Abstract
ErbB2 and ErbB3, members of the EGF receptor/ErbB family, form a heterodimer upon binding of a ligand, inducing the activation of Rac small G protein and Akt protein kinase for cell movement and survival, respectively. The enhanced ErbB3/ErbB2 signaling causes tumorigenesis, invasion, and metastasis. We found here that the ErbB3/ErbB2 signaling is regulated by immunoglobulin-like Necl-2, which is down-regulated in various cancer cells and serves as a tumor suppressor. The extracellular region of ErbB3, but not ErbB2, interacted in cis with that of Necl-2. This interaction reduced the ligand-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 and inhibited the consequent ErbB3-mediated activation of Rac and Akt, resulting in the inhibition of cancer cell movement and survival. These inhibitory effects of Necl-2 were mediated by the protein-tyrosine phosphatase PTPN13 which interacted with the cytoplasmic tail of Necl-2. We describe here this novel mechanism for silencing of the ErbB3/ErbB2 signaling by Necl-2.
ErbB2 and ErbB3 are members of the EGF receptor/ErbB family, which has ErbB1 and ErbB4 as additional members (1). ErbB2 and ErbB3 are also known as HER2/Neu and HER3, respectively. No ligands binding directly to ErbB2 have been identified yet, whereas heregulin (HRG)3-α and -β, also known as neuregulin-1 and -2, respectively, directly bind to ErbB3. ErbB2 and ErbB3 have kinase domains in their cytoplasmic tails, but that of ErbB3 lacks kinase activity. Therefore, the homodimer of ErbB3 formed by binding of HRG does not transduce any intracellular signaling. By contrast, ErbB2 heterophilically interacts in cis with HRG-occupied ErbB3 and phosphorylates nine tyrosine residues of ErbB3, causing recruitment and activation of the p85 subunit of phosphoinositide 3-kinase (PI3K) and the subsequent activation of Rac small G protein and Akt protein kinase (2). The activation of Rac enhances cell movement and that of Akt prevents cell apoptosis (3).
ErbB2 serves as an oncogenic protein (4), and amplification of the ErbB2 gene is observed in many types of cancers. For instance, it is amplified in ∼3% of lung cancers, ∼30% of breast cancers, ∼20% of gastric cancers, and ∼60% of ovarian cancers (5). Moreover, mutation of the ErbB2 gene is found in many types of cancers, namely, ∼10% of lung cancers, ∼4% of breast cancers, ∼5% of gastric cancers, and ∼3% of colorectal cancers (6). This gene amplification or mutation causes enhanced signaling for cell movement and survival, eventually resulting in tumorigenesis, invasiveness, and metastasis. On the basis of these properties of ErbB2, it has been recognized as a good target for cancer therapy; indeed, ErbB2-targeting drugs have already been developed and used clinically (7, 8). However, it remains unknown whether ErbB2 is involved in oncogenesis in cancers in which its gene is not amplified or mutated. In addition, it was recently reported that overexpression of ErbB3 is also involved in tumor malignancy (9), but it remains unknown how ErbB3 serves as an oncogenic protein in cancers in which it is not overexpressed.
The nectin-like molecule (Necl) family consists of five members, Necl-1, Necl-2, Necl-3, Necl-4, and Necl-5, and comprises a superfamily with the nectin family, which consists of four members, nectin-1, nectin-2, nectin-3, and nectin-4 (10). All members of this superfamily have similar domain structures: they have one extracellular region with three Ig-like loops, one transmembrane segment, and one cytoplasmic tail. We recently found that the extracellular region of Necl-5 directly interacts in cis with that of the platelet-derived growth factor (PDGF) receptor and that this interaction enhances the PDGF-induced cell proliferation and movement (11–14). Necl-5 is up-regulated in many types of cancer cells and causes at least partly enhanced movement and proliferation of cancer cells (11, 12). These earlier findings prompted us to study the potential interaction of other Necls with other growth factor receptors. Consequently, we found here that the extracellular region of Necl-2 directly interacts in cis with that of ErbB3, but not ErbB2, and reduces the HRG-induced signaling pathways of the ErbB3/ErbB2 heterodimer for cell movement and survival.
Necl-2 is known by many names: IgSF4a, RA175, SgIGSF, TSLC1, and SynCAM1 (15–19). Necl-2 was directly reported in GenBankTM in 1998; IgSF4a was identified as a candidate for a tumor suppressor gene in the loss of heterozygosity region of chromosome 11q23.2 (16); RA175 was identified as a gene highly expressed during the neuronal differentiation of embryonic carcinoma cells (19); SgIGSF was identified as a gene expressed in spermatogenic cells during the early stages of spermatogenesis (18); TSLC1 was identified as a tumor suppressor in human non-small cell lung cancer (17); and SynCAM1 was identified as a brain-specific synaptic adhesion molecule (15). In this study, we use the name “Necl-2,” because it was first reported.
Necl-2 shows Ca2+-independent homophilic cell-cell adhesion activity and Ca2+-independent heterophilic cell-cell adhesion activity with other members of the nectin and Necl families, Necl-1 and nectin-3, and another Ig-like molecule with two Ig-like loops, CRTAM (20–22). These cell-cell adhesion activities are mediated by their extracellular regions. Necl-2 is associated with many peripheral membrane proteins through its cytoplasmic tail. The juxtamembrane region of the cytoplasmic tail contains a band 4.1-binding motif and binds the tumor suppressor DAL-1, a band 4.1 family member, which connects Necl-2 to the actin cytoskeleton (23). In addition, the cytoplasmic tail contains a PDZ domain-binding motif in its C-terminal region and binds Pals2, Dlg3/MPP3, and CASK, which are MAGUK subfamily members having an L27 domain (15, 20, 24, 25). However, the exact roles of the binding of these molecules to Necl-2 remain unknown.
Necl-2 is widely expressed in various tissues and organs, and abundantly expressed in epithelial cells (20, 26). Its expression is down-regulated in many types of cancer cells owing to hypermethylation of the Necl-2 gene promoter and/or loss of heterozygosity of 11q23.2 (26). Its expression is also undetectable in fibroblasts, such as NIH3T3, Swiss3T3, and L cells (20). Necl-2 has been shown to be a tumor suppressor in human non-small cell lung cancer (17), but it remains unknown how it fulfills this role. The relationship between Necl-2 and the ErbB family remains unknown, either. In addition, the heterophilic interaction of Necl-2 with CRTAM enhances the cytotoxicity of NK cells and the secretion of γ-interferon from CD8+ T cells to attack the Necl-2-expressing cells (22, 27). Studies using Necl-2-deficient mice have revealed that Necl-2 in Sertoli cells is an important cell adhesion molecule for Sertoli-spermatid junctions during spermatogenesis (28–30). In the seminiferous tubules of Necl-2-deficient mice, round and elongating spermatids with a distorted shape are generated owing to failure of contact with Sertoli cells, resulting in male-specific infertility. In the present study, we focused on the role of Necl-2 as a tumor suppressor and clarified its mode of action.
EXPERIMENTAL PROCEDURES
Molecular Cloning of Mouse Necl-2, Necl-3, and Necl-4 cDNAs
To obtain the Necl-2, Necl-3, and Necl-4 cDNAs corresponding to GenBankTM/EMBL/DDBJ accession numbers AF434663, NM_178721, AY059394, respectively, we performed reverse transcription-PCR cloning. Reverse transcription-PCR from C57BL/6 mouse liver (for Necl-2) or brain (for Necl-3 and Necl-4) total RNA was performed using the Isogen RNA extraction kit (Nippon Gene, Tokyo, Japan), Ready-To-Go You-Prime FirstStrand Beads and pd(N)6 (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK), pyrobest DNA polymerase (TakaraBio, Shiga, Japan), and the specific primers for mouse Necl-2, Necl-3, and Necl-4. The reverse transcription-PCR products were cloned using a Zero Blunt TOPO PCR cloning kit (Invitrogen). DNA sequencing was performed by the dideoxy nucleotide termination method using a DNA sequencer (ABI Prism 3100 Genetic Analyzer, PE Biosystems, Foster City, CA).
