state or, in fact, measure protonation/deprotonation because of the relaxation of structural factors that depress the cysteine pKa value in the folded state. Thus, although it is clear that the N-terminal cysteine pKa values are considerably lower in DsbA-like proteins (including BdbD) than in reducing thioredoxin-like proteins (which have pKa values of >6.5), or for free cysteine in solution (pKa value ∼8.5), it is not clear that the pKa value for any of these proteins has been accurately determined.
Probing Redox Interactions of BdbD
To investigate the ability of sBdbD to oxidize a substrate, a 15-residue peptide containing the active site sequence CXXCH of B. subtilis apo-cytochrome c550 (CccA) was used. A significant body of evidence indicates that BdbD oxidizes apo-cytochrome c substrates upon their export to the outside of the cytoplasmic membrane before they are subsequently re-reduced by the reducing TDOR ResA immediately prior to covalent heme attachment (21, 38, 52). sBdbD was oxidized overnight by the addition of 10 mm GSSG, and a 2:1 excess of reduced peptide was added. The resulting 1H 15N-HSQC spectrum showed no evidence for the presence of a stable interaction between sBdbD and the peptide, consistent with the transient nature of the interaction of DsbA-like proteins with substrates, but it did reveal that sBdbD was present entirely in the reduced form (data not shown). An identical experiment with an equivalent peptide containing an SXXSH active site resulted in the observation of sBdbD only in the oxidized state. These experiments clearly demonstrate that oxidized sBdbD is able to transfer its disulfide bond to the apo-cytochrome c550 active site mimic peptide.
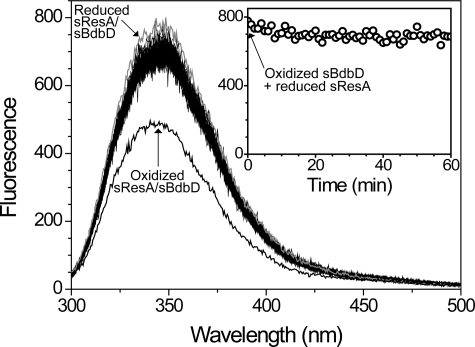
The data presented here (and previously) show that BdbD is an oxidizing extra-cytoplasmic TDOR. B. subtilis contains a number of other extra-cytoplasmic TDORs, two of which, ResA and StoA, have been shown to be reducing TDORs involved in cytochrome c maturation and endospore biogenesis, respectively (21–23). The existence of both oxidizing and reducing TDORs on the outside of the membrane raises questions about how unproductive redox cycling of these proteins and their substrates is avoided. We therefore sought to determine whether or not BdbD can directly oxidize reduced ResA in vitro. The fluorescence emission intensity of sResA at 353 nm is highly sensitive to the redox state of the protein, becoming significantly quenched upon oxidation (21). As noted above, the fluorescence intensity of sBdbD, on the other hand, is insensitive to the redox state of the protein. Therefore, fluorescence spectroscopy was used to investigate whether sBdbD and sResA can undergo a redox reaction with one another. Oxidized sBdbD was mixed with an equimolar concentration of reduced sResA and fluorescence spectra measured over a period of 1 h (Fig. 9). Initially, a small loss of intensity occurred, but overall significant changes in intensity were not observed. A similar slow loss of intensity was observed upon addition of a redox-inactive SXXSH cytochrome c550 active site mimic peptide, indicating that small intensity changes are observed even when electron transfer cannot take place (data not shown). Intensity changes are shown more clearly in the plot of fluorescence intensity at 345 nm as a function of time (Fig. 9, inset). As a control, sBdbD was also mixed in 1:1 ratio with oxidized sResA, verifying that oxidation of sResA results in a large decrease in intensity. The data indicate that, despite a significant thermodynamic driving force, the proteins do not undergo a redox reaction, at least in vitro.
FIGURE 9.
