Abstract
The preproteins targeted to the mitochondria are transported through the translocase of the outer membrane complex. Tom70/Tom71 is a major surface receptor of the translocase of the outer membrane complex for mitochondrial preproteins. The preproteins are escorted to Tom70/Tom71 by molecular chaperones Hsp70 and Hsp90. Here we present the high resolution crystal structures of Tom71 and the protein complexes between Tom71 and the Hsp70/Hsp90 C terminus. The crystal structures indicate that Tom70/Tom71 may exhibit two distinct states. In the closed state, the N-terminal domain of Tom70/Tom71 partially blocks the preprotein-binding pocket. In the open state, the N-terminal domain moves away, and the preprotein-binding pocket is fully exposed. The complex formation between the C-terminal EEVD motif of Hsp70/Hsp90 and Tom71 could lock Tom71 in the open state where the preprotein-binding pocket of Tom71 is ready to receive preproteins. The interactions between Hsp70/Hsp90 and Tom71 N-terminal domain generate conformational changes that may increase the volume of the preprotein-binding pocket. The complex formation of Hsp70/Hsp90 and Tom71 also generates significant domain rearrangement within Tom71, which may position the preprotein-binding pocket closer to Hsp70/Hsp90 to facilitate the preprotein transfer from the molecular chaperone to Tom71. Therefore, molecular chaperone Hsp70/Hsp90 may function to prepare the mitochondrial outer membrane receptor Tom71 for preprotein loading.
The mitochondrion plays important roles in cell physiology. The mitochondrion functions as the “cellular power house” by generating most of the supply of ATP for the cell. In addition, the mitochondrion is involved in a number of critical cellular processes including the synthesis of metabolites, lipid metabolism, free radical production, and metal ion homeostasis. The mitochondrion consists of four compartments, the outer membrane, the inner membrane, the intermembrane space, and the mitochondrial matrix. The mitochondrion contains a large number of proteins (1), but only a few of these are translated within the mitochondrion (2). Therefore, the majority of the mitochondrial proteins are synthesized in the cytosol and translocated into the mitochondrion.
The mitochondrial preproteins contain specific targeting signals to reach the correct compartments within the mitochondria. The mitochondrial matrix preproteins contain N-terminal targeting sequences that form the short amphipathic helices (2–6). On the other hand, some mitochondrial proteins of the inner and outer membrane contain internal targeting signals within the mature proteins (7). The mitochondrion has developed a set of delicate translocons to transport the preproteins into the mitochondrial compartments, one translocase of the outer membrane (TOM)2 and two translocases of the inner membrane (TIM23 and TIM22) (4, 5, 8). The TOM complex has two surface receptors, Tom20 and Tom70 (9, 10). Tom20 recognizes the N-terminal mitochondrial targeting signals from the preproteins, whereas Tom70 binds to internal targeting sequences of preproteins such as the multi-transmembrane carrier proteins residing in the mitochondrial membranes (9–12). The crystal structure of Saccharomyces cerevisiae Tom70 revealed that Tom70 contained 11 TPR motifs, and the TPR motifs were clustered into two domains. The three TPR motifs in the N-terminal domain of Tom70p form a peptide-binding groove for the C-terminal EEVD motif of Hsp70/Hsp90, whereas the C-terminal domain of Tom70p contains a large preprotein-binding pocket (13).
Molecular chaperones Hsp70 and Hsp90 play important roles in targeting the preproteins to TOM complex (14). Hsp70 and Hsp90 can protect these preproteins from aggregation in the cytosol (15). The C-terminal EEVD motifs of Hsp70/Hsp90 may interact directly with the N-terminal domain of Tom70p to target the preproteins to TOM complex (13, 14, 16). The C-terminal EEVD motif of Hsp70/Hsp90 has been indicated to bind several proteins containing TPR motifs including Hop and CHIP. The complex structures for the Hsp70/Hsp90 EEVD motif and Hop and CHIP TPR regions have been determined (17–21).
Tom71 (also known as Tom72) was identified as a homologue with Tom70 with high amino acid sequence identity (>50%) (22). Tom71 shares overlapping functions with Tom70 to transfer the preproteins and maintain the mitochondrial morphology (23, 24). In this study, we have determined the crystal structures of S. cerevisiae Tom71 and the complexes of Tom71 and Hsp70/Hsp90 C-terminal EEVD motifs. These structures suggest that the Hsp70/Hsp90 binding to Tom70/Tom71 may keep Tom70/Tom71 in the open state for receiving preproteins. The Hsp70/Hsp90 interactions may also increase the volume of the preprotein-binding pocket of Tom70/Tom71 and prepare Tom70/Tom71 for preprotein loading.
