Abstract
Available evidence shows that short amidated neuropeptides are widespread and have important functions within the nervous systems of all flatworms (phylum Platyhelminthes) examined, and could therefore represent a starting point for new lead drug compounds with which to combat parasitic helminth infections. However, only a handful of these peptides have been characterised, the rigorous exploration of the flatworm peptide signalling repertoire having been hindered by the dearth of flatworm genomic data. Through searches of both expressed sequence tags and genomic resources using the basic local alignment search tool (BLAST), we describe 96 neuropeptides on 60 precursors from 10 flatworm species. Most of these (51 predicted peptides on 14 precursors) are novel and are apparently restricted to flatworms; the remainder comprise nine recognised peptide families including FMRFamide-like (FLPs), neuropeptide F (NPF)-like, myomodulin-like, buccalin-like and neuropeptide FF (NPFF)-like peptides; notably, the latter have only previously been reported in vertebrates. Selected peptides were localised immunocytochemically to the Schistosoma mansoni nervous system. We also describe several novel flatworm NPFs with structural features characteristic of the vertebrate neuropeptide Y (NPY) superfamily, previously unreported characteristics which support the common ancestry of flatworm NPFs with the NPY-superfamily. Our dataset provides a springboard for investigation of the functional biology and therapeutic potential of neuropeptides in flatworms, simultaneously launching flatworm neurobiology into the post-genomic era.
Keywords: Bioinformatics, Helminth, Neuropeptide precursor, npp
1. Introduction
The nervous system occupies a position of pivotal importance in flatworm biology. In addition to carrying sensory and neuromuscular signals, it may be responsible for the systemic transmission of developmental and hormonal cues, because as acoelomates these organisms lack the body cavity and circulatory system which would otherwise contribute to such functions. Amidated neuropeptides display constitutive and widespread immunoreactivity in flatworms and are linked to roles in locomotion, reproduction, feeding and larval host-finding (see Marks and Maule, 2008 for review). Physiology studies demonstrate that neuropeptides have contractile and regulatory effects on flatworm muscle and other tissues (Day et al., 1994; Marks et al., 1997; Hrckova et al., 2004; Humphries et al., 2004; Kreshchenko et al., 2008). The breadth of these insights is remarkable given the paucity of structural data on endogenous flatworm neuropeptides – only eight native sequences have been described, defining two structural families: four FMRFamide-like peptides (FLPs; short bioactive peptides with C-terminal -RF.NH2 motifs) and four neuropeptide Fs (NPFs; 36–39 amino acid peptides with conserved C-terminal Y(−17)Y(−10)GRPRF.NH2 motif) (Curry et al., 1992; Maule et al., 1992, 1993, 1994; Johnston et al., 1995, 1996). FLPs and NPFs are functionally different, reflecting their distinct structures. Available evidence suggests that FLPs function at the flatworm neuromuscular synapse/junction, with a clear myoexcitatory effect on muscle strips and dispersed/individual muscle fibres in vitro (Day et al., 1994; Marks et al., 1997; Moneypenny et al., 2001). These monospecific effects suggest the presence of a single, muscle-based, FLP receptor (Day et al., 1994, 1997). Schistosome NPF displays a potent inhibition of forskolin-stimulated cAMP accumulation in schistosome homogenates (Humphries et al., 2004), indicating conservation of the archetypal neuropeptide Y (NPY) receptor signalling mechanism by NPF receptors. Additionally in regenerating turbellarians, NPF appears to stimulate the mitotic division of neoblasts, the “stem cells” involved in regenerating the head and associated neuromusculature after decapitation (Kreshchenko et al., 2008). This finding suggests that flatworm neuropeptides may have core functions in controlling neoblast proliferation during regeneration.
The importance of peptide signalling in flatworm biology implies that recently characterised neuropeptide receptors (G-protein coupled receptors, GPCRs) and associated signalling mechanisms (Omar et al., 2007) represent attractive targets for novel anthelmintics. Improved data on the primary structures of native neuropeptide ligands will provide a source of molecular templates to guide the development of peptidomimetics with potential as next-generation anthelmintics (Greenwood et al., 2005). Despite successful applications of mass spectrometry (MS) to identification of neuropeptides and neurohormones in several invertebrates (Husson et al., 2005; Behrens et al., 2008; Ma et al., 2008; Weaver and Audsley, 2008), no such studies have yet been performed on flatworms. MS-based neuropeptidomic studies are performed on either dissected neural cells/tissues, or gram-scale homogenisations of whole worms (Hummon et al., 2006). Currently, parasitic flatworms preclude both of these methods: the former because flatworms are acoelomate, compromising the selective recovery of neural tissues from surrounding parenchyma and thereby offering source tissues with a very low neural to non-neural tissue ratio; the latter because parasitic flatworms must be cultured in animal hosts, and are therefore extremely difficult to obtain in large quantities. Coupled to these technical difficulties, flatworm neurobiology has suffered from a scarcity of bioinformatic datasets such that knowledge of flatworm neuropeptide diversity has remained static for ~15 years. Only recently have exploitable genomic and transcriptomic datasets provided the opportunity to expand this knowledge-base.
The primary aim of this work was to improve our understanding of native flatworm neuropeptides using in silico analyses of neuropeptide-encoding transcripts. The approach to their discovery utilised web-based basic local alignment search tool (BLAST) searches to identify potential neuropeptide precursors in expressed sequence tag (EST) and genomic databases that are available for many parasitic species well as a number of free-living forms that serve as model organisms (Dugesia ryukyuensis, Macrostomum lignano and Schmidtea mediterranea). Our initial approach using BLAST searches was followed up by manual curation/inspection and led to the identification of 60 neuropeptide precursors incorporating 96 short peptide transmitters in 10 species of flatworm. Two of the predicted mature peptides were chosen for immunodetection and shown to be expressed in schistosomes.
2. Materials and methods
2.1. Bioinformatics
An approach based on using BLAST searches (Altschul et al., 1990) was employed to identify putative neuropeptides from phylum Platyhelminthes. In an initial specific approach, known invertebrate neuropeptides flanked by “KR” dibasic cleavage sites were used as search queries. These included adipokinetic and related hormones, allatostatins, bombesins, corazonins, diuretic and antidiuretic hormones, myosuppressins, pigment dispersing factor peptides, proctolins, prothoracicotropic hormones, pyrokinins, sulfakinins, tachykinins and related peptides, and vertebrate opioid peptides. All of these search sequences were derived from multiple source species (Supplementary Fig. S1), but provided no positive matches in flatworms. Latterly, a novel approach using BLAST searches was employed using “degenerate” search strings, designed around the core features of small amidated peptide precursors. Degenerate search strings were constructed according to the template KRX3–11GKRX3–11GKRX3–11GKRX3–11GKR, where KR represents prohormone convertase-specific cleavage sites, X3–11 represents the variable length (three to 11 amino acids) of small propeptides, while G represents the Glycine (Gly) residue which will be processed for C-terminal amidation. In both approaches, search strings were applied to tBLASTn searches of available platyhelminth ESTs in the Genbank database (www.ncbi.nlm.nih.gov) using the National Center for Biotechnology Information (NCBI) BLAST server, as well as the Schistosoma mansoni (www.genedb.org/genedb/smansoni) and S. mediterranea (http://smedgd.neuro.utah.edu) genome databases, and two M. lignano EST datasets curated in-house at the University of Innsbruck (http://flatworm.uibk.ac.at/mac-est/). Expect values >100,000 were employed to account for the short length of many of the queries. Returns were translated (http://www.expasy.ch/tools/dna.html) and manually examined for neuropeptide motifs and cleavage sites. Cleavage site prediction was supplemented by the ProP 1.0 server (http://www.cbs.dtu.dk/services/ProP/) (Duckert et al., 2004). Finally, each prepropeptide sequence was analysed for the presence of an N-terminal secretory signal peptide using the SignalP 3.0 online server (http://www.cbs.dtu.dk/services/SignalP) (Emanuelsson et al., 2007). Sequences were examined for similarity with other genes/proteins using BLASTp and BLASTx tools (http://www.ncbi.nlm.nih.gov/BLAST), as well as InterProScan (http://www.ebi.ac.uk/InterProScan) (Zdobnov and Apweiler, 2001). Finally, transcripts were scored out of five according to how many of the following criteria they fulfilled: (1) Incorporate single or multiple copies of peptide intermediates; (2) Are flanked by mono- or dibasic (K, R, KR, KK or RR) cleavage sites; (3) Possess a C-terminal Gly residue, indicative of post-translational processing to confer amidation in the mature peptide; (4) Possess a hydrophobic N-terminal secretory signal peptide sequence; (5) Display sequence similarity to characterised neuropeptides. Scores can be seen in Supplementary Figs. S2–S26.