Constructions
FLAG-tagged Necl-2, Necl-3, and Necl-4 expression vectors were constructed in pCAGI-puro. Various constructs of Necl-2, Necl-3, and Necl-4 contained the following aa: pCAGI-Puro-FLAG-Necl-2, aa 43–445 (deleting the signal peptide); pCAGI-Puro-FLAG-Necl-3, aa 25–404 (deleting the signal peptide); pCAGI-Puro-FLAG-Necl-4, aa 26–388 (deleting the signal peptide). The plasmids pCAGI-Puro-FLAG-Necl-2-ΔCP (deleting the cytoplasmic tail, aa 404–445), pCAGI-Puro-FLAG-Necl-2-ΔEC (deleting the extracellular region, aa 1–362), pCAGI-Puro-FLAG-Necl-2-ΔFERM (deleting the FERM domain-binding motif, aa 401–413), and pCAGI-Puro-FLAG-Necl-2-ΔC (deleting the PDZ domain-binding motif, aa 442–445) were constructed by PCR using pCAGI-Puro-FLAG-Necl-2 as template and PrimeSTAR HS DNA polymerase (TakaraBio). Expression vectors were kindly provided as follows: human ErbB2 (pME18s-c-erbB2) was from Dr. T. Yamamoto (University of Tokyo, Tokyo, Japan), and human ErbB3 (pRc/CMV-ErbB3) was from Dr. S. Higashiyama (Ehime University, Ehime, Japan). FLAG-tagged Necl-1 expression vector (pCAGIPuro-FLAG-Necl-1) was prepared as described (20). FLAG-tagged Necl-5 expression vector (pCAGIPuro-FLAG-Necl-5) was prepared as described (31).
Abs and Reagents
A rabbit anti-Necl-2 polyclonal antibody (pAb) was raised against the cytoplasmic tail of Necl-2 (aa 372–417). The following rabbit pAbs were purchased from commercial sources: anti-ErbB2, anti-ErbB3, and anti-PTPN13 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-FLAG (for Western blotting, Sigma), anti-ErbB4, anti-phospho-ErbB3 Y1289, anti-Akt, and anti-phospho-Akt T308 (Cell Signaling, Danvers, MA). A rabbit anti-ErbB2 monoclonal antibody (mAb) for immunoprecipitation was purchased from Epitomics (Burlingame, CA). The following mouse mAbs were purchased from commercial sources: anti-FLAG M2 (for immunoprecipitation, Sigma), anti-Rac1, and anti-ErbB1 (BD Biosciences, Franklin Lakes, NJ). Horseradish peroxidase-conjugated secondary Abs and Protein G-Sepharose 4 Fast Flow were purchased from Amersham Biosciences. Fatty acid-free BSA, trypsin inhibitor, phosphatase inhibitor mixture I, and HRG-β were purchased from Sigma. The following transfection plasmids were purchased from commercial sources: pAcGFP1-Mem (Clontech, Mountain View, CA) and pmaxGFP (Amaxa Biosystems, Cologne, Germany).
Cell Culture and Transfection
A549 cells were purchased from the American Type Culture Collection (Rockville, MD). A549 and HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. For transient expression of various constructs in A549 cells, the Nucleofector system (Amaxa Biosystems) was used, according to the manufacturer's protocol. For transient expression of various constructs in HEK293 cells, Lipofectamine 2000 reagent was used, according to the manufacturer's protocol. Caco-2 cells were purchased from the European Collection of Cell Cultures (Porton Down, UK). Caco-2 cells were maintained in DMEM supplemented with 10% fetal calf serum and 0.1 mm non-essential amino acids.
Co-immunoprecipitation Assay
HEK293 cells were co-transfected with various combinations of plasmids, cultured for 48 h, detached with 0.05% trypsin and 0.53 mm EDTA, and treated with a trypsin inhibitor. Cells were cultured in suspension with DMEM containing 0.5% fatty acid-free BSA for 30 min, collected by centrifugation, washed with Wash buffer (20 mm Tris-HCl at pH 8.0, 150 mm NaCl, and 1 mm Na3VO4), and lysed with Lysis buffer (20 mm Tris-HCl at pH 8.0, 150 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, 10% glycerol, 1% Nonidet P-40, 10 mm NaF, 1 mm Na3VO4, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 10 μm (p-amidinophenyl)methanesulfonyl fluoride, and phosphatase inhibitor mixture I). The lysates were rotated for 30 min and subjected to centrifugation at 12,000 × g for 15 min. The supernatant was pre-cleared with protein G-Sepharose 4 Fast Flow beads at 4 °C for 1 h. The anti-FLAG M2 mAb was preincubated with protein G-Sepharose beads at 4 °C for 2 h. The cell extracts were incubated with the anti-FLAG M2 mAb-conjugated Protein G-Sepharose beads at 4 °C for 4 h. After the beads were extensively washed with the lysis buffer, bound proteins were eluted by boiling the beads in an SDS sample buffer (60 mm Tris-HCl, pH 6.7, 3% SDS, 2% 2-mercaptoethanol, and 5% glycerol) for 5 min (32), and subjected to SDS-polyacrylamide gel electrophoresis, followed by Western blotting.
Small Interference RNA Experiments
For knockdown using a small interference RNA (siRNA) method, stealth RNAi duplexes against human Necl-2 and human PTPN13, and a stealth RNAi negative control duplex were purchased from Invitrogen. For knockdown of Necl-2, we used stealth RNAi duplexes of 5′-CCACAGGAAAGUUACACCACCAUCA-3′ (#61), 5′-GGUGAGGAGAUUGAAGUCAACUGCA-3′ (#62), and 5′-GGACGCGCUUGAGUUAACAUGUGAA-3′ (#63), and essentially the same results were obtained from the experiments using these three RNAi duplexes. For knockdown of PTPN13, we used stealth RNAi duplexes of 5′-UCACAUUUCUGAACCAACUAGACAA-3′ (#38), 5′-GACUCUGCCCAAAGAAUCUUAUAUA-3′ (#39), and 5′-GAAUGGCUGUGAAGAAUAUUGUGAA-3′ (#40), and essentially the same results were obtained from the experiments using these three RNAi duplexes. We showed the results by using #62 against Necl-2 and #40 against PTPN13 in this study. Transfection of cells with siRNA was performed using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's protocol.
Assay for Activation of ErbB3 and Akt
A549 cells transfected with various expression vector and/or stealth RNAi were plated at a density of 3 × 104 cells per square centimeter on dishes and cultured for 1 day. Caco-2 cells transfected with various stealth RNAi were plated at a density of 1 × 104 cells per square centimeter on dishes covered by a thin coat of Matrigel, cultured for 2 days. These cells were starved of serum with DMEM containing 0.5% fatty acid-free BSA for 20 h and then stimulated by DMEM containing 0.5% fatty acid-free BSA and 10 ng/ml HRG. Cells were washed with ice-cold phosphate-buffered saline several times and lysed with RIPA buffer (20 mm Tris-HCl at pH 7.5, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 10% glycerol, 137 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, 50 mm NaF, 1 mm Na3VO4, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 10 μm (p-amidinophenyl)methanesulfonyl fluoride, and phosphatase inhibitor mixture I). The lysates were then boiled in an SDS sample buffer for 5 min and subjected to SDS-PAGE, followed by Western blotting.
Assay for Activation of Rac
The assay for Rac activation was performed using glutathione-S-transferase-tagged p21-activated protein kinase-Cdc42/Rac interactive binding domain as described (33). Briefly, cells were prepared as described above and starved of serum with DMEM containing 0.5% fatty acid-free BSA for 20 h. After serum starvation, cells were stimulated by DMEM containing 0.5% fatty acid-free BSA and 10 ng/ml HRG, and then subjected to the assay for Rac activation.
Boyden Chamber Assay
Fluorescence blocking polyethylene terephthalate membrane cell culture inserts (8.0-μm pores, BD Falcon, Franklin Lakes, NJ) were coated with serum for 30 min. A549 cells co-transfected with pmaxGFP and empty vector or FLAG-Necl-2 and/or ErbB3 expression vector were detached with 0.05% trypsin and 0.53 mm EDTA, and then treated with a trypsin inhibitor. The cells were then re-suspended in DMEM containing 0.5% fatty acid-free BSA and seeded at a density of 5 × 104 cells per insert. The cells were incubated at 37 °C for 18 h in the presence or absence of 100 ng/ml HRG. HRG was added only to the bottom well to generate a concentration gradient. After incubation, the cells were fixed using 4% paraformaldehyde/phosphate-buffered saline. Transfected cells were identified based on the expression of GFP. The number of migrated GFP-positive cells in ten randomly chosen fields per filter was counted by fluorescence microscopic examination.