MATERIALS AND METHODS
Expression and Crystallization of Tom71, Tom71-Hsp70, and Tom71-Hsp90 Complexes
The gene encoding S. cerevisiae Tom71 cytosolic fragment (residues 107–639) was cloned into the vector pET28b. The Tom71 sequence was confirmed by DNA sequencing. The plasmid was then transformed into Escherichia coli strain BL21 (DE3) for protein expression. The BL21(DE3) cells were induced with 0.2 mm isopropyl-β-d-thiogalactopyranoside at A600 of 0.5–0.6. After shaking overnight at 15 °C, the cells were harvested and sonicated in a buffer with 20 mm Tris (pH 7.9), NaCl 500 mm, imidazole 5 mm. The supernatant was collected after centrifugation at 15,000 × g for 1 h and loaded to nickel-nitrilotriacetic acid affinity beads (GE Healthcare). The protein was stripped by buffer containing 20 mm Tris (pH 7.9), 500 mm NaCl, 50 mm EDTA. After thrombin cleavage overnight at room temperature, the protein was concentrated and purified by Superdex 200 (GE Healthcare) in a buffer with 10 mm Tris (pH 7.5), 150 mm NaCl. The peak fraction was collected, concentrated to 20 mg ml−1, and subjected to crystallization trials. Large rod-shaped crystals (0.5 × 0.1 × 0.1 mm) were obtained by the hanging drop vapor diffusion method at room temperature. The well solution consisted of 1 ml of 100 mm Tris buffer (pH 7.5), 25% (w/v) polyethylene glycol 4000, 0.2 m NaCl. The crystals of Tom71 complexed with yeast Hsp70 Ssa1 C-terminal peptide (PEAEGPTVEEVD) were grown by the hanging drop vapor diffusion method by mixing Tom71 and Hsp70 peptide using a 1:1.2 molar ratio in buffer 10 mm Tris (pH 7.5), 100 mm NaCl. The well solution contains 1 ml of 100 mm Tris buffer (pH 7.5), 20% polyethylene glycol 6000, 10% ethylene glycol. The crystals of yeast Tom71 and yeast Hsp90 Hsp82 C-terminal peptide (EVPADTEMEEVD) complex were obtained using similar protocols as for the Tom71 and Hsp70 complexes. The peptides were synthesized in Genscript.
Structure Determinations
The Tom71 crystals diffracted x-ray to 2.0 Å in the beamline Southeast Regional Collaborative Access Team at Advanced Photon Source. The atomic coordinates of the N- and C-terminal domains of yeast Tom70 were used individually to search for molecular replacement solutions for the crystals of Tom71 by the program PHASER (27). The initial model was built by use of WARP/ARP at 2.0 Å resolution (28), followed by manual model building using COOT (29). The final model was refined by REFMAC5 to and Rfactor of 18.7% and an Rfree of 22.7% (30). Tom71 complexed with Hsp70 crystals diffracted x-ray to 2.15 Å in the beamline Southeast Regional Collaborative Access Team at Advanced Photon Source. The refined atomic coordinates of Tom71 were used as a searching model to obtain molecular replacement solution. The initial model building was completed by use of WARP/ARP at 2.15 Å resolution, followed by manual model building using COOT. The final model was refined by REFMAC5 to an Rfactor of 20.8% and an Rfree of 26.1%. Tom71 complexed with yeast Hsp90 (Hsp82) crystals diffracted x-ray to 2.0 Å in the beamline Southeast Regional Collaborative Access Team at Advanced Photon Source. The structure of yeast Tom71 and yeast Hsp90 (Hsp82) C-terminal peptide complex was solved using similar protocols as for Tom71-Hsp70 complex. The complex structure was refined with an Rfactor of 19.9% and an Rfree of 23.6%.
Sequence Conservation Drawing
To generate the sequence conservation drawing, program ClustalW was utilized to align the Tom71 sequences from S. cerevisiae with those from Candida albicans and Tom70 from S. cerevisiae, Homo sapiens, and Drosophila melanogaster. The aligned sequences in multiple sequence alignment formats were converted into a property file by use of the program ProSkin. The property file was then visualized by using the program Pymol.