2.2. Immunocytochemistry (ICC)
Polyclonal primary antisera were generated by Genosphere, France (anti-GFVRIamide), EZBiolab, USA (anti-AAYMDLPWamide), or in-house (anti-FMRFamide, and anti-NPF, the latter raised against the C-terminal decapeptide, YFAIIGRPRFamide, of Moniezia expansa NPF) in New Zealand white rabbits. Adult S. mansoni, supplied by Dr. Fred Lewis, Biomedical Research Institute, Rockville, MD, USA, were flat-fixed between microscope slides immersed in 4% (w/v) paraformaldehyde (PFA) in 0.1 M PBS (pH7.4), for 4 h at 4°C, then stored in antibody diluent (AbD, 0.1 M PBS containing 0.1% (v/v) Triton X-100, 0.1% (w/v) BSA and 0.1% (w/v) NaN3) at 4°C. Fixed worms were incubated in primary antisera (diluted 1/200– 1/1600 in AbD) for 96 h, followed by a 24 h wash in AbD. Secondary antiserum (goat-anti-rabbit IgG, conjugated to FITC, Sigma Aldrich), diluted 1/100 in AbD, was applied for 48 h, followed by another 24 h wash in AbD. Finally, worms were incubated in phalloidin-TRITC (Sigma Aldrich) diluted 1/100 in AbD for 72 h. After a final 24 h wash, worms were mounted in PBS/glycerol (1:9 (v/v)) and viewed on a Leica TCS AOBS SP2 confocal scanning laser microscope. All incubation and wash steps were performed at 4°C. Controls included omission of primary antisera and pre-adsorption of primary antisera with relevant antigens. Cross reactivity controls were performed by preadsorbing antisera with similar antigens. Images were labelled and plates assembled using Adobe Photoshop software.
3. Results
3.1. Overview
Using a search strategy based on the BLAST algorithm, we believe this is the first study to describe a global dataset of neuropeptide-encoding transcripts from phylum Platyhelminthes. We identified 60 prepropeptide transcripts (detailed in Supplementary Figs. S2–S26) from 10 species of parasitic and free-living flatworms, which we have grouped by sequence similarity into 25 sequelogs (Table 1). Sequelogs are genes which encode peptides of similar sequence among different species (Varshavsky, 2004), recognising the sequence similarity of peptides present in multiple species, without implying (or dismissing) any functional or evolutionary relationship between peptides. This system has previously been used to describe nematode FLPs and neuropeptide-like proteins (NLPs) (McVeigh et al., 2005, 2008). From the precursors we predicted a total of 96 distinct novel neuropeptides in the phylum as a whole, representing nine structural families of mature peptide (Tables 1 and 2). In individual species, peptide complements range from single peptides in Echinococcus granulosus, Opisthorchis viverrini, and Clonorchis sinensis to 33 peptides identified in M. lignano.
Table 1.
Neuropeptide precursor (NPP) complement of the phylum Platyhelminthes. Distributions of npp genes across flatworm species are detailed, as well as the motifs and proposed similarities of NPP peptides. Numbers in species columns indicate transcript score out of the five criteria described in Materials and methods. Sequences are derived from expressed sequence tags (ESTs) except where indicated.
| Structural family |
npp gene |
Encoded peptide motifa | Most similarity |
Schistosoma mansoni |
Schistosoma japonicum |
Macrostomum lignano |
Schmidtea mediterranea |
Dugesia ryukyuensis |
Dugesia japonica |
Echinococcus granulosus |
Opisthorchis viverrini |
Taenia solium |
Clonorchis sinensis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FMRFamide-like peptides(FLPs) | 3b | AI[L/V]LTR[F/Y]G | 4 | ||||||||||
| 4b | -[V/A]FR[F/Y]G | Cestode GNFFRFamide | 5 | ||||||||||
| 11 | -N[T/M]RW[P/H/T]SR[L/F/W][G/L] | 3 | |||||||||||
| 13c | -F[M/V]PQRFG | Vertebrate neuropeptide FF | 5 | ||||||||||
| 19b;c | FXoF[N/D]L[R/K]DTRWG NXXIYXXESGXRXNXGXRFG | 3 | 4 | 3 | 3 | ||||||||
| 23 | YMRFG | Turbellarian YIRFamide; Molluscan FMRFamide | 5 | ||||||||||
| L/M/Iamides | 1c | -[S/G/A][F/Y]VR[L/M/I]G | 4 | 4 | 4 | 4 | |||||||
| 2b;c | RGXoIG | 4 | 4 | 3 | 4 | ||||||||
| 6b;c | [A/L]XX[L/S]XoRLG | Molluscan myomodulin | 5 | 5 | 4 | 4 | 5 | 5 | |||||
| 7 | EWQ[R/L]GSRLG | 4 | |||||||||||
| 9 | GAYSGFLG | Molluscan buccalin | 5 | ||||||||||
| 14c | GLRNMRMG | 3 | 4 | ||||||||||
| 15c | VQFLRLG SAYPYVG | 4 | 4 | ||||||||||
| 16c | -YLWD[V/T]RLG | 3 | 4 | ||||||||||
| 22 | A[K/S]Y[F/I]R[L/M]G | 4 | 4 | ||||||||||
| 24 | -[G/R]LLG | 4 | |||||||||||
| 25 | A[Y/W]YXSPRLG | 4 | |||||||||||
| Unamidated peptides | 21 | APFW | Molluscan APGWamide | 4 | |||||||||
| 10 | -TPS[Y/D]SW | 2 | |||||||||||
| Neuropeptide F (NPF) | 20b;cd | Y(−17)Y(−10)GRPR[F/Y]G | NPF/Neuropeptide Y superfamily | 5 | 5 | 5 | 5 | 5 | |||||
| PWamides | 5b;c | -[A/N][Y/W]X[D/V][M/L/V]PWG | 4 | 4 | 4 | 4 | 4 | ||||||
| SGFamides | 8b | -Q[R/A]W[S/Y]SG[F/Y]G | 3 | 3 | |||||||||
| D5amides | 17b;c | DX3[G/F]G | 3 | 3 | |||||||||
| Amidated tripeptides | 12b | NY[Y/F]G | 3 | 3 | |||||||||
| VVamides | 18b | GAEFFIRRVVG | 3 | 4 |
Single letter annotation for amino acids: X, variable amino acid; Xo, hydrophobic amino acid; residues in parentheses are alternatives for that position; subscript numbers indicate the number of intervening amino acids; a hyphen at N-terminus indicates variable (sequence/length) extensions.