DNA Synthesis Assay
The DNA synthesis assay was performed by using BrdUrd labeling and detection kit I (Roche Diagnostics, Basel, Switzerland). A549 cells co-transfected with pmaxGFP and empty vector or FLAG-Necl-2 expression vector (2 × 104 cells) were plated on coverslips in 24-well culture plates. Cells were starved of serum for 24 h and stimulated by 10 ng/ml HRG in the presence of BrdUrd. After 14 h of incubation, cells were fixed and BrdUrd-positive cells were detected according to the manufacturer's protocol. DNA was visualized with 4′,6-diamidino-2-phenylindole. Transfected cells were identified based on the expression of GFP. The samples were analyzed by fluorescence microscopic examination.
TUNEL Assay
Apoptotic cells were detected by TUNEL assay using the DeadEnd Colorimetric TUNEL System (Promega, Madison, WI) and streptavidin-Cy3 (Jackson ImmunoResearch, West Grove, PA). A549 cells co-transfected with pmaxGFP and empty vector or FLAG-Necl-2 expression vector (2 × 104 cells) were plated on coverslips in 24-well culture plates and cultured with DMEM containing 0.5% fatty acid-free BSA in the presence or absence of 10 ng/ml HRG. After 3 days, a TUNEL assay was carried out according to the manufacturer's protocol, and apoptotic cells were detected by streptavidin-Cy3. DNA was visualized with 4′,6-diamidino-2-phenylindole. Transfected cells were identified based on the expression of GFP. The samples were analyzed by fluorescence microscopic examination.
Anoikis Assay
To prevent cell adhesion, plates were coated with a solution of polyhydroxyethylmethacrylate (Sigma) dissolved at 25 mg/ml in ethanol. To coat plates, 100 μl of polyhydroxyethylmethacrylate solution was added to each plate per square centimeter, and the plates were kept at 37 °C for 2 days until the solvent had completely evaporated. The coated surfaces were sterilized by UV irradiation for 16 h and gently washed twice with phosphate-buffered saline before use. A549 cells co-transfected with pmaxGFP and empty vector or FLAG-Necl-2 and/or ErbB3 expression vector were cultured in suspension using a polyhydroxyethylmethacrylate-coated dish with DMEM containing 10 ng/ml HRG and 0.5% fatty acid-free BSA for 48 h. Cells were collected and attached to Matsunami adhesive silane-coated slide glass (Matsunami Glass, Osaka, Japan) using a Cytospin 4 Cytocentrifuge (Thermo Scientific, Waltham, MA), and apoptotic cells were detected by TUNEL assay using the DeadEnd Colorimetric TUNEL System and streptavidin-Cy3. DNA was visualized with 4′,6-diamidino-2-phenylindole. Transfected cells were identified based on the expression of GFP. The samples were analyzed by fluorescence microscopic examination.
RESULTS
Interaction of ErbB3 with Necl-2
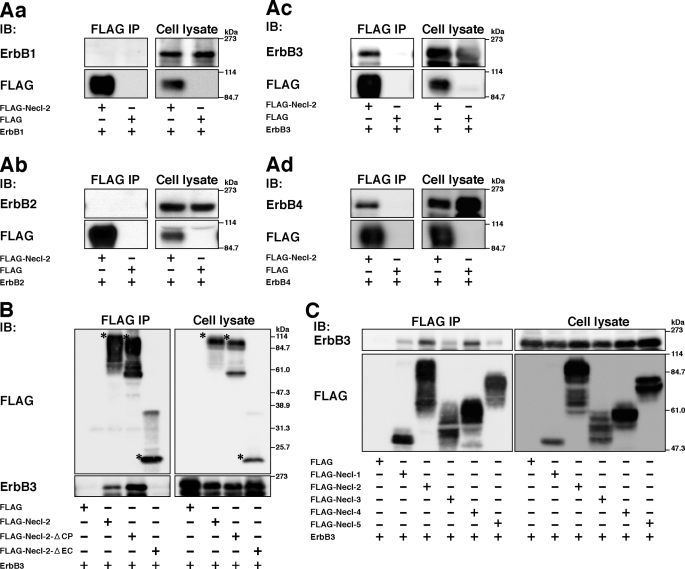
We first examined whether Necl-2 physically interacts with members of the ErbB family. FLAG-tagged Necl-2 (FLAG-Necl-2) or FLAG alone and each member of the ErbB family were co-expressed in HEK293 cells, and the cells were cultured in suspension. The reason why a suspension culture was used was that it enabled the detection of a possible cis-interaction between Necl-2 and members of the ErbB family on the plasma membrane. When FLAG-Necl-2 was immunoprecipitated from each cell lysate using the anti-FLAG mAb, ErbB3 and ErbB4 were co-immunoprecipitated with FLAG-Necl-2, but ErbB1 or ErbB2 was not (Fig. 1, Aa–Ad). When FLAG alone was immunoprecipitated from each cell lysate using the anti-FLAG mAb, no ErbB family members were co-immunoprecipitated (Fig. 1, Aa–Ad). A role for ErbB3 in tumorigenesis has been reported, but it is not known whether ErbB4 is involved in tumorigenesis (34). Therefore, in the present study we focused on ErbB3.
FIGURE 1.
Interaction of ErbB family members with Necl-2. Co-immunoprecipitation assay using HEK293 cells. Cells were co-transfected with various combinations of indicated plasmids and cultured in suspension. Each FLAG-tagged Necl family member was immunoprecipitated using the anti-FLAG mAb, and samples were subjected to Western blotting using the indicated Abs. A, assay for co-immunoprecipitation of ErbB family members with Necl-2. Aa, FLAG-Necl-2 and ErbB1; Ab, FLAG-Necl-2 and ErbB2; Ac, FLAG-Necl-2 and ErbB3; Ad, FLAG-Necl-2 and ErbB4. B, interaction of Necl-2 with ErbB3 through its extracellular region. Asterisks, full-length or mutants of Necl-2. C, assay for co-immunoprecipitation of ErbB3 with Necl family members. The results shown are representative of three independent experiments.
We next examined whether the extracellular region and/or cytoplasmic tail of Necl-2 are necessary for the interaction with ErbB3. FLAG-Necl-2 of which the cytoplasmic tail or extracellular region was deleted (FLAG-Necl-2-ΔCP or FLAG-Necl-2-ΔEC, respectively), and ErbB3 were co-expressed in HEK293 cells, and a co-immunoprecipitation assay was performed. Before the co-immunoprecipitation assay, we checked the intracellular localization of FLAG-Necl-2 and its mutants by confocal immunofluorescence microscopy. All the signals for FLAG-Necl-2, FLAG-Necl-2-ΔCP, and FLAG-Necl-2-ΔEC, determined by the anti-FLAG mAb, were observed at the cell periphery and their immunofluorescence intensities were at similar levels (supplemental Fig. S1). This result suggests that the similar amounts of these molecules were sorted on the plasma membrane. ErbB3 was co-immunoprecipitated with FLAG-Necl-2-ΔCP, whereas it was not co-immunoprecipitated with FLAG-Necl-2-ΔEC (Fig. 1B). Two bands of ErbB3 shown in this figure might be produced by the glycosylation as previously described (35). The amount of ErbB3, that was co-immunoprecipitated with FLAG-Necl-2-ΔCP, was about four times as much as that co-immunoprecipitated with full-length FLAG-Necl-2. These results indicate that ErbB3 physically interacts in cis with Necl-2 through their extracellular regions and that the cytoplasmic tail of Necl-2 is not essential for this interaction but inhibits the interaction of the extracellular region of ErbB3 with Necl-2.
We then examined whether other members of the Necl family also interact with ErbB3. When FLAG-tagged members of this family were co-expressed with ErbB3 in HEK293 cells, a co-immunoprecipitation assay was performed. ErbB3 was co-immunoprecipitated with FLAG-Necl-4 in addition to FLAG-Necl-2, but not with FLAG-Necl-1, Necl-3, or Necl-5 (Fig. 1C). These results indicate that ErbB3 interacts specifically with both Necl-2 and Necl-4.