RESULTS AND DISCUSSION
Tom71 May Exhibit Open and Closed States
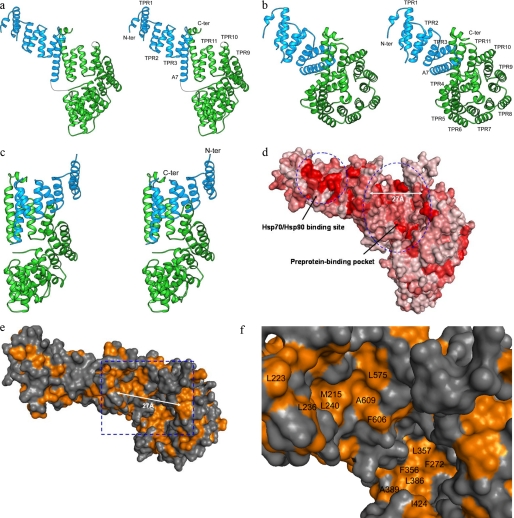
The S. cerevisiae Tom71 (639 amino acids) contains a short N-terminal transmembrane fragment anchored in the mitochondrial outer membrane and a large C-terminal fragment located in cytosol. The crystal structure of yeast Tom71 (residues 107–639) was determined to 2.0 Å resolution by a molecular replacement method using the yeast Tom70 structure as the searching model (Table 1). The structure of the Tom71 monomer consists of 28 α-helices (A0–A27) and no β-strands (Fig. 1, a and b). The electron densities for the loops between helices A1 and A2, A7 and A8, and A22 and A23 are missing. A majority of the helices of Tom71 form 11 TPR motifs (TPR1–TPR11). Two domains can be clearly identified from the Tom71 structure. The N-terminal domain covers helices A0–A7 (TPR1–TPR3), and the C-terminal domain includes helices A8–A27 (TPR4–TPR11). The N-terminal domain of Tom71 contains the Hsp70/Hsp90-binding site, and the C-terminal domain constitutes the preprotein-binding pocket. The two domains are connected by a long helix A7. Most of the helices in the C-terminal domain (A10–A26) stack together to form one turn of the “superhelix” (Fig. 1, a and b).
TABLE 1.
Data collection and refinement statistics for crystal structures of yeast Tom71, Tom71-Hsp70 complex, and Tom71-Hsp90 complex
Statistics in the highest resolution shell are shown in parentheses.
| Tom71 | Tom71-Hsp70 | Tom71-Hsp90 | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions: a, b, c (Å) | 76.90, 83.42, 109.11 | 47.82, 116.03, 150.65 | 47.87, 116.29, 150.74 |
| Wavelength (Å) | 1.000 | 1.000 | 0.9718 |
| Resolution (Å) | 2.0 | 2.15 | 2.0 |
| Rsym or Rmerge | 0.053 (0.226) | 0.078 (0.506) | 0.088 (0.446) |
| I/σI | 47.1 (9.2) | 19.2 (2.3) | 25.0 (2.47) |
| Completeness (%) | 99.8 (99.9) | 90.3 (77.7) | 97.2 (86.1) |
| Redundancy | 7.2 (7.0) | 5.0 (3.9) | 5.0 (2.5) |
| Refinement | |||
| No. reflections | 47,008 | 42,316 | 57,803 |
| Rwork/Rfree | 18.9 (21.9)/22.7 (28.3) | 20.8 (26.1)/26.1 (35.3) | 19.9 (24.7)/23.6 (27.9) |
| No. atoms protein (water) | 4735 (573) | 4427 (268) | 4559 (451) |
| B factors | 34.55 | 17.85 | 36.59 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.018 | 0.023 | 0.018 |
| Bond angels (°) | 1.46 | 1.787 | 1.524 |
FIGURE 1.
The yeast Tom71 structure. a, ribbon drawing of the yeast Tom71 monomer structure (open state) in side-by-side stereo mode (31). The N- and C-terminal domains of Tom71 are shown in light blue and green, respectively. The N (N-ter) and C termini (C-ter) are labeled. TPR1–TPR3 and TPR9–TPR11 are labeled. The missing residues in the electron density map are shown in dotted lines. b, ribbon drawing of the yeast Tom71 monomer structure in side-by-side stereo mode. The Tom71 structure in this figure is rotated ∼90° along the horizontal axis from that in a. The N and C termini of the structure are labeled. TPR1–TPR11 are labeled. c, ribbon drawing of the yeast Tom70 monomer structure (closed state) in side-by-side stereo mode. The coloring of the N- and C-terminal domains of yeast Tom70 is the same as a. d, the sequence conservation drawing for Tom71. Tom71 in this figure is in similar orientation as that in a. The sequence conservation score obtained by sequence alignment (see Fig. 2 for details) is mapped to the Tom71 molecular surface by Pymol. The red color denotes the conserved regions. The Hsp70/Hsp90-binding site and the preprotein-binding pocket located on the Tom71 surface are indicated by dotted circles. To indicate the size of the preprotein-binding pocket of Tom71, the distance (27 Å) between Pro234 and Phe496 is shown by an arrow. e, the hydrophobicity drawing of Tom71 monomer by Pymol. Gold color denotes hydrophobic regions. Tom71 in this figure is rotated ∼90° along the horizontal axis from that in d and is in similar orientation as that in b. The distance (27 Å) between Pro234 and Phe496 is shown by an arrow. The blue box covers the preprotein-binding pocket, which will be amplified in f. f, this figure shows the magnified version of the area within the blue box in the e. Some conserved hydrophobic residues involved in forming the preprotein-binding pocket are labeled.