Redox interaction between sBdbD and sResA. Fluorescence spectra of reduced sResA (0.5 μm) at increasing time points following the addition of oxidized sBdbD (0.5 μm) in 0.1 m Tris-HCl, pH 8.0. A plot of fluorescence intensity at 345 nm as a function of time is shown in the inset. Spectra of reduced (in gray) and oxidized sResA/sBdbD (0.5 μm in each) are shown for reference, as indicated.
Concluding Remarks
The requirement for disulfide bonds in many proteins located on the outside of the cytoplasmic membrane has led to the evolution of a range of thiol-disulfide exchange systems in many bacteria (15). Although most of these involve proteins closely related to the DsbA/DsbB system of E. coli, there are significant variations. M. tuberculosis, for example, utilizes a protein, DsbE, more closely related to thioredoxin than DsbA to generate extra-cytoplasmic disulfide bonds, and S. aureus does not contain a DsbB homologue and therefore must re-oxidize its DsbA protein by another pathway. Members of the genus Bacillus, with few exceptions, contain BdbD/BdbC, which are homologues of DsbA/DsbB. Here our studies of BdbD from B. subtilis have revealed further evidence of variation, in that the protein possesses an intrinsic metal site not present in other DsbA-like proteins. Upon isolation, the site is fully occupied by a Ca2+ ion, which commonly plays a structural role in proteins. In this case, the function of the site appears to be more subtle as removal of the metal does not result in unfolding, or indeed any major structural changes. It does have an effect on conformational stability, however, and, intriguingly, this is specific to the reduced state of the protein, such that presence of Ca2+ at the site significantly affects the redox properties of the protein, despite being some 14 Å away from the enzyme active site. Electrostatics may be a significant contributor to the observed effect. In simple terms, binding of a positively charged calcium ion would be expected to stabilize the reduced form of the active site, in which Cys69 is negatively charged at physiological pH. On the other hand, no stabilization would be expected to result from Ca2+ binding to the neutral oxidized (disulfide-bonded) state. This provides a qualitative explanation for the selective stabilization by Ca2+ of the reduced state of BdbD, which would be predicted to result in an increase in reduction potential, as observed.
Some TDORs, such as human glutaredoxin 2, are iron-sulfur cluster-containing proteins (70, 71). However, in such cases, the cluster cofactor is coordinated by at least one of the active site cysteines, and therefore TDOR activity and cluster coordination are mutually exclusive properties. To the best of our knowledge, B. subtilis BdbD is the first example of a TDOR protein that contains a metal cofactor remote from the CXXC active site, and it represents an important example in which a non-redox active metal affects the redox properties of a protein. This is somewhat reminiscent of peroxidase enzymes in which Ca2+ has an important effect on the properties of the active site heme (72). For BdbD, the effect of Ca2+ is to increase the reduction potential, and the magnitude of the effect is of the same order as that observed for XX dipeptide substitutions at a TDOR CXXC active site (38). This leads us to propose that the role of the metal site is to provide a boost to the oxidizing power of BdbD. A further possibility that remains to be explored is that modulation of the active site reduction potential is important for the re-oxidation of the BdbD active site by its integral membrane partner BdbC.
Supplementary Material
Acknowledgments
We thank University of East Anglia and the Wolfson Foundation for funding the UEA 800 MHz NMR facility. We thank Ingrid Stål for technical assistance; the beam line staff at the European Synchrotron Radiation Facility for assistance in x-ray data collection; Dr. Mark Banfield of the John Innes Centre for collecting x-ray data sets for the EDTA-treated BdbD crystals; Dr. Gaye White for assistance with ICP-AE measurements; and Dr. Arthur Oubrie for useful discussions and help with initial crystallization experiments.
This work was supported by Grant 076017/Z/04/Z from the Wellcome Trust and by Grant 621-2007-6094 from the Swedish Research Council.
The atomic coordinates and structure factors (codes 3EU3, 3EU4, 3GH9, and 3GHA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, additional references, Figs. S1 and S2, and Table S1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, additional references, Figs. S1 and S2, and Table S1.