No Effect of Necl-2 on the HRG-induced Formation of the ErbB3/ErbB2 Heterodimer
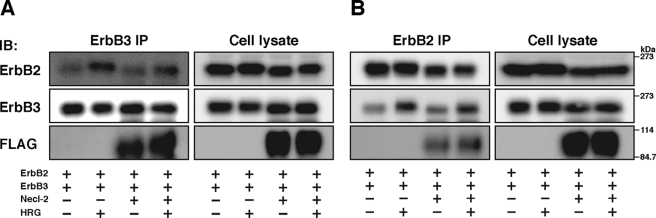
We next examined the effect of Necl-2 on the formation of the ErbB3/ErbB2 heterodimer induced by binding of HRG to ErbB3. ErbB3 and ErbB2 were co-expressed in HEK293 cells, which were incubated in the presence and absence of HRG. When ErbB3 was immunoprecipitated using the anti-ErbB3 pAb, ErbB2 was co-immunoprecipitated with ErbB3 (Fig. 2A). The amount of ErbB2 co-immunoprecipitated with ErbB3 in the presence of HRG was larger than that in the absence of HRG. Similarly, when ErbB2 was immunoprecipitated using the anti-ErbB2 mAb, ErbB3 was co-immunoprecipitated with ErbB2 (Fig. 2B). The amount of ErbB3 co-immunoprecipitated with ErbB2 in the presence of HRG was also larger than that in the absence of HRG. These results are consistent with earlier observations (36). FLAG-Necl-2, ErbB3, and ErbB2 were co-expressed in HEK293 cells and incubated in the presence and absence of HRG. When ErbB3 was immunoprecipitated using the anti-ErbB3 pAb, FLAG-Necl-2 and ErbB2 were co-immunoprecipitated with ErbB3 (Fig. 2A). As before, the amount of ErbB2 co-immunoprecipitated with ErbB3 in the presence of HRG was larger than that in the absence of HRG and did not change compared with that in the absence of Necl-2. The amount of FLAG-Necl-2 co-immunoprecipitated with ErbB3 in the presence of HRG was similar to that co-immunoprecipitated in the absence of HRG. When ErbB2 was immunoprecipitated using the anti-ErbB2 mAb, ErbB3 and FLAG-Necl-2 were co-immunoprecipitated with ErbB2 (Fig. 2B). The amounts of FLAG-Necl-2 and ErbB3 co-immunoprecipitated with ErbB2 in the presence of HRG were larger than those in the absence of HRG and that of ErbB3 did not change compared with that in the absence of Necl-2. These results indicate that Necl-2 does not affect the HRG-induced formation of the ErbB3/ErbB2 heterodimer and that it forms a ternary complex with ErbB3 and ErbB2.
FIGURE 2.
No effect of Necl-2 on the HRG-induced formation of the ErbB3/ErbB2 heterodimer. HEK293 cells were co-transfected with various combinations of the indicated plasmids. ErbB2 and ErbB3 were immunoprecipitated using the anti-ErbB2 mAb and ErbB3 pAb, respectively, and samples were subjected to Western blotting using the indicated Abs. A, immunoprecipitation of ErbB2; B, immunoprecipitation of ErbB3. The results shown are representative of three independent experiments.
Inhibition by Necl-2 of the HRG-induced, ErbB2-catalyzed Tyrosine Phosphorylation of ErbB3 and Its Signaling in A549 Cells
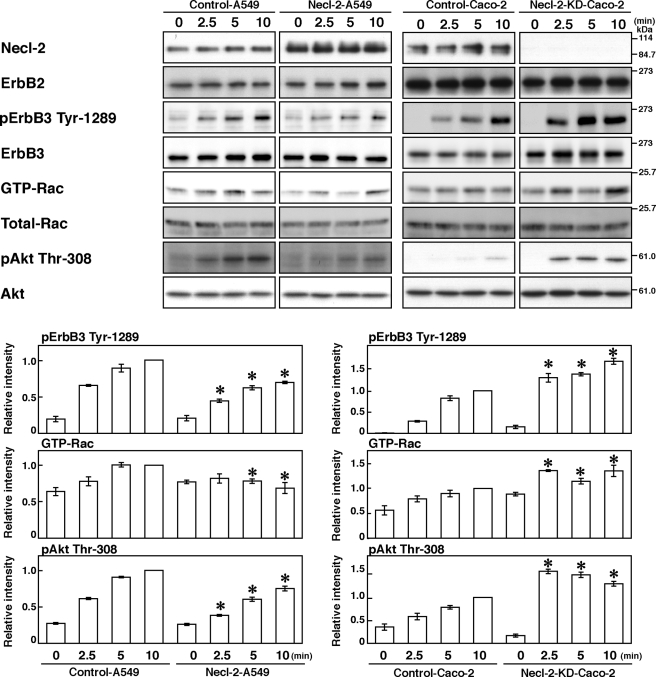
In a human lung cancer cell line, A549, Necl-2 was down-regulated by loss of heterozygosity on chromosome 11q23 where the Necl-2 gene is located (17) (Fig. 3). In this cell line, ErbB2 and ErbB3 were abundantly expressed as easily detected by Western blotting (Fig. 3), whereas the expression of ErbB4 was undetectable (data not shown). We therefore decided to use this cell line to examine whether exogenous expression of Necl-2 affects the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 and subsequent activation of Rac and Akt. A549 cells transiently expressing empty vector (Control-A549 cells) and A549 cells transiently expressing Necl-2 (Necl-2-A549 cells) were starved of serum and stimulated by HRG for various periods of time. The tyrosine 1289 residue of ErbB3 was phosphorylated in a time-dependent manner in response to HRG in both types of cells, but the phosphorylation level was much less in Necl-2-A549 cells than in Control-A549 cells (Fig. 3). Rac and Akt were also activated in time-dependent manners in response to HRG in both types of cells, but the levels of activated Rac and Akt were much lower in Necl-2-A549 cells than in Control-A549 cells (Fig. 3). These results indicate that Necl-2 inhibits the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 and subsequent activation of Rac and Akt.
FIGURE 3.
Inhibition by Necl-2 of the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 and its signaling. A549 cells were transfected with empty vector or FLAG-Necl-2 expression vector (Control A549 and Necl-2-A549 cells, respectively). Caco-2 cells were transfected with negative control stealth RNAi and stealth RNAi against Necl-2 (Control-Caco2 and Necl-2-KD-Caco-2 cells, respectively). Cells were starved of serum for 20 h and stimulated by 10 ng/ml HRG for the indicated periods of time. Samples were subjected to a Rac pulldown assay or Western blotting using the indicated Abs. Bars in the graph represent the relative band intensity of activated ErbB3, Rac, and Akt, normalized for the total amount of ErbB3, Rac, and Akt, respectively, as compared with a value of control cells treated with HRG for 10 min, which is expressed as 1. Error bars indicate the means ± S.E. of three independent experiments. *, p < 0.05 versus control cells.
Enhancement by Necl-2 Knockdown of the HRG-induced, ErbB2-catalyzed Tyrosine Phosphorylation of ErbB3 and Its Signaling in Caco-2 Cells
We further investigated whether Necl-2 knockdown could enhance the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 and its signaling in an epithelial-like human colon carcinoma cell line, Caco-2 cells. The reason why we used this cell line was that it expressed Necl-2, ErbB2, and ErbB3 to high levels enough to be detected by Western blotting. Necl-2 was knocked down in Caco-2 cells by the transient transfection with an siRNA against Necl-2 (Necl-2-KD-Caco-2 cells). As a control, Caco-2 cells were transfected with a negative control of siRNA (Control-Caco-2 cells). In this cell line, the amount of the Necl-2 protein was decreased to <5% of that in Control-Caco-2 cells as estimated by Western blotting (Fig. 3). Control-Caco-2 and Necl-2-KD-Caco-2 cells were starved of serum and stimulated by HRG. The tyrosine 1289 residue of ErbB3 was phosphorylated in a time-dependent manner in response to HRG in both types of cells, but the phosphorylation level was much higher in Necl-2-KD-Caco-2 cells than in Control-Caco-2 cells (Fig. 3). Rac and Akt were also activated in time-dependent manners in both types of cells in response to HRG, but the activation levels were much higher in Necl-2-KD-Caco-2 cells than in Control-Caco-2 cells (Fig. 3). These results provide another line of evidence that Necl-2 inhibits the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 and its signaling.