When the crystal structure of yeast Tom71 is compared with that of Tom70, we found that the individual N- and C-terminal domains of Tom71 resemble the folds from those of Tom70 (Fig. 1, a and c) (13). Surprisingly, the N- and C-terminal domains of Tom71 are arranged in a very different fashion from that in the Tom70 crystal structure. In the crystal structure of Tom71, the N-terminal domain is swung away from the C-terminal domain, which renders the Tom71 structure an “open” state (Fig. 1a). In the yeast Tom70 structure, the two domains are packed together to form an elongated molecule as a superhelix, which represents a “closed” state (Fig. 1c). The N-terminal domain in the Tom71 structure (open state) moves away from that in the Tom70 structure (closed state) along helix A7. A putative preprotein-binding pocket was identified in the Tom70 C-terminal domain (13). In the Tom70 structure, the preprotein-binding pocket is partially blocked by the N-terminal domain. However, in the Tom71 structure, the preprotein-binding pocket is dramatically enlarged and fully exposed and accessible to the solvent (Fig. 1). In the closed state, the N-terminal domain of Tom70 is connected to the C-terminal domain by polar interactions between A7 and A25. In the open state, the N-terminal domain of Tom71 is associated to the C-terminal domain by hydrophobic interactions between A6, A7, and A26.
Because yeast Tom70 and Tom71 share high protein sequence homology (53% sequence identity) and have overlapping cellular functions, we reason that Tom70 and Tom71 may adopt both open and closed states in solution and on the mitochondrial membrane. In the closed state, the preprotein-binding pocket of Tom70/Tom71 is blocked and not ready to receive preprotein, whereas in the open state, the pocket is exposed for preprotein loading. Previous biophysical and thermodynamics studies showed that Tom70 exhibited substantial structural flexibility and may undergo multiple conformational changes at physiological temperature (25). This is consistent with our structural observations.
In the open state, the N-terminal domain of Tom71 moves away, which creates significantly more space for the preprotein-binding pocket. In the open state, the pocket has an estimated dimension of ∼25 × 35 × 20 Å (width × length × depth), which more than doubles the size of pocket in the closed state. The preprotein-binding pocket is large enough to accommodate a polypeptide with secondary structures. A pair of α-helices can be easily modeled into the pocket. Helices A12, A14, A16, A18, A20, A22, A24, and A26 from the C-terminal domain of Tim71 constitute the inner surface of the preprotein-binding pocket. Helices A6 and A7 from the N-terminal domain of Tom71 are also involved in forming one side of the wall for the pocket (Fig. 1).
The conservation drawing of Tom71 structure showed that the residues involved in forming the preprotein-binding pocket surface are quite conserved among Tom70 and Tom71 families, indicating that Tom70 and Tom71 proteins may share similar specificity for preproteins (Fig. 1d). A large portion of the conserved areas in the preprotein-binding pocket are generated by hydrophobic residues. On the upper part of the pocket, residues Met215, Leu223, Leu236, Leu240, Leu575, Phe606, and Ala609 form a long hydrophobic strip. On the lower half of the pocket, residues Pro264, Phe272, Phe356, Leu357, Leu386, Ala389, Phe423, Ile424, Tyr447, and Phe485 form most of the bottom of the pocket (Fig. 1, e and f). The hydrophobicitiy of these residues is nicely conserved as shown by sequence alignment (Fig. 2). This structural feature is consistent with the hypothesis that Tom70/Tom71 recognizes the hydrophobic internal targeting sequences from the preproteins (12, 16).
FIGURE 2.
Sequence alignment of the Tom71/Tom70 family members. Program ClustalW was utilized to align the Tom71 sequences from S. cerevisiae (Sc Tom71) with that from C. albicans (Ca Tom71) and Tom70 from S. cerevisiae (Sc Tom70), H. sapiens (Hs Tom70), and D. melanogaster (Dm Tom70). The amino acid residues of yeast Tom71 are numbered above the alignment. The eleven TPR motifs (TPR1–TPR11) within Tom71 are labeled. Helices A0–A27 are labeled. The conserved residues involved in binding the Hsp70/Hsp90 C-terminal EEVD motifs are labeled with blue bars. The conserved residues involved in mediating the conformational changes generated by Hsp70/Hsp90 binding are marked with pink bars. The conserved hydrophobic residues forming the Tom70p preprotein-binding pocket among the family are marked with green bars.