- TDOR
- thiol:disulfide oxidoreductase
- badan
- 6-bromoacetyl-2-dimethylaminonaphthalene
- Ches
- N-cyclohexyl-2-aminoethanesulfonic acid
- DTT
- dithiothreitol
- ICP-AE
- inductively coupled plasma-atomic emission
- Mes
- 2-(N-morpholino)ethanesulfonate
- MALDI-TOF
- Matrix-assisted laser desorption/ionization-time of flight
- Mops
- 3-morpholinopropanesulfonate
- PEG
- polyethylene glycol
- HSQC
- heteronuclear single quantum coherence
- NHE
- normal hydrogen electrode.
REFERENCES
- 1.Dailey F. E., Berg H. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90,1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamanaka H., Kameyama M., Baba T., Fujii Y., Okamoto K. (1994) J. Bacteriol. 176,2906–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J., Webb H., Hirst T. R. (1992) Mol. Microbiol. 6,1949–1958 [DOI] [PubMed] [Google Scholar]
- 4.Heras B., Shouldice S. R., Totsika M., Scanlon M. J., Schembri M. A., Martin J. L. (2009) Nat. Rev. Microbiol. 7,215–225 [DOI] [PubMed] [Google Scholar]
- 5.Aslund F., Beckwith J. (1999) J. Bacteriol. 181,1375–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin J. L. (1995) Structure 3,245–250 [DOI] [PubMed] [Google Scholar]
- 7.Bardwell J. C., McGovern K., Beckwith J. (1991) Cell 67,581–589 [DOI] [PubMed] [Google Scholar]
- 8.Martin J. L., Bardwell J. C., Kuriyan J. (1993) Nature 365,464–468 [DOI] [PubMed] [Google Scholar]
- 9.Wunderlich M., Glockshuber R. (1993) Protein Sci. 2,717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zapun A., Bardwell J. C., Creighton T. E. (1993) Biochemistry 32,5083–5092 [DOI] [PubMed] [Google Scholar]
- 11.Nelson J. W., Creighton T. E. (1994) Biochemistry 33,5974–5983 [DOI] [PubMed] [Google Scholar]
- 12.Bardwell J. C., Lee J. O., Jander G., Martin N., Belin D., Beckwith J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90,1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bader M., Muse W., Ballou D. P., Gassner C., Bardwell J. C. (1999) Cell 98,217–227 [DOI] [PubMed] [Google Scholar]
- 14.Kadokura H., Bader M., Tian H., Bardwell J. C., Beckwith J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97,10884–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutton R. J., Boyd D., Berkmen M., Beckwith J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,11933–11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heras B., Kurz M., Jarrott R., Shouldice S. R., Frei P., Robin G., Cemazar M., Thöny-Meyer L., Glockshuber R., Martin J. L. (2008) J. Biol. Chem. 283,4261–4271 [DOI] [PubMed] [Google Scholar]
- 17.Goulding C. W., Apostol M. I., Gleiter S., Parseghian A., Bardwell J., Gennaro M., Eisenberg D. (2004) J. Biol. Chem. 279,3516–3524 [DOI] [PubMed] [Google Scholar]
- 18.Kouwen T. R., van der Goot A., Dorenbos R., Winter T., Antelmann H., Plaisier M. C., Quax W. J., van Dijl J. M., Dubois J. Y. (2007) Mol. Microbiol. 64,984–999 [DOI] [PubMed] [Google Scholar]
- 19.Meima R., Eschevins C., Fillinger S., Bolhuis A., Hamoen L. W., Dorenbos R., Quax W. J., van Dijl J. M., Provvedi R., Chen I., Dubnau D., Bron S. (2002) J. Biol. Chem. 277,6994–7001 [DOI] [PubMed] [Google Scholar]
- 20.Erlendsson L. S., Hederstedt L. (2002) J. Bacteriol. 184,1423–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erlendsson L. S., Acheson R. M., Hederstedt L., Le Brun N. E. (2003) J. Biol. Chem. 278,17852–17858 [DOI] [PubMed] [Google Scholar]