Inhibition by Necl-2 of Movement and Anchorage-independent Survival of A549 Cells
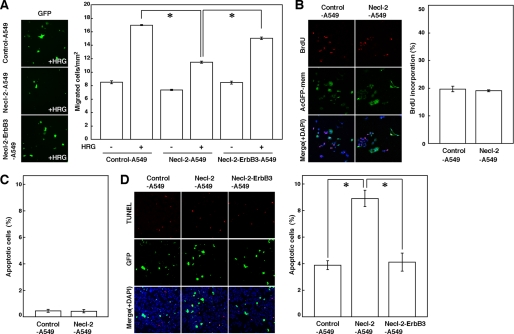
We then examined whether exogenous expression of Necl-2 could suppress cell movement, proliferation, and anchorage-dependent and -independent survival in A549 cells. Control-A549 and Necl-2-A549 cells were cultured on filter inserts with 8-μm pores in the presence or absence of HRG (Fig. 4A). In the absence of HRG, the degree of cell movement was similar between Control-A549 and Necl-2-A549 cells, whereas in the presence of HRG, Control-A549 cells moved more rapidly than Necl-2-A549 cells. When ErbB3 was additively expressed in Necl-2-A549 cells (Necl-2-ErbB3-A549 cells), the inhibitory effect of Necl-2 on cell movement was canceled. The reason for this cancellation was probably that the amount of Necl-2-free ErbB3 was increased again. These results indicate that Necl-2 suppresses the HRG-induced movement of A549 cells by its interaction with ErbB3. For analysis of cell proliferation, BrdUrd incorporation was measured. The level of DNA synthesis was comparable between Control-A549 and Necl-2-A549 cells (Fig. 4B), indicating that Necl-2 does not affect the proliferation of A549 cells, consistent with an earlier observation (37). In the analysis of anchorage-dependent cell survival, hardly any apoptotic cells were observed between Control-A549 and Necl-2-A549 cells (Fig. 4C). However, in the analysis of anchorage-independent cell survival, the number of apoptotic Necl-2-A549 cells was greater than the number of apoptotic Control-A549 cells, consistent with an earlier observation of cell growth in soft agar (38). In Necl-2-ErbB3-A549 cells, the stimulatory effect of Necl-2 on apoptosis was reduced (Fig. 4D). These results indicate that Necl-2 suppresses anchorage-independent, but not anchorage-dependent, cell survival of A549 cells by its interaction with ErbB3.
FIGURE 4.
Inhibition by Necl-2 of movement and anchorage-independent survival of A549 cells. A, measurement of cell movement using a Boyden chamber assay. A549 cells were co-transfected with empty vector and pmaxGFP (Control-A549 cells), FLAG-Necl-2 expression vector and pmaxGFP (Necl-2-A549 cells), or FLAG-Necl-2, ErbB3 expression vector, and pmaxGFP (Necl-2-ErbB3-A549 cells), and were plated on cell culture inserts (8.0-μm pore membrane filters) in the presence or absence of 100 ng/ml HRG in the bottom well for 18 h. The EGFP-positive migrated cells were counted by microscopic examination. Error bars indicate the means ± S.E. of three independent experiments. *, p < 0.001. B, measurement of cell proliferation. A549 cells which were co-transfected either with empty vector and pAcGFP-mem or with FLAG-Necl-2 expression vector and pAcGFP-mem (Control-A549 and Necl-2-A549 cells, respectively) were starved of serum and stimulated by HRG in the presence of BrdUrd. After 14 h of incubation, cells were fixed, and then double stained with the anti-BrdUrd mAb and 4′,6-diamidino-2-phenylindole (DAPI). AcGFP-mem-positive cells were measured. The left panels show representative fields of three independent experiments. Error bars indicate the means ± S.E. of three independent experiments. C, measurement of anchorage-dependent apoptotic cells. A549 cells which were co-transfected either with empty vector and pmaxGFP or with FLAG-Necl-2 expression vector and pmaxGFP (Control-A549 and Necl-2-A549 cells, respectively) were starved of serum for 48 h and subjected to a TUNEL assay. Error bars indicate the means ± S.E. of three independent experiments. D, measurement of anchorage-independent apoptotic cells. A549 cells that were co-transfected with empty vector and pmaxGFP (Control-A549 cells), FLAG-Necl-2 expression vector and pmaxGFP (Necl-2-A549 cells), or FLAG-Necl-2, ErbB3 expression vector, and pmaxGFP (Necl-2-ErbB3-A549 cells) were cultured in suspension in the presence of 0.5% fatty acid-free BSA and 10 ng/ml HRG. After 48 h of incubation, the cells were subjected to a TUNEL assay. Error bars indicate the means ± S.E. of three independent experiments. *, p < 0.02.
Involvement of the Cytoplasmic Tail of Necl-2 in Its Inhibitory Effect
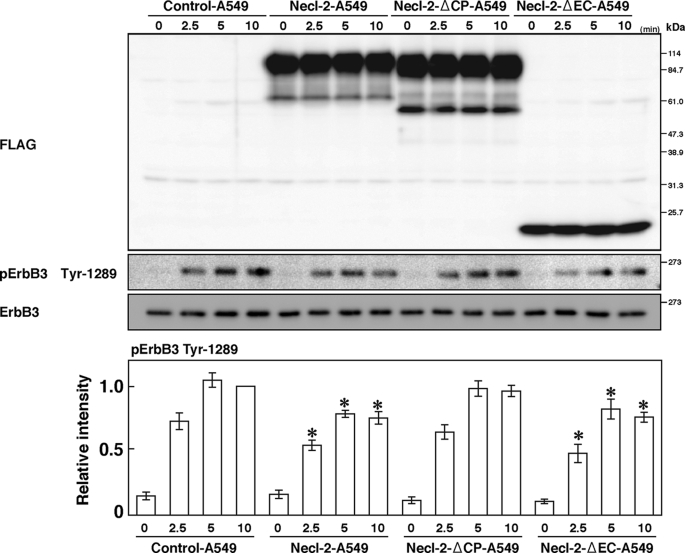
We investigated how Necl-2 inhibits the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3. Expression of Necl-2-ΔEC, but not Necl-2-ΔCP, in A549 cells inhibited the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 (Fig. 5). This result indicates that the cytoplasmic tail of Necl-2, but not the extracellular region, is necessary to exert its inhibitory effect, even though the extracellular region, but not the cytoplasmic tail, is necessary for the interaction with ErbB3.
FIGURE 5.
Involvement of the cytoplasmic tail of Necl-2 in its inhibitory effect. A549 cells were transfected with empty vector, FLAG-Necl-2, FLAG-Necl-2-ΔCP, or FLAG-Necl-2-ΔEC expression vector (Control-A549, Necl-2-A549, Necl-2-ΔCP-A549, or Necl-2-ΔEC-A549 cells, respectively). Cells were starved of serum for 20 h and stimulated by 10 ng/ml HRG for the indicated periods of time. Samples were subjected to Western blotting using the indicated Abs. Bars in the graph represent the relative band intensity of phosphorylated ErbB3, assessed as described in Fig. 3. Error bars indicate the means ± S.E. of three independent experiments. *, p < 0.05 versus Control-A549 cells. The results shown are representative of three independent experiments.
Involvement of PTPN13 in the Inhibitory Effect of Necl-2
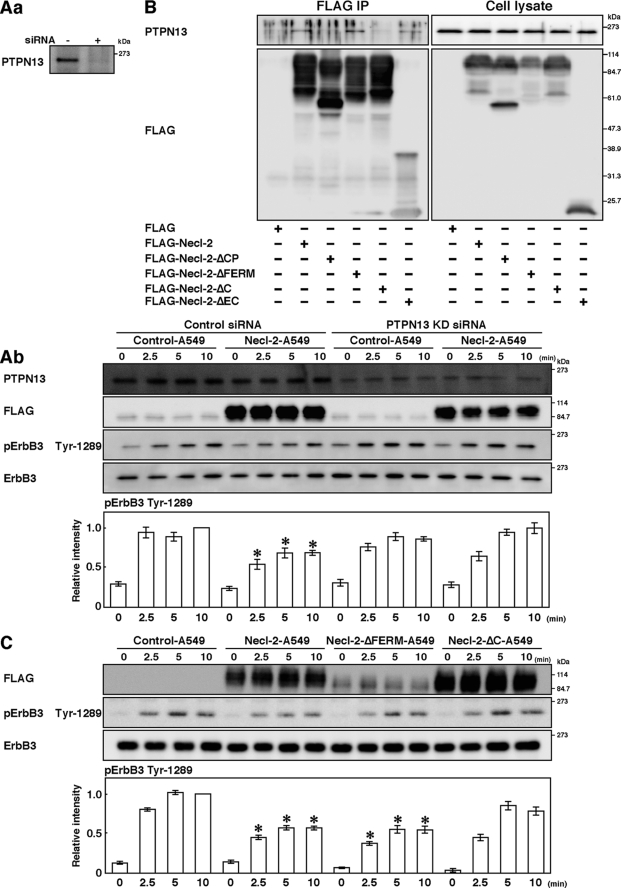
It was reported that the protein-tyrosine phosphatase PTPN13 (also called PTPL1, FAP-1, PTP-BAS, and PTP1E), a known tumor suppressor (39), inhibits the ErbB2 activity by dephosphorylating the signal domain of ErbB2 and plays a role in attenuating the invasiveness and metastasis of ErbB2-overactive tumors (40). PTPN13 is a non-receptor type phosphatase with one FERM domain and five PDZ domains in addition to the catalytic domain (39). We therefore examined whether PTPN13 is responsible for the inhibitory effect of Necl-2 on the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3. Western blotting showed the presence of PTPN13 in A549 cells (Fig. 6, Aa). Knockdown of this protein by siRNA reduced the amount of this protein in A549 cells (Fig. 6, Aa) and prevented the reduction by exogenous expression of Necl-2 of the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 (Fig. 6, Ab). These results indicate that PTPN13 is involved in the inhibitory effect of Necl-2 on the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3.