The Interactions between Tom71 and Hsp70/Hsp90 C Terminus
Molecular chaperone Hsp70/Hsp90 can target the preproteins to Tom70/Tom71 by anchoring the Hsp70/Hsp90 C-terminal EEVD motif to the N-terminal domain of Tom70/Tom71 (13, 14). Here we present the crystal structure of yeast Tom71 complexed with yeast Hsp70 (Ssa1) C-terminal peptide PEAEGPTVEEVD to 2.15 Å resolution. The crystal structure of yeast Tom71 complexed with yeast Hsp90 (Hsp82) C-terminal peptide EVPADTEMEEVD was determined to 2.0 Å resolution. Residues PTVEEVD (residues −6 to 0) of Hsp70 and residues MEEVD (residues −4 to 0) of Hsp90 are clearly visible in the electron density map. Both bound peptides exhibit similar conformations and are located in a groove formed within the N-terminal domain of Tom71 (Fig. 3). The Hsp70/Hsp90 C-terminal peptide binds to a region of Tom71 that is overall quite basic. In both complex structures, the Asp at position 0 of Hsp70/90 makes polar interactions with residues Lys131, Asn135, and Lys196 from Tom71. The main chain nitrogen of this Asp residue forms a hydrogen bond with the side chain of Asn166 of Tom71. The Val at position −1 makes hydrophobic interactions with Phe138. The carbonyl oxygen of Glu at position −2 forms strong hydrogen bonds with the side chains of Lys196 and Arg200 of Tom71. The Val at position −4 of Hsp70 makes hydrophobic interactions with Leu199 of Tom71. The Met at position −4 of Hsp90 contacts Leu199 and the hydrophobic side chain of Lys196 from Tom71. These residues (Lys131, Asn135, Phe138, Asn166, Lys196, Leu199, and Arg200) of Tom71 involved in binding the C-terminal EEVD motif of Hsp70/Hsp90 are very nicely conserved for Tom71 and Tom70 among species, suggesting that the binding mechanism is shared within the Tom70/Tom71 family members (Fig. 2). These residues form a conserved surface at the Hsp70/90-binding site located on the N-terminal domain of Tom71 (Fig. 1d).
FIGURE 3.
The complex structures of Tom71 and the Hsp70/Hsp90 C-terminal EEVD motif. a, the surface potential drawing of Tom71 complexed with Hsp70 C-terminal peptide PTVEEVD. Tom71 is shown in surface potential drawing generated by Pymol and Apbs. Blue and red denote positively and negatively electrostatic potentials, respectively. The bound Hsp70 peptide is shown in a rod model. In the rod model, carbon atoms are shown in green, oxygen atoms are shown in red, and nitrogen atoms are shown in blue. To indicate the enlargement of the preprotein-binding pocket of Tom71 after Hsp70 binding, the distance (32 Å) between Pro234 and Phe496 of Tom71 is shown. b, the surface potential drawing of Tom71 N-terminal domain interacting with the Hsp70 C-terminal peptide PTVEEVD in a rod model. The residues of Tom71 involved in binding Hsp70 are labeled in green, and the residues of Hsp70 peptide PTVEEVD are labeled in black. c, ribbon drawing of Tom71 N-terminal domain complexed with Hsp70 C terminus in stereo mode. Tom71 is in a silver ribbon drawing, and the Hsp70 C-terminal peptide is in a solid rod model. The residues of Tom71 involved in binding Hsp70 are drawn in dotted rod model and labeled in blue. The residues of Hsp70 peptide are labeled in black. d, the surface potential drawing of Tom71 N-terminal domain interacting with Hsp90 C-terminal peptide MEEVD in a rod model. The residues of Tom71 involved in binding Hsp90 are labeled in green, and the residues of Hsp90 peptide MEEVD are labeled in black.
The hydrophobic interactions between the Val/Met at the −4 position of Hsp70/Hsp90 with Leu199 of Tom71 make the bound Hsp70/Hsp90 peptide to form a turn at the upstream of the EEVD fragment of Hsp70/Hsp90 (Fig. 3, b and d). The turn of the bound peptide indicated that the upstream part of the Hsp70/Hsp90 C terminus does not make specific contacts with Tom70/Tom71. This phenomenon is also observed in the complex structure of CHIP and the Hsp90 C-terminal EEVD motif (20). This may provide flexibility for Hsp70/Hsp90 to deliver the bound preprotein to the Tom70/Tom71 preprotein-binding pocket.