Sequelog represented in S. mediterranea genome database.
Sequelog represented in S. mansoni genome database.
NPF-like peptides are encoded on multiple genes in S. mansoni, M. lignano and S. mediterranea, see Supplementary Figs. S2–S26 for details.
Table 2.
Mature peptides predicted from flatworm neuropeptide precursor (npp) genes. Peptides were predicted from the prepropeptides detailed in Supplementary Figs. S2–S26. Where there was ambiguity in cleavage site identification, these were selected according to criteria described by McVeigh et al. (2005). Note that NPP-20 peptides are omitted; these are detailed in Fig. 1
| Schistosoma mansoni | Sj-npp-15 | Ml-npp-21 | Smed-npp-12 | ||||
| Sm-npp-1 | VQFLRLamide | 13x | APFW | 3x | NYFamide | ||
| AFVRLamide | SAYPYVamide | 2x | DPFW | Smed-npp-18 | |||
| 2x | GFVRIamide | Sj-npp-16 | Ml-npp-22 | 2x | GAEFFIRRVVamide | ||
| Sm-npp-2 | NYLWDTRLamide | 8x | ASYIRMamide | Smed-npp-19 | |||
| GMIamide | SYLWDVRLamide | Ml-npp-23 | FYFDLRDTRWamide | ||||
| Sm-npp-5 | Sj-npp-17 | 10x | YMRFamide | Smed-npp-22 | |||
| AAYMDLPWamide | DDFRGamide | Ml-npp-24 | 3x | AKYFRLamide | |||
| AAYIDLPWamide | DHRPFamide | GAYYGLLamide | Dugesia ryukyuensis | ||||
| Sm-npp-6 | Sj-npp-19 | GPYHRLLamide | Dr-npp-2 | ||||
| AVRLMRLamide | FLFNLRDTRWamide | 6x | AGYRGLLamide | 5x | GLIamide | ||
| Sm-npp-13 | NADIYESESGPRHNIGNRFamide | 4x | AGFRGLLamide | 2x | GMIamide | ||
| HFMPQRFamide | Macrostomum lignano | VYQRLLamide | Dr-npp-5 | ||||
| YTRFVPQRFamide | Ml-npp-1 | Ml-npp-25 | PNWKDMPWamide | ||||
| Sm-npp-14 | PSFVRMamide | AYYASPRLamide | 3x | SAWRDMPWamide | |||
| GLRNMRMamide | ASYVRMamide | 7x | AWYVSPRLamide | GAWRDMPWamide | |||
| Sm-npp-15 | Ml-npp-5 | AYYKSPRLamide | NAWRDMPWamide | ||||
| VQFLRLamide | 7x | AYGVVPWamide | Schmidtea mediterranea | Dr-npp-10 | |||
| SAYPYVamide | AYGAVPWamide | Smed-npp-1 | 5x | TMTPSYSW | |||
| Sm-npp-16 | Ml-npp-6 | SFVRLamide | 3x | TPSYSW | |||
| NYLWDTRLamide | 3x | AMRLMRLamide | 3x | ASFVRLamide | 4x | NTMTPSYSW | |
| Sm-npp-17 | AMPLMRLamide | Smed-npp-2 | NTMTPSYSWI | ||||
| DDFRGamide | Ml-npp-7 | 7x | GLIamide | 4x | ITMTPSDSW | ||
| DHRAFamide | NAWERGSRLamide | Smed-npp-3 | Dr-npp-18 | ||||
| Sm-npp-19 | 4x | EWQRGSRLamide | AILLTRYamide | GAEFFIRRVVamide | |||
| FLFNLKDTRWamide | EWQRGSRLS | AIVLTRFamide | Dugesia japonica | ||||
| NHGSRFamide | EWQLGSRLS | Smed-npp-4 | Dj-npp-12 | ||||
| Schistosoma japonicum | Ml-npp-8 | SSVFRFamide | NYYamide | ||||
| Sj-npp-1 | 2x | PDQRWSSGFamide | SVAFRFamide | NYFamide | |||
| AFVRLamide | 3x | AGQRWSSGFamide | RGVAFRFamide | Echinococcus granulosus | |||
| GFVRLamide | AGQRWSSGFV | GSVFRYamide | Eg-npp-6 | ||||
| GFVRIamide | Ml-npp-9 | QSVFRYamide | AIRSLRLamide | ||||
| Sj-npp-2 | 11x | GAYSGFLamide | Smed-npp-5 | opisthorchis viverrini | |||
| GMIamide | Ml-npp-11 | PNWKDMPWamide | ov-npp-6 | ||||
| GFMamide | WNTRWTSRLamide | 5x | SAWRDMPWamide | GLRQLMRLamide | |||
| Sj-npp-5 | 3x | WNTRWPSRLamide | Smed-npp-6 | clonorchis sinensis | |||
| AAYMDLPWamide | WNTRWPSRSL | AYRLMRMamide | cs-npp-19 | ||||
| AAYIDLPWamide | WNMRWHSRFamide | AVRLMRMamide | FFFDLRDTRWamid | ||||
| Sj-npp-6 | 3x | NTRWPSRFamide | AVRLMRLamide | ||||
| AVRLMRLamide | 2x | NMRWPSRLamide | Smed-npp-8 | ||||
| Sj-npp-14 | WNTRWPSRFamide | DPRFSDQVWHSGYamide | |||||
| GLRNMRMamide | WNTRWPSRWL | NYYNRFDGAWYSGYamide | |||||
| NTRWPSRLamide | NDAFDGAWYSGYamide | ||||||
| NYYNRFDGQAWYSGYamide | |||||||
| NDAFDGQAWYSGYamide | |||||||
We propose the title neuropeptide precursor (npp) for flatworm neuropeptide genes, and have numbered them consecutively as sequelogs, with a species-specific prefix (e.g. Sm for S. mansoni, Smed for S. mediterranea; these are detailed in Supplementary Figs. S2–S26). Our numbering system reflects only the order of discovery, and is open-ended to acknowledge the possibility of future discoveries. Since we consider the phylum as a whole, this system imposes a situation where gene numbers are currently not contiguous in most species, but we anticipate that further gene discovery/genome sequencing will fill many of the gaps in Table 1. All candidate secretory peptide precursors were assessed using the five criteria described in Materials and methods. The strongest candidates have a score of 5/5, with >70% of transcripts fulfilling at least four of the five criteria. All similarity and identity values that follow are expressed as the mean percentage value across each comparable amino acid position between all predicted peptides. Precursors of all the peptides referred to below are illustrated in Supplementary Figs. S2–S26, together with their database accession numbers.