- 22.Erlendsson L. S., Möller M., Hederstedt L. (2004) J. Bacteriol. 186,6230–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crow A., Liu Y., Möller M. C., Le Brun N. E., Hederstedt L. (2009) J. Biol. Chem. 284,10056–10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schägger H., von Jagow G. (1987) Anal. Biochem. 166,368–379 [DOI] [PubMed] [Google Scholar]
- 25.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4,2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62,48–57 [DOI] [PubMed] [Google Scholar]
- 27.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62,72–82 [DOI] [PubMed] [Google Scholar]
- 28.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60,2288–2294 [DOI] [PubMed] [Google Scholar]
- 29.Morris R. J., Perrakis A., Lamzin V. S. (2003) Methods Enzymol. 374,229–244 [DOI] [PubMed] [Google Scholar]
- 30.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60,2126–2132 [DOI] [PubMed] [Google Scholar]
- 31.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53,240–255 [DOI] [PubMed] [Google Scholar]
- 32.Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26,283–291 [Google Scholar]
- 33.Kleywegt G. J., Jones T. A. (1996) Acta Crystallogr. D Biol. Crystallogr. 52,826–828 [DOI] [PubMed] [Google Scholar]
- 34.Kleywegt G. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63,94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chau M. H., Nelson J. W. (1991) FEBS Lett. 291,296–298 [DOI] [PubMed] [Google Scholar]
- 36.Lewin A., Crow A., Hodson C. T., Hederstedt L., Le Brun N. E. (2008) Biochem. J. 414,81–91 [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6,277–293 [DOI] [PubMed] [Google Scholar]
- 38.Lewin A., Crow A., Oubrie A., Le Brun N. E. (2006) J. Biol. Chem. 281,35467–35477 [DOI] [PubMed] [Google Scholar]
- 39.Monika E. M., Goldman B. S., Beckman D. L., Kranz R. G. (1997) J. Mol. Biol. 271,679–692 [DOI] [PubMed] [Google Scholar]
- 40.Käll L., Krogh A., Sonnhammer E. L. (2004) J. Mol. Biol. 338,1027–1036 [DOI] [PubMed] [Google Scholar]
- 41.Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., Ito K. (2006) Cell 127,789–801 [DOI] [PubMed] [Google Scholar]
- 42.Guddat L. W., Bardwell J. C., Martin J. L. (1998) Structure 6,757–767 [DOI] [PubMed] [Google Scholar]
- 43.Chivers P. T., Prehoda K. E., Raines R. T. (1997) Biochemistry 36,4061–4066 [DOI] [PubMed] [Google Scholar]
- 44.Grauschopf U., Winther J. R., Korber P., Zander T., Dallinger P., Bardwell J. C. A. (1995) Cell 83,947–955 [DOI] [PubMed] [Google Scholar]
- 45.Moutevelis E., Warwicker J. (2004) Protein Sci. 13,2744–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren G., Stephan D., Xu Z., Zheng Y., Tang D., Harrison R. S., Kurz M., Jarrott R., Shouldice S. R., Hiniker A., Martin J. L., Heras B., Bardwell J. C. (2009) J. Biol. Chem. 284,10150–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stirnimann C. U., Rozhkova A., Grauschopf U., Grütter M. G., Glockshuber R., Capitani G. (2005) Structure 13,985–993 [DOI] [PubMed] [Google Scholar]
- 48.Qin J., Clore G. M., Kennedy W. M., Huth J. R., Gronenborn A. M. (1995) Structure 3,289–297 [DOI] [PubMed] [Google Scholar]
- 49.Qin J., Clore G. M., Kennedy W. P., Kuszewski J., Gronenborn A. M. (1996) Structure 4,613–620 [DOI] [PubMed] [Google Scholar]