The binding of the Hsp70/Hsp90 C-terminal EEVD motif to Tom71 may lock Tom71 in the open state. The bound EEVD motif of Hsp70/Hsp90 may cause severe collisions with a loop region between A22 and A23 of Tom71 C-terminal domain if the N-terminal domain of Tom71 moves back into the closed state. Tom71 has to stay in the open state after molecular chaperone Hsp70/Hsp90 binding, which is the favored state for Hsp70/Hsp90 to load the preproteins.
The Conformational Changes of Tom71 Generated by Hsp70/Hsp90 Binding
The binding of the Hsp70/Hsp90 C-terminal EEVD motif to Tom71 generates significant conformational changes in the Tom71 structure (Fig. 4). When Hsp70/Hsp90 binds the Tom71 N-terminal domain, Lys196 and Arg200 from helix A5 form a salt bridge and strong hydrogen bonds with Asp at position 0 and Glu at position −2 of Hsp70/Hsp90 C terminus. Leu199 of helix A5 forms hydrophobic interactions with the Val at position −4 of Hsp70 (or Met at the position −4 of Hsp90). All of these interactions pull helix A5 of Tom71 toward the bound Hsp70/Hsp90 C terminus peptide by ∼1.5 Å after Hsp70/Hsp90 binding (Fig. 4a). Helix A5 of Tom71 is associated with helices A6 and A7 by a hydrophobic cluster formed by Leu198, Leu199, and Ala202 from A5; Leu218, Leu221, and Val220 from A6; and Met235 from the N terminus of the long helix A7. The salt bridges formed by Arg201 from A5, Asp217 from A6, Glu206 from A5, and Arg238 from A7 also contribute to the association of helices A5, A6, and A7. The binding between the Hsp70/Hsp90 C-terminal EEVD motifs and Tom71 cause helix A5 of Tom71 to move to the bound Hsp70/Hsp90 peptide by ∼1.5 Å. Because of the association of helices A5, A6, and A7, the conformational changes are translated to A6 and A7 and, surprisingly, significantly amplified within helices A6 and A7. After Hsp70/Hsp90 binding to Tom71, helix A6 of Tom71 moves toward the bound Hsp70/Hsp90 peptide by ∼2.5 Å. Helix A7 moves toward the center of N-terminal domain by ∼4.5 Å. More importantly, the pulling at the N terminus of helix A7 causes the long helix A7 to rotate ∼20° after Hsp70/Hsp90 binding (Fig. 4a). The residues of Tom71 involved in mediating the conformational changes (Lys196, Leu198, Leu199, Arg200, Arg201, Ala202, Glu206, Asp217, Leu218, Val220, Leu221, Met235, and Arg238) are well conserved among the Tom70/Tom71 family (Fig. 2).
FIGURE 4.
The conformational changes of Tom71 generated by Hsp70 binding. a, the N-terminal domain of Tom71 is superimposed with that in the Tom71-Hsp70 complex and they are shown by a ribbon drawing. The Tom71 N-terminal domain is in light blue. The Tom71 N-terminal domain within the Tom71-Hsp70 complex is in silver. The bound Hsp70 C-terminal peptide is in red. Helices A5, A6, and A7 are labeled in blue. Some residues of Tom71 involved in generating the conformational changes are labeled in black. Residues Lys196, Arg200, and Leu199 of Tom71 involved in binding Hsp70 are labeled. The residues forming hydrophobic cluster to associate A5, A6, and A7 are labeled. Glu206 and Arg238 linking A5 and A7 by forming a salt bridge are also labeled. b, Cα trace drawings of yeast Tom71 structure and the Tom71-Hsp70 complex structure. The N-terminal domain of Tom71 is superimposed with that in the Tom71-Hsp70 complex structure. The molecules in this figure are in a similar orientation as in a. The uncomplexed Tom71 structure is in purple. In the Tom71-Hsp70 C terminus complex, Tom71 is in green, and the Hsp70 C terminus is in red. The N- and C-terminal domains of Tom71 are labeled. Helix A7 acting as the hinge to connect the N-and C-terminal domains of Tom71 is labeled.
The N-terminal domain of Tom70/Tom71 may function as a sensor to detect the presence of the molecular chaperone Hsp70/Hsp90. Lys196 and Arg200 of Tom71 play central roles to bind Hsp70/90 and induce the subsequent conformational changes. Lys196 and Arg200 protrude out of the molecular surface of Tom71 to form strong hydrogen bonds and salt bridges with the Hsp70/90 C-terminal EEVD motif (Fig. 3). The interactions between Lys196/Arg200 and Hsp70/Hsp90 function as the major driving force for the downstream conformational changes. The functions of the conserved Lys196 and Arg200 can be defined as a “Lys/Arg switch.”