3.2. Some NPPs resemble characterised flatworm, molluscan and/or deuterostome neuropeptides
Two npp transcripts encoded peptides remarkably similar to some previously characterised flatworm neuropeptides. A transcript from M. lignano (Ml-npp-23) encoded 10 copies of YMRFamide (Table 2; Supplementary Fig. S24), a peptide almost identical to both the planarian FLP, YIRFamide, and molluscan FMRFamide (Price and Greenberg, 1977; Johnston et al., 1996). Additionally, a set of peptides bearing a [V/A]FR[F/Y]amide motif, encoded by Smed-npp-4 in S. mediterranea (Table 2; Supplementary Fig. S5), bears notable similarity (57% identity, 86% similarity) to GNFFRFamide, a cestode FLP from M. expansa (Maule et al., 1993), although these also resemble molluscan LFRFamides (53% identity, 80% similarity) (Hoek et al., 2005). One of our FLP-encoding transcripts from S. mansoni (Sm-npp-13) contained two peptides with a C-terminal PQRFamide signature (Table 2; Supplementary Fig. S14) identical to that of vertebrate NPFF and related peptides. Since our methods did not identify similar peptides in any other invertebrate species, this represents the first description of NPFF-like peptides in any invertebrate.
Supplementing the known NPF-encoding cDNAs from S. mansoni and Schistosoma japonicum (Humphries et al., 2004), and characterised peptides from M. expansa (Maule et al., 1992) and Arthurdendyus triangulatus (formerly Arthioposthia triangulata) (Dougan et al., 2002), we identified novel NPF precursors in the cyclophyllidean cestode Taenia solium (Ts-npp-20), and the turbellarians M. lignano (Ml-npp-20) and S. mediterranea (Smed-npp-20) (Table 1; Fig. 1). In total, NPF-encoding npp-20 transcripts were identified in seven species. Interestingly, we found multiple NPFs in three species (Fig. 1; Supplementary Fig. S21): these include three distinct NPF-like transcripts in S. mediterranea (Smed-npp-20a, -20b and -20c), two distinct NPF-like precursors in M. lignano (Ml-npp-20a and -20b) and a novel second NPF precursor in S. mansoni (Sm-npp-20b). These represent the first descriptions of multiple NPF genes per se in any invertebrate; they are quite distinct from the so-called ‘short NPFs’ of arthropods which are less than 12 amino-acids long and lack the key features of NPY superfamily peptides (Hewes and Taghert, 2001; Huybrechts et al., 2004; Garczynski et al., 2007; Christie, 2008; Ma et al., 2008). All of the NPF peptides that we identified were 36-39 amino acids long and displayed a C-terminal Y(−17)Y(−10)RPR[F/Y]amide motif. Additionally, Ml-NPP-20a, Ml-NPP-20b, Sm-NPP-20b and Smed-NPP-20c displayed an N-terminal PP[E/A][R/K]P motif, resembling the polyprolyl PXXPXXP domain of vertebrate NPY superfamily peptides. The six available genomic DNA sequences from S. mansoni, S. mediterranea and M. expansa all displayed conserved phase 2 introns within the codons of the penultimate arginine residues of the mature peptides, another feature common to vertebrate NPY superfamily genes (Fig. 1).
Fig. 1.
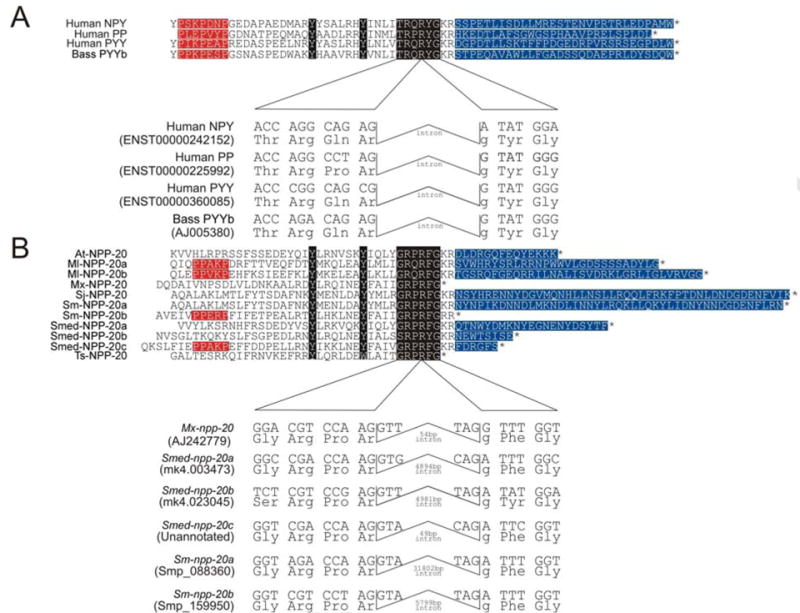
Similarities of flatworm neuropeptide Fs (NPFs) with the vertebrate neuropeptide Y (NPY) superfamily. (A) Alignment of NPY superfamily members from selected species; PP, pancreatic polypeptide; PYY, peptide tyrosine-tyrosine; PY, peptide tyrosine. Position of conserved intron is indicated. Human sequences obtained from Ensembl (www.ensembl.org). (B) Alignment of all 11 known flatworm NPFs (Neuropeptide Precursor-20, NPP-20) identified by this study and previous work (Mair et al., 2000; Dougan et al., 2002). In (A) and (B), predicted mature peptide sequences are shown, as are C-terminal cleavage recognition sites and/or translation stop signals (*), as well as the C-flanking peptide of NPY (CPON, denoted by blue or dark grey highlighting) where present. Conserved residues characteristic of NPY superfamily members are highlighted in black; polyprolyl domains are highlighted in red (or underlined). Intronic architecture of available genomic npp-20 sequences is displayed. Genomic accession numbers are provided; relevant databases are detailed in Materials and methods. Moniezia expansa sequence reported in Mair et al. (2000). Intron sizes and positions are indicated. At, Arthurdendyus triangulatus (formerly Artioposthia triangulata); Ml, Macrostomum lignano; Mx, Moniezia expansa; Smed, Schmidtea mediterranea; Sj, Schistosoma japonicum; Sm, Schistosoma mansoni; Ts, Taenia solium.
We identified npp-6 in five species, encoding peptides with [A/L]XX[L/S]XoRLamide motifs (where X denotes a variable residue, and Xo denotes a hydrophobic residue) displaying structural similarities with molluscan myomodulin (Tables 2; Supplementary Fig. S7). Myomodulin was discovered in molluscs (Cropper et al., 1987), but similar peptides have been recognised in other invertebrates (Christie et al., 1994; Takahashi et al., 1994; Wang et al., 1998; Nathoo et al., 2001). We believe our study provides the first description of myomodulin-like peptides in flatworms. Conservative amino acid differences from molluscan myomodulin (Lymnaea stagnalis -MLRLamides) distinguish the NPP-6 peptides from M. lignano (AM[R/P]LMRLamide; 46% identity, 80% similarity), S. mansoni and S. japonicum (AVRLMRLamide; 38% identity, 88% similarity), E. granulosus (AIRSLRLamide; 55% identity, 70% similarity) and O. viverrini (GLRQLMRLamide; 36 % identity, 64 % similarity). Buccalin is another peptide first identified in molluscs but recognised in other invertebrates (Cropper et al., 1988; Nathoo et al., 2001). The M. lignano Ml-npp-9 precursor incorporates 11 copies of a single peptide (GAYSGFLamide) (Table 2; Supplementary Fig. S10), which resembles Aplysia californica buccalin (GMDSLAFSGGLamide, 42% identity, 67% similarity). We were unable to identify npp-9 from any other flatworm species. We believe this finding represents the first description of a putative buccalin-like peptide in phylum Platyhelminthes.