FIGURE 6.
Involvement of PTPN13 in the inhibitory effect of Necl-2. A, knockdown of PTPN13. A549 cells were transfected with negative control stealth RNAi and stealth RNAi against PTPN13 (Control-A549 and PTPN13-KD-A549 cells, respectively). Aa, confirmation of knockdown of PTPN13. Samples were subjected to Western blotting using the anti-PTPN13 pAb. Ab, assay for the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3. Cells were starved of serum for 20 h and stimulated by 10 ng/ml HRG for the indicated periods of time and lysed with RIPA buffer. Samples were subjected to Western blotting using the indicated Abs. Bars in the graph represent the relative band intensity of phosphorylated ErbB3, assessed as described in Fig. 3. Error bars indicate the means ± S.E. of three independent experiments. *, p < 0.02 versus Control-A549 cells. B, assay for the co-immunoprecipitation of PTPN13 with Necl-2. HEK293 cells were transfected with FLAG-Necl-2, FLAG-Necl-2-ΔCP, FLAG-Necl-2-ΔFERM, FLAG-Necl-2-ΔC, or FLAG-Necl-2-ΔEC. Each FLAG-Necl-2 mutant was immunoprecipitated using the anti-FLAG mAb, and samples were subjected to Western blotting using the indicated Abs. C, assay for the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3 using Necl-2 mutants. A549 cells were transfected with empty vector, FLAG-Necl-2, FLAG-Necl-2-ΔFERM, or FLAG-Necl-2-ΔC expression vector (Control-A549, Necl-2-A549, Necl-2-ΔFERM-A549, or Necl-2-ΔC-A549 cells, respectively). Cells were starved of serum for 20 h and stimulated by 10 ng/ml HRG for the indicated periods of time. Samples were subjected to Western blotting using indicated Abs. Bars in the graph represent the relative band intensity of phosphorylated ErbB3, assessed as described in Fig. 3. Error bars indicate the means ± S.E. of three independent experiments. *, p < 0.01 versus Control-A549 cells. The results shown are representative of three independent experiments.
We then examined whether Necl-2 interacts with PTPN13, because the cytoplasmic tail of Necl-2 has the FERM domain-binding motif at its juxtamembrane region and the PDZ domain-binding motif at its C-terminal region and PTPN13 has one FERM domain and five PDZ domains. FLAG-Necl-2 or FLAG alone was expressed in HEK293 cells. When FLAG-Necl-2 was immunoprecipitated from each cell lysate using the anti-FLAG mAb, endogenous PTPN13 was co-immunoprecipitated with FLAG-Necl-2 (Fig. 6B). When FLAG was immunoprecipitated, endogenous PTPN13 was not co-immunoprecipitated with FLAG. When FLAG-Necl-2-ΔEC, FLAG-Necl-2-ΔCP, FLAG-Necl-2 of which the FERM domain-binding motif was deleted (FLAG-Necl-2-ΔFERM), or FLAG-Necl-2 of which the C-terminal PDZ domain-binding motif was deleted (FLAG-Necl-2-ΔC) was expressed in HEK293 cells and immunoprecipitated using the anti-FLAG mAb, PTPN13 was not co-immunoprecipitated with FLAG-Necl-2-ΔCP or FLAG-Necl-2-ΔC, whereas it was co-immunoprecipitated with FLAG-Necl-2-ΔEC and FLAG-Necl-2-ΔFERM (Fig. 6B). These results indicate that Necl-2 interacts with PTPN13 through the C-terminal PDZ domain-binding motif.
Finally, we examined the effects of expression of Necl-2-ΔFERM and Necl-2-ΔC on the HRG-induced activation of the tyrosine phosphorylation of ErbB3. Expression of Necl-2-ΔFERM, but not Necl-2-ΔC, in A549 cells inhibited the HRG-induced tyrosine phosphorylation of ErbB3 (Fig. 6C). Taken together, these results indicate that Necl-2 interacts with PTPN13, dephosphorylates the ErbB3 phosphorylated by the action of ErbB2, and induces the inactivation of Akt and Rac.
DISCUSSION
We first showed here that ErbB3 and ErbB4, but not ErbB1 or ErbB2, of the ErbB family physically interact with the Ig-like cell adhesion molecule Necl-2. ErbB3 does not interact with other members of the Necl family, except for Necl-4. Phylogenic tree analyses revealed that of the ErbB family members, ErbB3 and ErbB4 have the most homologous amino acid sequences and that, among the Necl family members, Necl-2 and Necl-4 have the most homologous amino acid sequences. The interaction of ErbB4 with Necl-2 and the interaction of ErbB3 with Necl-4 will be studied in detail in the future. The extracellular region of Necl-2, but not the cytoplasmic tail, is essential for the interaction with ErbB3, but the cytoplasmic tail may suppress the interaction of the extracellular region of Necl-2 with presumably the extracellular region of ErbB3, because the amount of ErbB3 that was co-immunoprecipitated with the extracellular region of Necl-2 was about four times as much as that co-immunoprecipitated with full-length Necl-2.
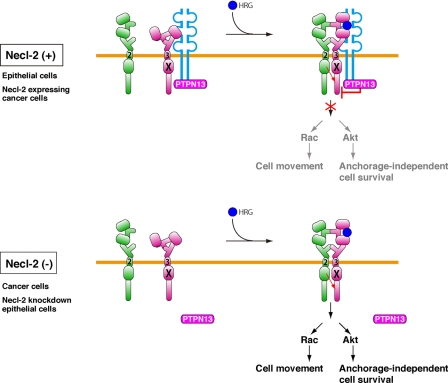
It was shown that, upon binding of HRG to ErbB3, an ErbB3/ErbB2 heterodimer is formed and ErbB2 phosphorylates ErbB3 at nine tyrosine residues (1). Most of these phosphorylated tyrosine residues then recruit the p85 subunit of PI3K, which consequently induces the activation of PI3K. The activation of PI3K induces the activation of Rac and Akt. The activation of Rac enhances cell movement and that of Akt enhances cell survival (3). We found here that the interaction of Necl-2 with ErbB3 does not inhibit the formation of ErbB3/ErbB2 heterodimer, but rather leads to the formation of a heterotrimer. Theoretically, in the absence of HRG, ErbB3 and ErbB2 are present diffusely on the plasma membrane as their respective monomers, homodimers, and/or heterodimers, and these states of ErbB3 and ErbB2 are in equilibrium, depending on their respective affinities. When cells are stimulated by HRG, HRG binds to ErbB3, which shifts this equilibrium to produce more ErbB3/ErbB2 heterodimers. Our present results indicate that, in cells expressing Necl-2, ErbB3 forms a complex with Necl-2 that exists diffusely on the plasma membrane without stimulation by HRG, because Necl-2 interacts with ErbB3 irrespective of the presence or absence of HRG. When cells are stimulated by HRG, HRG binds to ErbB3 of the ErbB3/Necl-2 heterodimer to produce the ErbB3/ErbB2-Necl-2 heterotrimers (Fig. 7).
FIGURE 7.
A model mechanism for the regulation of ErbB3/ErbB2 signaling by Necl-2. See “Discussion” for details.