The structural consequence of the Hsp70/Hsp90 EEVD motif binding to Tom71 is that the volume of the preprotein-binding pocket located on the C-terminal domain of Tom71 is significantly increased. Because helices A6 and A7 are involved in forming one side of the preprotein-binding pocket, the translations of helices A6 and A7 toward the center of the N-terminal domain after Hsp70/Hsp90 binding widens up the pocket located at the C-terminal domain substantially (Fig. 4a). In addition, the rotation of helix A7 after EEVD motif binding may also contribute to the opening up of the pocket. The width of the pocket increases to ∼30 Å after Hsp70/Hsp90 binding from ∼25 Å before binding. For example, residues Pro234 and Phe496 are located at the opposite side on the preprotein-binding pocket of Tom71. The distance between the Cα atoms of Pro234 and Phe496 increases to 32 Å after complex formation, whereas the distance was 27 Å before Hsp70/Hsp90 binding. (Figs. 1d and 3, a and d). An enlarged preprotein-binding pocket may facilitate efficient preprotein loading from Hsp70/Hsp90 to Tom70/Tom71. As a similar comparison, the cavity inside of chaperonin GroEL is also significantly increased after GroES binding (26).
Helix A7 is positioned at the joint of Tom71 N-terminal and C-terminal domains and acts as a hinge to link the two domains together. The rotation of helix A7 (∼20°) of Tom71 after Hsp70/Hsp90 binding is transduced to the entire C-terminal domain and generates significant rearrangement of the N-terminal and C-terminal domains of Tom71. The C-terminal domain of Tom71 is swung from the open state to the closed state by ∼20° after the Hsp70/Hsp90 binding (Fig. 4a). The 20° rotation of the Tom71 C-terminal domain after the Hsp70/Hsp90 binding can bring the preprotein-binding pocket much closer to the molecular chaperone Hsp70/Hsp90, which may facilitate the preprotein transfer from Hsp70/Hsp90 to Tom71 (Fig. 4b). Therefore the domain rearrangement within Tom71 molecule may have significant functional benefit for Tom71 to receive the preproteins from Hsp70/Hsp90.
The crystal structures of yeast Tom71 and Tom70 suggest that the mitochondrial translocon receptor Tom70/Tom71 may exhibit the open and closed states. In the closed state, the N-terminal domain of Tom70/Tom71 folds on top of the C-terminal domain and partially covers the preprotein-binding pocket. This may block the preproteins from entering the pocket. In the open state, the N-terminal domain moves away, and the preprotein-binding pocket is dramatically enlarged and exposed. When molecular chaperone Hsp70/Hsp90 escorts preproteins to approach the TOM complex, the Hsp70/Hsp90 C-terminal EEVD motif will bind the Tom71 N-terminal domain. The interactions will lock Tom71 in the open state. The binding between Hsp70/Hsp90 and Tom71 could generate significant conformational changes in Tom71 structure, which could further increase the volume of the preprotein-binding pocket and rotate the pocket closer to Hsp70/Hsp90. Thus, the molecular chaperone Hsp70/Hsp90 could ensure that Tom70/Tom71 only receive the preproteins escorted by molecular chaperones (Fig. 5). The cooperativity of Tom70/Tom71 and the molecular chaperone Hsp70/Hsp90 provides a delicate gating system for Tom70/Tom71 to recognize the correct preprotein substrates.
FIGURE 5.
The cartoon drawing for the mechanism how Hsp70/Hsp90 prepares Tom70/Tom71 for preprotein loading. a, Tom70/Tom71 may exhibit two distinct states: the open and closed state. The Tom70/Tom71 molecule is shown in blue. The N- and C-terminal domains are labeled. The mitochondria outer membrane is shown in orange. b, the interactions between the Hsp70/Hsp90 C-terminal EEVD motif will lock the Tom70/Tom71 in the open state. The Hsp70/Hsp90 is shown in gold. The binding between Hsp70/Hsp90 and Tom71 could increase the volume of the preprotein-binding pocket. The complex formation might rotate the Tom71 C-terminal domain ∼20° back toward the closed state and therefore position the preprotein-binding pocket closer to the Hsp70/Hsp90. The Hsp70/Hsp90 EEVD motif is shown as a red arrow. The preprotein is shown as a green triangle. c, Hsp70/Hsp90 will then load the preprotein into the enlarged preprotein-binding pocket of Tom70/Tom71.