A single transcript from M. lignano (Ml-npp-21) encoded 15 copies of the tetrapeptide [A/D]PFW (Table 2; Supplementary Fig. S22). These peptides, although not predicted to be amidated since they lack the C-terminal Gly donor, resemble the APGWamides (50% identity, 70% similarity) of the molluscan central nervous system, which are themselves similar to crustacean red pigment concentrating hormone (RPCH) (Kuroki et al. 1990).
3.3. npps are represented in available genomic datasets
We used BLAST searches to interrogate all of our EST-derived NPP sequences against S. mansoni and S. mediterranea genomic data, which represented the two flatworm genome sequences available at the time of our searches (LoVerde et al., 2004; Robb et al., 2008). Using the BLAST tools hosted on the respective genome websites we were able to identify genomic sequences for several of our precursors (Table 1). A total of 11 NPP precursors were identified in S. mansoni, including three which were solely genome-derived (Sm npp-14 through -16), with no representative ESTs (although these sequences are represented by S. japonicum ESTs). The remaining eight npp sequences had dual representation in the S. mansoni genome and on ESTs. S. mediterranea has 13 npp sequelogs, three of which (Smed-npp-1, -21, and -22) are only found on ESTs, and five of which are solely represented in the genome (Smed-npp-5, -6, -12, -17, and -18). The remaining five Schmidtea sequelogs display both genomic and EST representatives.
3.4. GFVRIamide and AAYMDLPWamide localise to the schistosome nervous system
To verify the neuropeptide status of our predicted peptides, we raised polyclonal antisera to two novel schistosome peptides – Sm-NPP-1 (GFVRIamide) and Sm-NPP-5 (AAYMDLPWamide) – and used those in standard ICC approaches. Both sera demonstrated strong and specific immunoreactivities (IR) in elements of the S. mansoni nervous system (Fig. 2). GFVRIamide (Fig. 2A) displayed restricted expression, localising to a pair of CNS neurones within the brain ganglia which projected posteriorly along the main nerve cords, but was not apparent in any of the peripheral innervation to the somatic muscle or organs. In contrast, AAYMDLPWamide-IR (Fig. 2B) was almost exclusively peripheral in location, with IR visibly branching from the ventral nerve cords to innervate the somatic muscle on the worm flanks, as well as the oral and ventral suckers, the oesophagus and the female reproductive system.
Fig. 2.
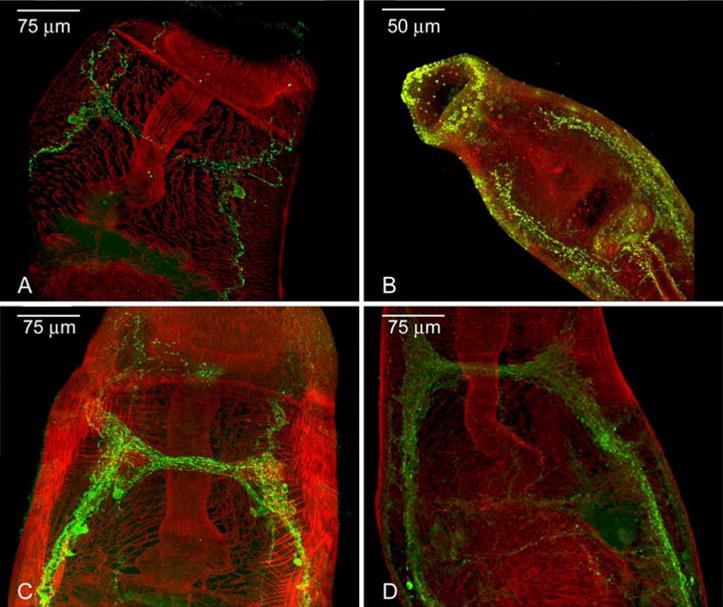
Immunocytochemical localization of neuropeptides in anterior of adult Schistosoma mansoni. (A) GFVRIamide-immunoreactivity (IR) is restricted to two brain neurons which project across the cerebral commissure, anteriorly towards the oral sucker, and posteriorly along the main nerve cords; (B) AAYMDLPWamide-IR appears in peripheral nerves innervating the somatic musculature, oesophagus and oral and ventral suckers; both neuropeptide F-IR (C) and FMRFamide-IR (D) are apparent in the brain, central and peripheral nerves. C and D are shown for comparison of A and B with known neuropeptide expression patterns. In all images, anterior is towards the top of the page; green staining (FITC) represents peptide immunoreactivity, while red staining represents filamentous actin in muscle detected by TRITC-conjugated phalloidin.
4. Discussion
We believe that this study reports the first description of new flatworm neuropeptides in ~15 years and represents a major expansion in our knowledge of flatworm neuropeptide biology. We have described 25 npp sequelogs in phylum Platyhelminthes which encode putative neuropeptide precursor proteins grouped into nine distinct families of neuropeptide (Table 1). While most of the predicted peptides are completely novel, showing little similarity with peptides from other genera, several NPPs appear similar to characterised neuropeptides from molluscs and even vertebrates. Although none of the previously characterised flatworm FLPs were identified, some similar peptides were predicted. As well as Smed-npp-4 peptides (-[V/A]FR[F/Y]amides), which display some C-terminal similarity to the M. expansa peptide GNFFRFamide (Maule et al., 1993), we discovered YMRFamide, an M. lignano peptide encoded by Ml-npp-24. Previous structure-activity studies on YIRFamide (one of the most potently myoactive peptides tested in flatworm neuromuscular bioassays) showed that substitution of Isoleucine for Methionine at position 2 did not significantly alter the peptide’s potency on S. mansoni dispersed muscle fibres (Day et al., 1997), suggesting that YMRFamide should retain a myoexcitatory function. While YIRFamide/YMRFamide have not yet been identified outside of the Turbellaria, YIRFamide’s potent contractile effect on schistosome muscle fibres cannot be ignored, and suggests that a similar peptide could reside within the S. mansoni genome. In a rational approach to drug discovery, this peptide’s muscle-based receptor represents an attractive drug target. The absence of a YIRFamide-like peptide from the S. mansoni genome, as well as discrepancies in Schmidtea, (where Smed-npp-1, -22, and -23 are represented by ESTs but do not appear in the current genome database), raise an important point regarding the current levels of genome coverage in these projects. Despite the reported 7× coverage of the S. mansoni genome (LoVerde et al., 2004; Haas et al., 2007), and 10× coverage of the S. mediterranea genome (Cantarel et al., 2008), our data suggest that some stretches of these genomes remain unsequenced. It is possible that further npp genes will be uncovered as genome curation continues to shed new light on these areas.