We showed here that the HRG-induced, ErbB2-catalyzed tyrosine phosphorylation of ErbB3, subsequent signaling, and cell responses, such as cell movement and survival, are inhibited by Necl-2, indicating that, although the HRG-ErbB3/ErbB2 complex is active in signaling, the HRG-ErbB3/ErbB2-Necl-2 complex is inactive in signaling. The extracellular region together with the transmembrane segment of Necl-2 showed no inhibitory effect, whereas the cytoplasmic tail together with the transmembrane segment of Necl-2 showed inhibitory effects. These results, together with the results of the co-immunoprecipitation assay showing that the affinity of the extracellular region together with the transmembrane segment of Necl-2 for ErbB3 is higher than that for full-length molecules, indicate that the cytoplasmic tail is involved in both the affinity of Necl-2 for ErbB3 and the inhibitory effect of Necl-2.
The C-terminal region of Necl-2 has a PDZ domain-binding motif, through which the protein-tyrosine phosphatase PTPN13 binds. The deletion mutant of this PDZ domain-binding motif did not show inhibitory effect and knockdown of PTPN13 abolished the inhibitory effect of Necl-2. On the basis of these observations, we propose here the following regulatory mechanisms for the ErbB3/ErbB2 signaling by Necl-2 (Fig. 7). In cells expressing Necl-2, 1) Necl-2, PTPN13, and ErbB3 form a ternary complex irrespective of the presence or absence of HRG; 2) the binding of HRG to ErbB3 causes the ErbB3/ErbB2 dimer formation; 3) activation of the tyrosine kinase domain of ErbB2 tyrosine-phosphorylates ErbB3; 4) because PTPN13 is active by the action of Necl-2, tyrosine-phosphorylated ErbB3 is immediately dephosphorylated by PTPN13; and 5) ErbB3 signaling is attenuated. In cells in which Necl-2 is down-regulated, 1) the binding of HRG to ErbB3 causes the ErbB3/ErbB2 dimer formation; 2) activation of the tyrosine kinase domain of ErbB2 tyrosine-phosphorylates ErbB3; 3) because PTPN13 is inactive in the absence of Necl-2, tyrosine-phosphorylated ErbB3 is not dephosphorylated by PTPN13; and 4) ErbB3 signaling is transduced.
Thus changes in the amount of Necl-2 on the plasma membrane owing to its expression and down-regulation regulate ErbB3/ErbB2 signaling. In many types of cancer cells, including A549 cells used in this study, Necl-2 is down-regulated and has been proposed to serve as a tumor suppressor (17). The enhanced ErbB3/ErbB2 signaling caused by amplification or mutation of the ErbB2 gene has been shown to be responsible for tumorigenesis, invasion, and metastasis, in many, but not all, cancers (4). In addition to the amplification of the ErbB2 gene, ErbB3 preferentially associates with ErbB2, and the ErbB3/ErbB2 heterodimer is the most effective complex for activation of downstream pathways, such as PI3K-Rac and PI3K-Akt pathways, resulting in the malignancy of cancer cells (9, 41). However, it remains unknown whether ErbB2 is involved in oncogenesis in cancers in which its gene is not amplified or mutated or how ErbB3 serves as an oncogenic protein in cancers in which it is not overexpressed. Although the ErbB2 gene is not amplified or mutated and ErbB3 is not overexpressed in A549 cancer cells used here (42, 43), we have shown here that the down-regulation of Necl-2 causes the activation of ErbB3/ErbB2 signaling for cell movement and anchorage-independent cell survival. Therefore, both the amplification/mutation of the ErbB2 gene and the down-regulation of Necl-2 would give more malignant properties to cancer cells than either one of them.
Cancer cells show enhanced cell proliferation. We showed here that Necl-2 does not affect the growth of A549 cells. Necl-5 is up-regulated in many types of cancer cells and causes at least partly enhanced movement and proliferation of cancer cells (11, 12, 44–47). We recently found that Necl-5 physically and functionally interacts in cis with the PDGF receptor and enhances the PDGF-induced activation of Ras and subsequent cell proliferation (11–14). The Necl-5-PDGF receptor complex may be involved in enhancement of the proliferation of A549 cells. It is of crucial importance to investigate the expression levels of ErbB2, Necl-2, and Necl-5 in many types of cancer cells.
In embryonic development, individually moving and proliferating mesenchymal cells first form primordial cell-cell contacts through the collisions of cells. They are transformed into epithelial cells and form specialized cell-cell junction complexes, such as adherens junctions and tight junctions. This phenomenon is called mesenchymal-epithelial transition (MET). By contrast, in embryonic development, epithelial cells lose their connections to neighboring cells and become free, which increases cell migration and proliferation. This opposite phenomenon is correlated with epithelial-mesenchymal transition (EMT), which is characterized by a change to a fibroblastoid spindle-shaped cell morphology, loss of epithelial markers, including E-cadherin, induction of mesenchymal markers, and acquisition of motility machinery. The dynamic regulation of MET and EMT is crucial for multicellular organisms to develop and survive. Necl-2 is abundantly expressed in epithelial cells but is not expressed in fibroblasts. Therefore, it is likely that Necl-2 is down-regulated during EMT and that its inhibitory effect on cell movement is released and cells start to move in response to ligands after epithelial cells are transformed into mesenchymal cells. Necl-2 is up-regulated during MET, inhibits cell movement, and induces anchorage-independent apoptosis after mesenchymal cells are transformed into epithelial cells. Mesenchymal cells do not grow in soft agar in an anchorage-independent manner, even though the inhibitory effect of Necl-2 in anchorage-independent cell survival is released. Thus, Necl-2 plays an important role in expression of the cell phenotypes observed in both EMT and MET.
Supplementary Material
Acknowledgments
We thank Drs. T. Yamamoto and S. Higashiyama for their generous gifts of reagents.
This work was supported by grants-in-aid for Scientific Research and for Cancer Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (2007, 2008).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HRG
- heregulin
- PI3K
- phosphoinositide 3-kinase
- Necl
- nectin-like molecule
- PDGF
- platelet-derived growth factor
- pAb
- polyclonal antibody
- mAb
- monoclonal antibody
- DMEM
- Dulbecco's modified Eagle's medium
- siRNA
- small interference RNA
- MET
- mesenchymal-epithelial transition
- EMT
- epithelial-mesenchymal transition
- BrdUrd
- bromodeoxyuridine
- aa
- amino acid(s)
- BSA
- bovine serum albumin
- RNAi
- RNA interference
- GFP
- green fluorescent protein
- TUNEL
- terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- KD
- knockdown.