Acknowledgments
We are grateful to the staff scientists in Advanced Photon Source beamline Southeast Regional Collaborative Access Team for help with data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK56203 and R01 GM65959. This work was also supported by Army Research Office Grant 51894LS.
The atomic coordinates and structure factors (codes 3FP2, 3FP3, and 3FP4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TOM
- translocase of the outer membrane.
REFERENCES
- 1.Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., Schönfisch B., Perschil I., Chacinska A., Guiard B., Rehling P., Pfanner N., Meisinger C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray M. W., Burger G., Lang B. F. (1999) Science 283,1476–1481 [DOI] [PubMed] [Google Scholar]
- 3.Neupert W. (1997) Annu. Rev. Biochem. 66,863–917 [DOI] [PubMed] [Google Scholar]
- 4.Neupert W., Brunner M. (2002) Nat. Rev. Mol. Cell Biol. 3,555–565 [DOI] [PubMed] [Google Scholar]
- 5.Pfanner N. (2000) Curr. Biol. 10,R412–415 [DOI] [PubMed] [Google Scholar]
- 6.Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. (2000) Cell 100,551–560 [DOI] [PubMed] [Google Scholar]
- 7.Rehling P., Model K., Brandner K., Kovermann P., Sickmann A., Meyer H. E., Kühlbrandt W., Wagner R., Truscott K. N., Pfanner N. (2003) Science 299,1747–1751 [DOI] [PubMed] [Google Scholar]
- 8.Koehler C. M. (2004) Annu. Rev. Cell Dev. Biol. 20,309–335 [DOI] [PubMed] [Google Scholar]
- 9.Söllner T., Pfaller R., Griffiths G., Pfanner N., Neupert W. (1990) Cell 62,107–115 [DOI] [PubMed] [Google Scholar]
- 10.Söllner T., Griffiths G., Pfaller R., Pfanner N., Neupert W. (1989) Cell 59,1061–1070 [DOI] [PubMed] [Google Scholar]
- 11.Brix J., Dietmeier K., Pfanner N. (1997) J. Biol. Chem. 272,20730–20735 [DOI] [PubMed] [Google Scholar]
- 12.Chan N. C., Likiæ V. A., Waller R. F., Mulhern T. D., Lithgow T. (2006) J. Mol. Biol. 358,1010–1022 [DOI] [PubMed] [Google Scholar]
- 13.Wu Y., Sha B. (2006) Nat. Struct. Mol. Biol. 13,589–593 [DOI] [PubMed] [Google Scholar]
- 14.Young J. C., Hoogenraad N. J., Hartl F. U. (2003) Cell 112,41–50 [DOI] [PubMed] [Google Scholar]
- 15.Beddoe T., Lithgow T. (2002) Biochim. Biophys. Acta 1592,35–39 [DOI] [PubMed] [Google Scholar]
- 16.Brix J., Ziegler G. A., Dietmeier K., Schneider-Mergener J., Schulz G. E., Pfanner N. (2000) J. Mol. Biol. 303,479–488 [DOI] [PubMed] [Google Scholar]
- 17.Demand J., Lüders J., Höhfeld J. (1998) Mol. Cell. Biol. 18,2023–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Mol. Cell. Biol. 19,4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) Nat. Cell Biol. 3,93–96 [DOI] [PubMed] [Google Scholar]
- 20.Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Mol. Cell 20,525–538 [DOI] [PubMed] [Google Scholar]
- 21.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101,199–210 [DOI] [PubMed] [Google Scholar]
- 22.Bömer U., Pfanner N., Dietmeier K. (1996) FEBS Lett. 382,153–158 [DOI] [PubMed] [Google Scholar]
- 23.Koh J. Y., Hájek P., Bedwell D. M. (2001) Mol. Cell. Biol. 21,7576–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo-Okamoto N., Shaw J. M., Okamoto K. (2008) EMBO Rep. 9,63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beddoe T., Bushell S. R., Perugini M. A., Lithgow T., Mulhern T. D., Bottomley S. P., Rossjohn J. (2004) J. Biol. Chem. 279,46448–46454 [DOI] [PubMed] [Google Scholar]
- 26.Chen L., Sigler P. B. (1999) Cell 99,757–768 [DOI] [PubMed] [Google Scholar]
- 27.CCP4 ( 1994) Acta Crystallogr. D Biol. Crystallogr. 50,760–763 [DOI] [PubMed] [Google Scholar]
- 28.Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Prot. 3,1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60,2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57,122–133 [DOI] [PubMed] [Google Scholar]
- 31.Carson M. (1997) Methods Enzymol. 277,493–505 [PubMed] [Google Scholar]