The overwhelming presence of C-terminal glycine residues on the peptides in our dataset (Table 2) suggests that the vast majority will be C-terminally amidated. We deliberately focused our search protocol on amidated peptides because previous experiments illustrate that C-terminal amidation appears to be one of the key structural components required for maintenance of bioactivity of flatworm neuropeptides (Day et al., 1997). However, in other organisms (particularly Caenorhabditis elegans), many non-amidated peptides are present and appear to play important roles in worm biology. While the identifications of Ml-npp-22 and Dr-npp-10, which appear to lack an amidation site, illustrate that our search protocol was capable of uncovering at least some non-amidated peptides, one should note the possibility that further non-amidated peptides await discovery in flatworms.
While several NPPs resemble neuropeptides originally described in flatworms, molluscs and even vertebrates, the majority of predicted peptides (52 putative peptides encoded by 15 sequelogs) are novel and show little or no obvious sequence similarity with neuropeptides from other genera (Tables 1 and 2). Such pathogen-specific neuropeptides, in their absence from host species, represent particularly promising avenues for drug development. Provided that their endogenous receptors are ligand selective, drugs targeting these receptors should be pathogen-specific and less likely to exhibit host-toxicity through non-specific interactions with host receptors. All of these transcripts score at least 3/5 on our criteria-based system (completely novel peptides can only score a maximum of 4/5, see Materials and methods section), indicating that they are strong candidates for consideration as secretory peptide precursors. The nematode C. elegans represents an analogous system which illustrates the validity of our method of bioinformatics-based gene identification/peptide prediction. Most of the currently known C. elegans neuropeptide genes were initially identified by similarity-based sequence searches similar to the methods used in the current study (Nelson et al., 1998; Li et al., 1999a, 1999b). To date, the majority of these have been validated by neuronal localisation, knockout analysis and/or peptide physiology – none of those original discoveries have been proven to represent “false positives”. Most of our genome-derived peptides are also represented by ESTs, strengthening the validity of their prediction. Even S. mansoni genes Sm-npp-14 through -18, while not represented by S. mansoni ESTs, are indirectly suppported by S. japonicum ESTs, suggesting that these genes are probably transcribed.
The main issue encountered in our analyses was the absence of identifiable signal peptides from some precursors (npp-8, -10, -11, -12, and -17). In all of these cases, the associated ESTs represent 3′-directed (or otherwise partial) sequences which do not support full-length open reading frames (ORFs) with an N-terminal signal peptide. Mono- and di-basic cleavage sites were used to predict mature peptide sequences, using both manual annotation and the ProP 1.0 server (http://www.cbs.dtu.dk/services/ProP/) (Duckert et al., 2004). It should be noted that until MS methods are developed for, and applied to, flatworms to confirm the presence of ions corresponding to our predicted peptides, those predictions remain equivocal. In the absence of an established neuropeptide comparator with which to validate their status at the level of primary structure, other methods must be used to confirm their identity as neuropeptides. To this end, ICC or in situ hybridisation experiments are required to localise expression of npp peptides or transcripts to secretory tissues. As a first step, we raised Sm-NPP-1 (GFVRIamide) and Sm-NPP-5 (AAYMDLPWamide) antisera; both revealed strong and specific IRs in the schistosome nervous system, with distinct staining patterns, confirming these motifs as neuropeptides and strengthening the validity of our approach to neuropeptide discovery and annotation.
Schistosome Sm-npp-13 encoded two peptides with 100% C-terminal identity (PQRFamide) to NPFF-like peptides. In vertebrates, NPFF (and the closely-related peptides NPAF and NPSF) are thought to be involved in nociception and opiate-induced analgesia (Yang et al., 2008). Peptides with similar motifs exist throughout vertebrates, including mammalian RFamide-related peptides (RFRPs), gonadotropin inhibitory hormone (GnIH) and frog growth hormone releasing peptide (fGHRP), all of which possess PQRFamide or similar motifs (LPXRFamide). Prior to this study, PQRFamides had only been identified in vertebrates, including species belonging to ancient vertebrate lineages (e.g. the agnathan Petromyzon marinus (Osugi et al. 2006)). Our identification of PQRFamides in phylum Platyhelminthes represents the first description of these peptides in invertebrates. In light of current opinion on the molecular phylogeny of animal genera (Ruiz-Trillo et al., 1999; Philippe et al., 2005), the NPFF progenitor may have arisen in a shared ancestor of Lophotrochozoa and Deuterostomia, which would entail the loss of NPFF from Ecdysozoans. Precedent exists for such a relationship; genes have been described which are shared between S. mansoni and Deuterostomia but absent from Ecdysozoa (Venancio et al., 2007), these are thought to have been “co-opted” by schistosomes, perhaps by horizontal gene transfer, to perform a role in host-parasite interactions. It is interesting to consider such a role for NPP-13 in schistosomes since NPFF has been shown to stimulate or inhibit T-cell proliferation at low or high concentrations, respectively (Lecron et al., 1992; Minault et al., 1995). The ability to modulate the proliferation of T-cells by the release of Sm-NPP-13 would have obvious survival benefits for an intravascular parasite. However, until Sm-NPP-13 expression is demonstrated in a locale consistent with excretory/secretory release from schistosomes, such a hypothesis remains speculative.
The vertebrate NPY superfamily encompasses four similar, highly conserved peptides that share a common evolutionary origin – NPY, peptide YY (PYY) and pancreatic polypeptide (PP) in tetrapods, and pancreatic peptide Y (PY; now considered to be a duplicate of PYY and renamed PYYb) in fish (Sundstrom et al., 2008). Many invertebrates possess similar peptides which most commonly terminate in phenylalaninamide and are therefore known as NPFs. All NPFs exhibit structural correlates with NPY superfamily peptides - most notably, their C-terminal R[P/Q]R[F/Y]amide resembles the terminal RXR[Y/F]amide motif of the NPY family, while both NPF and NPY-like peptides display conserved tyrosyls at positions 10 and 17 from the C-terminus. At the N-terminus, not all identified flatworm NPFs share the characteristic and functionally important triprolyl (PXXPXXP) domain found in almost all NPY superfamily peptides (Day and Maule, 1999), clouding the relationship of NPF with NPY so that their common ancestry has been considered uncertain. Here, we identified NPF-like genes and peptides in four species of flatworm, four features of which support a common ancestry for NPF and NPY family peptides (Fig. 1): (i) Flatworms possess multiple NPF genes. The NPY family is thought to have arisen by sequential gene duplication events, which in a first instance generated NPY and PYY genes from a single ancestral coding sequence in teleost fishes followed by a local expansion of PYY to PY and PP (Sundstrom et al., 2008). We found three genes encoding NPF-like peptides in S. mediterranea (Smed-NPP-20a, -20b, and -20c), while S. mansoni and M. lignano both possessed two (Sm-NPP-20a, -20b and Ml-NPP-20a, -20b, respectively). All seven of the associated transcripts were amplified by reverse transcriptase (RT)-PCR, supporting their expression in adult worms (data not shown). While some evidence exists for a multiplicity of short peptides that resemble NPF in some invertebrates (four peptide YFs [PYFs] in the tiger prawn (Sithigorngul et al., 2002), and multiple variants of “short NPFs” in insects (Hewes and Taghert, 2001; Huybrechts et al., 2004; Garczynski et al., 2007; Christie, 2008; Ma et al., 2008), we believe our data represent the first description of multiple NPF-encoding genes or NPF-like peptides in any invertebrate. (ii) Some flatworm NPFs display N-terminal polyprolyl domains. S. mansoni Sm-NPP-20b, Schmidtea Smed-NPP-20c and Macrostomum Ml-NPP-20a and -20b, all display an N-terminal PPXXP motif, which resembles the PXXPXXP domain of NPY-family peptides. This domain is important in allowing NPY-family peptides to assume a functionally-important tertiary structure known as the PP fold (Glover et al., 1984). This tertiary structure has not yet been demonstrated in any NPF. (iii) Flatworm NPF genes share gene structure with the NPY family. All known NPY family peptides have a phase 2 intron within the codon for their penultimate arginine residue, which was also identified in the M. expansa NPF-encoding gene, but failed to amplify in a PCR-based approach in A. triangulatus and S. mansoni (Mair et al., 2000; Dougan et al., 2002; Humphries et al., 2004). Here, we have identified a further five genomic NPF sequences from the S. mansoni and S. mediterranea genomes, all of which display introns of variable size in the canonical position at phase 2 in the codon for the penultimate arginine (Fig. 1). (iv) Another typical feature of NPY family genes is the presence of a C-terminal extension of the NPF precursor protein (CPON, C-flanking peptide of NPY), immediately following the NPF peptide. Of the nine flatworm NPFs reported in this study, seven display a CPON domain on their precursor. These four factors, allied with the knowledge that schistosome NPF displays functional conservation of NPY’s archetypal cAMP-based signalling mechanism (Humphries et al., 2004), lend strong support to the hypothesis that vertebrate NPY family peptides are evolutionarily related to NPF.