REFERENCES
- 1.Olayioye M. A., Neve R. M., Lane H. A., Hynes N. E. (2000) EMBO J. 19,3159–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivanco I., Sawyers C. L. (2002) Nat. Rev. Cancer 2,489–501 [DOI] [PubMed] [Google Scholar]
- 3.Prigent S. A., Gullick W. J. (1994) EMBO J. 13,2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarden Y., Sliwkowski M. X. (2001) Nat. Rev. Mol. Cell Biol. 2,127–137 [DOI] [PubMed] [Google Scholar]
- 5.Schraml P., Kononen J., Bubendorf L., Moch H., Bissig H., Nocito A., Mihatsch M. J., Kallioniemi O. P., Sauter G. (1999) Clin. Cancer Res. 5,1966–1975 [PubMed] [Google Scholar]
- 6.Lee J. W., Soung Y. H., Seo S. H., Kim S. Y., Park C. H., Wang Y. P., Park K., Nam S. W., Park W. S., Kim S. H., Lee J. Y., Yoo N. J., Lee S. H. (2006) Clin. Cancer Res. 12,57–61 [DOI] [PubMed] [Google Scholar]
- 7.de Bono J. S., Rowinsky E. K. (2002) Trends Mol. Med. 8,S19–S26 [DOI] [PubMed] [Google Scholar]
- 8.Yip Y. L., Ward R. L. (2002) Cancer Immunol. Immunother. 50,569–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee-Hoeflich S. T., Crocker L., Yao E., Pham T., Munroe X., Hoeflich K. P., Sliwkowski M. X., Stern H. M. (2008) Cancer Res. 68,5878–5887 [DOI] [PubMed] [Google Scholar]
- 10.Takai Y., Miyoshi J., Ikeda W., Ogita H. (2008) Nat. Rev. Mol. Cell Biol. 9,603–615 [DOI] [PubMed] [Google Scholar]
- 11.Ikeda W., Kakunaga S., Takekuni K., Shingai T., Satoh K., Morimoto K., Takeuchi M., Imai T., Takai Y. (2004) J. Biol. Chem. 279,18015–18025 [DOI] [PubMed] [Google Scholar]
- 12.Kakunaga S., Ikeda W., Shingai T., Fujito T., Yamada A., Minami Y., Imai T., Takai Y. (2004) J. Biol. Chem. 279,36419–36425 [DOI] [PubMed] [Google Scholar]
- 13.Minami Y., Ikeda W., Kajita M., Fujito T., Amano H., Tamaru Y., Kuramitsu K., Sakamoto Y., Monden M., Takai Y. (2007) J. Biol. Chem. 282,18481–18496 [DOI] [PubMed] [Google Scholar]
- 14.Amano H., Ikeda W., Kawano S., Kajita M., Tamaru Y., Inoue N., Minami Y., Yamada A., Takai Y. (2008) Genes Cells 13,269–284 [DOI] [PubMed] [Google Scholar]
- 15.Biederer T., Sara Y., Mozhayeva M., Atasoy D., Liu X., Kavalali E. T., Südhof T. C. (2002) Science 297,1525–1531 [DOI] [PubMed] [Google Scholar]
- 16.Gomyo H., Arai Y., Tanigami A., Murakami Y., Hattori M., Hosoda F., Arai K., Aikawa Y., Tsuda H., Hirohashi S., Asakawa S., Shimizu N., Soeda E., Sakaki Y., Ohki M. (1999) Genomics 62,139–146 [DOI] [PubMed] [Google Scholar]
- 17.Kuramochi M., Fukuhara H., Nobukuni T., Kanbe T., Maruyama T., Ghosh H. P., Pletcher M., Isomura M., Onizuka M., Kitamura T., Sekiya T., Reeves R. H., Murakami Y. (2001) Nat. Genet. 27,427–430 [DOI] [PubMed] [Google Scholar]
- 18.Wakayama T., Ohashi K., Mizuno K., Iseki S. (2001) Mol. Reprod. Dev. 60,158–164 [DOI] [PubMed] [Google Scholar]
- 19.Urase K., Soyama A., Fujita E., Momoi T. (2001) Neuroreport 12,3217–3221 [DOI] [PubMed] [Google Scholar]
- 20.Shingai T., Ikeda W., Kakunaga S., Morimoto K., Takekuni K., Itoh S., Satoh K., Takeuchi M., Imai T., Monden M., Takai Y. (2003) J. Biol. Chem. 278,35421–35427 [DOI] [PubMed] [Google Scholar]
- 21.Masuda M., Yageta M., Fukuhara H., Kuramochi M., Maruyama T., Nomoto A., Murakami Y. (2002) J. Biol. Chem. 277,31014–31019 [DOI] [PubMed] [Google Scholar]
- 22.Galibert L., Diemer G. S., Liu Z., Johnson R. S., Smith J. L., Walzer T., Comeau M. R., Rauch C. T., Wolfson M. F., Sorensen R. A., Van der Vuurst de Vries A. R., Branstetter D. G., Koelling R. M., Scholler J., Fanslow W. C., Baum P. R., Derry J. M., Yan W. (2005) J. Biol. Chem. 280,21955–21964 [DOI] [PubMed] [Google Scholar]
- 23.Yageta M., Kuramochi M., Masuda M., Fukami T., Fukuhara H., Maruyama T., Shibuya M., Murakami Y. (2002) Cancer Res. 62,5129–5133 [PubMed] [Google Scholar]
- 24.Fukuhara H., Masuda M., Yageta M., Fukami T., Kuramochi M., Maruyama T., Kitamura T., Murakami Y., Masvuda M. (2003) Oncogene 22,6160–6165 [DOI] [PubMed] [Google Scholar]
- 25.Kakunaga S., Ikeda W., Itoh S., Deguchi-Tawarada M., Ohtsuka T., Mizoguchi A., Takai Y. (2005) J. Cell Sci. 118,1267–1277 [DOI] [PubMed] [Google Scholar]
- 26.Fukami T., Satoh H., Fujita E., Maruyama T., Fukuhara H., Kuramochi M., Takamoto S., Momoi T., Murakami Y. (2002) Gene 295,7–12 [DOI] [PubMed] [Google Scholar]
- 27.Boles K. S., Barchet W., Diacovo T., Cella M., Colonna M. (2005) Blood 106,779–786 [DOI] [PubMed] [Google Scholar]
- 28.van der Weyden L., Arends M. J., Chausiaux O. E., Ellis P. J., Lange U. C., Surani M. A., Affara N., Murakami Y., Adams D. J., Bradley A. (2006) Mol. Cell. Biol. 26,3595–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita E., Kouroku Y., Ozeki S., Tanabe Y., Toyama Y., Maekawa M., Kojima N., Senoo H., Toshimori K., Momoi T. (2006) Mol. Cell. Biol. 26,718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada D., Yoshida M., Williams Y. N., Fukami T., Kikuchi S., Masuda M., Maruyama T., Ohta T., Nakae D., Maekawa A., Kitamura T., Murakami Y. (2006) Mol. Cell. Biol. 26,3610–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikeda W., Kakunaga S., Itoh S., Shingai T., Takekuni K., Satoh K., Inoue Y., Hamaguchi A., Morimoto K., Takeuchi M., Imai T., Takai Y. (2003) J. Biol. Chem. 278,28167–28172 [DOI] [PubMed] [Google Scholar]
- 32.Laemmli U. K. (1970) Nature 227,680–685 [DOI] [PubMed] [Google Scholar]
- 33.Kawakatsu T., Shimizu K., Honda T., Fukuhara T., Hoshino T., Takai Y. (2002) J. Biol. Chem. 277,50749–50755 [DOI] [PubMed] [Google Scholar]
- 34.Witton C. J., Reeves J. R., Going J. J., Cooke T. G., Bartlett J. M. (2003) J. Pathol. 200,290–297 [DOI] [PubMed] [Google Scholar]
- 35.Yokoe S., Takahashi M., Asahi M., Lee S. H., Li W., Osumi D., Miyoshi E., Taniguchi N. (2007) Cancer Res. 67,1935–1942 [DOI] [PubMed] [Google Scholar]
- 36.Sliwkowski M. X., Schaefer G., Akita R. W., Lofgren J. A., Fitzpatrick V. D., Nuijens A., Fendly B. M., Cerione R. A., Vandlen R. L., Carraway K. L., 3rd (1994) J. Biol. Chem. 269,14661–14665 [PubMed] [Google Scholar]
- 37.Murakami Y. (2002) Oncogene 21,6936–6948 [DOI] [PubMed] [Google Scholar]
- 38.Mao X., Seidlitz E., Ghosh K., Murakami Y., Ghosh H. P. (2003) Cancer Res. 63,7979–7985 [PubMed] [Google Scholar]
- 39.Abaan O. D., Toretsky J. A. (2008) Cancer Metastasis Rev. 27,205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J. H., Chen R., Yi W., Cantin G. T., Fearns C., Yang Y., Yates J. R., 3rd, Lee J. D. (2008) Oncogene 27,2525–2531 [DOI] [PubMed] [Google Scholar]
- 41.Citri A., Skaria K. B., Yarden Y. (2003) Exp. Cell Res. 284,54–65 [DOI] [PubMed] [Google Scholar]
- 42.Amann J., Kalyankrishna S., Massion P. P., Ohm J. E., Girard L., Shigematsu H., Peyton M., Juroske D., Huang Y., Stuart Salmon J., Kim Y. H., Pollack J. R., Yanagisawa K., Gazdar A., Minna J. D., Kurie J. M., Carbone D. P. (2005) Cancer Res. 65,226–235 [PubMed] [Google Scholar]
- 43.Engelman J. A., Zejnullahu K., Gale C. M., Lifshits E., Gonzales A. J., Shimamura T., Zhao F., Vincent P. W., Naumov G. N., Bradner J. E., Althaus I. W., Gandhi L., Shapiro G. I., Nelson J. M., Heymach J. V., Meyerson M., Wong K. K., Jänne P. A. (2007) Cancer Res. 67,11924–11932 [DOI] [PubMed] [Google Scholar]
- 44.Masson D., Jarry A., Baury B., Blanchardie P., Laboisse C., Lustenberger P., Denis M. G. (2001) Gut 49,236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gromeier M., Lachmann S., Rosenfeld M. R., Gutin P. H., Wimmer E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97,6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chadéneau C., LeMoullac B., Denis M. G. (1994) J. Biol. Chem. 269,15601–15605 [PubMed] [Google Scholar]
- 47.Sloan K. E., Eustace B. K., Stewart J. K., Zehetmeier C., Torella C., Simeone M., Roy J. E., Unger C., Louis D. N., Ilag L. L., Jay D. G. (2004) BMC Cancer 4,73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.