- 50.Kadokura H., Tian H., Zander T., Bardwell J. C., Beckwith J. (2004) Science 303,534–537 [DOI] [PubMed] [Google Scholar]
- 51.Chivers P. T., Raines R. T. (1997) Biochemistry 36,15810–15816 [DOI] [PubMed] [Google Scholar]
- 52.Crow A., Acheson R. M., Le Brun N. E., Oubrie A. (2004) J. Biol. Chem. 279,23654–23660 [DOI] [PubMed] [Google Scholar]
- 53.Harding M. M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62,678–682 [DOI] [PubMed] [Google Scholar]
- 54.Dominguez D. C. (2004) Mol. Microbiol. 54,291–297 [DOI] [PubMed] [Google Scholar]
- 55.Norris V., Grant S., Freestone P., Canvin J., Sheikh F. N., Toth I., Trinei M., Modha K., Norman R. I. (1996) J. Bacteriol. 178,3677–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rigden D. J., Jedrzejas M. J., Moroz O. V., Galperin M. Y. (2003) Trends Microbiol. 11,295–297 [DOI] [PubMed] [Google Scholar]
- 57.Rigden D. J., Jedrzejas M. J., Galperin M. Y. (2003) FEMS Microbiol. Lett. 221,103–110 [DOI] [PubMed] [Google Scholar]
- 58.Welch R. A. (2001) Curr. Top. Microbiol. Immunol. 257,85–111 [DOI] [PubMed] [Google Scholar]
- 59.Bauche C., Chenal A., Knapp O., Bodenreider C., Benz R., Chaffotte A., Ladant D. (2006) J. Biol. Chem. 281,16914–16926 [DOI] [PubMed] [Google Scholar]
- 60.Meier R., Drepper T., Svensson V., Jaeger K. E., Baumann U. (2007) J. Biol. Chem. 282,31477–31483 [DOI] [PubMed] [Google Scholar]
- 61.Mitchell E., Houles C., Sudakevitz D., Wimmerova M., Gautier C., Pérez S., Wu A. M., Gilboa-Garber N., Imberty A. (2002) Nat. Struct. Biol. 9,918–921 [DOI] [PubMed] [Google Scholar]
- 62.Wilson J. J., Matsushita O., Okabe A., Sakon J. (2003) EMBO J. 22,1743–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fülöp V., Ridout C. J., Greenwood C., Hajdu J. (1995) Structure 3,1225–1233 [DOI] [PubMed] [Google Scholar]
- 64.Piotukh K., Kosslick D., Zimmermann J., Krause E., Freund C. (2007) Free Radic. Biol. Med. 43,1263–1270 [DOI] [PubMed] [Google Scholar]
- 65.Huber-Wunderlich M., Glockshuber R. (1998) Fold. Des. 3,161–171 [DOI] [PubMed] [Google Scholar]
- 66.Aslund F., Berndt K. D., Holmgren A. (1997) J. Biol. Chem. 272,30780–30786 [DOI] [PubMed] [Google Scholar]
- 67.Zapun A., Missiakas D., Raina S., Creighton T. E. (1995) Biochemistry 34,5075–5089 [DOI] [PubMed] [Google Scholar]
- 68.Siedler F., Rudolph-Böhner S., Doi M., Musiol H. J., Moroder L. (1993) Biochemistry 32,7488–7495 [DOI] [PubMed] [Google Scholar]
- 69.Charbonnier J. B., Belin P., Moutiez M., Stura E. A., Quéméneur E. (1999) Protein Sci. 8,96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lillig C. H., Berndt C., Holmgren A. (2008) Biochim. Biophys. Acta 1780,1304–1317 [DOI] [PubMed] [Google Scholar]
- 71.Lillig C. H., Berndt C., Vergnolle O., Lönn M. E., Hudemann C., Bill E., Holmgren A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102,8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettigrew G. W., Echalier A., Pauleta S. R. (2006) J. Inorg. Biochem. 100,551–567 [DOI] [PubMed] [Google Scholar]
- 73.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 74.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22,4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nicholas K. B., Nicholas H. B., Jr., Deerfield D. W., 2nd (1997) EMBNEW. News 4,14 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.