Due to their etiological importance as disease-causing pathogens it is certainly the parasitic members of phylum Platyhelminthes that are of greatest concern to most humans. Therefore, an important application for these data is as a resource for those interested in anthelmintic discovery, since this is, to our knowledge, the first study to describe multiple neuropeptide ligands from flatworms which can be used to “deorphanise” peptide receptors (such as the recently-reported Girardia tigrina GPCR (Omar et al., 2007)) following their heterologous expression. Given the importance of neuropeptides in the core neuromuscular functions of parasitic flatworms, we anticipate that some neuropeptide receptors could represent attractive, rational targets against which new, non-peptide, anthelmintic drugs could be directed (Mousley et al., 2004); indeed, a rich seam of potential receptors await exploitation, since preliminary screening suggests that both S. mansoni and S. mediterranea genomes contain at least 20 neuropeptide-like GPCR genes (M. Zamanian, unpublished data). Attention must be paid to investigating the bioactivity of new neuropeptides if we are to validate and prioritise their receptors in terms of drug target potential. Various methods are available for functional characterisation of bioactive peptides, including established dispersed muscle-fibre bioassays (Day et al., 1994; Moneypenny et al., 2001), as well as newer techniques being investigated by our laboratories such as computer-based behavioural analyses of the effects of peptides on whole worms. Silencing of npp expression using RNA interference (RNAi) is another potential avenue of investigation since RNAi is now an established technique in several of the species represented in our dataset (Sanchez Alvarado and Newmark, 1999; Krautz-Peterson et al., 2007; Pfister et al., 2008), however, our attempts at silencing neuronal genes in numerous flatworm species have shown, at best, limited success (Atkinson L, McVeigh P, Pierson L, unpublished data). Nevertheless, RNAi remains the most powerful method available for investigating gene function in flatworms, and large-scale RNAi-based screening of neuropeptide and receptor genes must be attempted if we are to begin to dissect the intricacies of the flatworm nervous system. In describing 51 flatworm-specific putative neuropeptides, our study has significantly widened the scope for such functional investigations. We hope that the data presented in this study will catalyse much needed further research into the roles of individual neuropeptides and receptors in these important pathogens.
Supplementary Material
Acknowledgments
We wish to acknowledge the laboratories responsible for generating the publicly available genomic and transcriptomic resources for flatworms, without which this study would not have been possible. This work was funded by The National Institutes of Health grant ROI-AI49162. A research studentship (for LA) from the Department of Education and Learning for Northern Ireland and a Wellcome Trust equipment grant (069411) is also acknowledged.
Footnotes
NOTE: Supplementary data is associated with this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Behrens HL, Chen R, Li L. Combining microdialysis, NanoLC-MS, and MALDI-TOF/TOF to detect neuropeptides secreted in the crab, Cancer borealis. Anal Chem. 2008;80:6949–6958. doi: 10.1021/ac800798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE. Neuropeptide discovery in Ixodoidea: An in silico investigation using publicly accessible expressed sequence tags. Gen Comp Endocrinol. 2008 doi: 10.1016/j.ygcen.2008.03.027. [DOI] [PubMed] [Google Scholar]
- Christie AE, Hall C, Oshinsky M, Marder E. Buccalin-like and myomodulin-like peptides in the stomatogastric ganglion of the crab Cancer borealis. J Exp Biol. 1994;193:337–343. doi: 10.1242/jeb.193.1.337. [DOI] [PubMed] [Google Scholar]
- Cropper EC, Miller MW, Tenenbaum R, Kolks MA, Kupfermann I, Weiss KR. Structure and action of buccalin: A modulatory neuropeptide localized to an identified small cardioactive peptide-containing cholinergic motor neuron of Aplysia californica. Proc Natl Acad Sci U S A. 1988;85:6177–6181. doi: 10.1073/pnas.85.16.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropper EC, Tenenbaum R, Kolks MA, Kupfermann I, Weiss KR. Myomodulin: A bioactive neuropeptide present in an identified cholinergic buccal motor neuron of Aplysia. Proc Natl Acad Sci U S A. 1987;84:5483–5486. doi: 10.1073/pnas.84.15.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry WJ, Shaw C, Johnston CF, Thim L, Buchanan KD. Neuropeptide F: Primary structure from the tubellarian, Artioposthia triangulata. Comp Biochem Physiol C. 1992;101:269–274. doi: 10.1016/0742-8413(92)90272-9. [DOI] [PubMed] [Google Scholar]
- Day TA, Maule AG. Parasitic peptides! the structure and function of neuropeptides in parasitic worms. Peptides. 1999;20:999–1019. doi: 10.1016/s0196-9781(99)00093-5. [DOI] [PubMed] [Google Scholar]
- Day TA, Maule AG, Shaw C, Halton DW, Moore S, Bennett JL, Pax RA. Platyhelminth FMRFamide-related peptides (FaRPs) contract Schistosoma mansoni (trematoda: Digenea) muscle fibres in vitro. Parasitology. 1994;109:455–459. doi: 10.1017/s0031182000080707. [DOI] [PubMed] [Google Scholar]
- Day TA, Maule AG, Shaw C, Pax RA. Structure-activity relationships of FMRFamide-related peptides contracting Schistosoma mansoni muscle. Peptides. 1997;18:917–921. doi: 10.1016/s0196-9781(97)00073-9. [DOI] [PubMed] [Google Scholar]
- Dougan PM, Mair GR, Halton DW, Curry WJ, Day TA, Maule AG. Gene organization and expression of a neuropeptide Y homolog from the land planarian Arthurdendyus triangulatus. J Comp Neurol. 2002;454:58–64. doi: 10.1002/cne.10440. [DOI] [PubMed] [Google Scholar]
- Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Garczynski SF, Crim JW, Brown MR. Characterization and expression of the short neuropeptide F receptor in the african malaria mosquito, Anopheles gambiae. Peptides. 2007;28:109–118. doi: 10.1016/j.peptides.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover ID, Barlow DJ, Pitts JE, Wood SP, Tickle IJ, Blundell TL, Tatemoto K, Kimmel JR, Wollmer A, Strassburger W. Conformational studies on the pancreatic polypeptide hormone family. Eur J Biochem. 1984;142:379–385. doi: 10.1111/j.1432-1033.1984.tb08298.x. [DOI] [PubMed] [Google Scholar]
- Greenwood K, Williams T, Geary T. Nematode neuropeptide receptors and their development as anthelmintic screens. Parasitology. 2005;131(Suppl):S169–77. doi: 10.1017/S003118200500819X. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Berriman M, Hirai H, Cerqueira GG, Loverde PT, El-Sayed NM. Schistosoma mansoni genome: Closing in on a final gene set. Exp Parasitol. 2007;117:225–228. doi: 10.1016/j.exppara.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11:1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek RM, Li KW, van Minnen J, Lodder JC, de Jong-Brink M, Smit AB, Van Kesteren RE. LFRFamides: A novel family of parasitation-induced -RFamide neuropeptides that inhibit the activity of neuroendocrine cells in Lymnaea stagnalis. J Neurochem. 2005;92:1073–1080. doi: 10.1111/j.1471-4159.2004.02927.x. [DOI] [PubMed] [Google Scholar]
- Hrckova G, Velebny S, Halton DW, Day TA, Maule AG. Pharmacological characterisation of neuropeptide F (NPF)-induced effects on the motility of Mesocestoides corti (syn. Mesocestoides vogae) larvae Int J Parasitol. 2004;34:83–93. doi: 10.1016/j.ijpara.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Hummon AB, Amare A, Sweedler JV. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- Humphries JE, Kimber MJ, Barton YW, Hsu W, Marks NJ, Greer B, Harriott P, Maule AG, Day TA. Structure and bioactivity of neuropeptide F from the human parasites Schistosoma mansoni and Schistosoma japonicum. J Biol Chem. 2004;279:39880–39885. doi: 10.1074/jbc.M405624200. [DOI] [PubMed] [Google Scholar]
- Husson SJ, Clynen E, Baggerman G, De Loof A, Schoofs L. Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem Biophys Res Commun. 2005;335:76–86. doi: 10.1016/j.bbrc.2005.07.044. [DOI] [PubMed] [Google Scholar]
- Huybrechts J, De Loof A, Schoofs L. Diapausing colorado potato beetles are devoid of short neuropeptide F I and II. Biochem Biophys Res Commun. 2004;317:909–916. doi: 10.1016/j.bbrc.2004.03.136. [DOI] [PubMed] [Google Scholar]
- Johnston RN, Shaw C, Halton DW, Verhaert P, Baguna J. GYIRFamide: A novel FMRFamide-related peptide (FaRP) from the triclad turbellarian, Dugesia tigrina. Biochem Biophys Res Commun. 1995;209:689–697. doi: 10.1006/bbrc.1995.1554. [DOI] [PubMed] [Google Scholar]
- Johnston RN, Shaw C, Halton DW, Verhaert P, Blair KL, Brennan GP, Price DA, Anderson PA. Isolation, localization, and bioactivity of the FMRFamide-related neuropeptides GYIRFamide and YIRFamide from the marine turbellarian Bdelloura candida. J Neurochem. 1996;67:814–821. doi: 10.1046/j.1471-4159.1996.67020814.x. [DOI] [PubMed] [Google Scholar]
- Krautz-Peterson G, Radwanska M, Ndegwa D, Shoemaker CB, Skelly PJ. Optimizing gene suppression in schistosomes using RNA interference. Mol Biochem Parasitol. 2007;153:194–202. doi: 10.1016/j.molbiopara.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Kreshchenko ND, Sedelnikov Z, Sheiman IM, Reuter M, Maule AG, Gustafsson MK. Effects of neuropeptide F on regeneration in Girardia tigrina (platyhelminthes) Cell Tissue Res. 2008;331:739–750. doi: 10.1007/s00441-007-0519-y. [DOI] [PubMed] [Google Scholar]
- Kuroki Y, Kanda T, Kubota I, Fujisawa Y, Ikeda T, Miura A, Minamitake Y, Muneoka Y. A molluscan neuropeptide related to the crustacean hormone, RPCH. Biochem Biophys Res Commun. 1990;167:273–279. doi: 10.1016/0006-291x(90)91761-g. [DOI] [PubMed] [Google Scholar]
- Lecron JC, Minault M, Allard M, Goube de Laforest P, Gombert J, Simonnet G. Modulation of human lymphocyte proliferation by FLFQPQRFamide, a FMRFamide-like peptide with anti-opiate properties. J Neuroimmunol. 1992;38:1–8. doi: 10.1016/0165-5728(92)90084-x. [DOI] [PubMed] [Google Scholar]
- Li C, Kim K, Nelson LS. FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Brain Res. 1999a;848:26–34. doi: 10.1016/s0006-8993(99)01972-1. [DOI] [PubMed] [Google Scholar]
- Li C, Nelson LS, Kim K, Nathoo A, Hart AC. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann N Y Acad Sci. 1999b;897:239–252. doi: 10.1111/j.1749-6632.1999.tb07895.x. [DOI] [PubMed] [Google Scholar]
- LoVerde PT, Hirai H, Merrick JM, Lee NH, El-Sayed N. Schistosoma mansoni genome project: An update. Parasitol Int. 2004;53:183–192. doi: 10.1016/j.parint.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Ma M, Chen R, Sousa GL, Bors EK, Kwiatkowski MA, Goiney CC, Goy MF, Christie AE, Li L. Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the american lobster Homarus americanus. Gen Comp Endocrinol. 2008;156:395–409. doi: 10.1016/j.ygcen.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair GR, Halton DW, Shaw C, Maule AG. The neuropeptide F (NPF) encoding gene from the cestode, Moniezia expansa. Parasitology. 2000;120 ( Pt 1):71–77. doi: 10.1017/s0031182099005296. [DOI] [PubMed] [Google Scholar]
- Marks NJ, Maule AG. Neuropeptides in helminths: occurrence and distribution. In: Geary TG, Maule AG, editors. Neuropeptide systems as targets for parasite and pest control. Landes Bioscience/Eurekah.com; Georgetown, TX, USA: 2008. [Google Scholar]
- Marks NJ, Maule AG, Halton DW, Geary TG, Shaw C, Thompson DP. Pharmacological effects of nematode FMRFamide-related peptides (FaRPs) on muscle contractility of the trematode, Fasciola hepatica. Parasitology. 1997;114:531–539. [PubMed] [Google Scholar]
- Maule A, Shaw C, Halton D, Thim L. GNFFRFamide: A novel FMRFamide-immunoreactive peptide isolated from the sheep tapeworm, Moniezia expansa. Biochem Biophys Res Commun. 1993;193:1054–1060. doi: 10.1006/bbrc.1993.1732. [DOI] [PubMed] [Google Scholar]
- Maule AG, Shaw C, Halton DW, Brennan GP, Johnston CF, Moore S. Neuropeptide F (Moniezia expansa): Localization and characterization using specific antisera. Parasitology. 1992;105:505–512. doi: 10.1017/s0031182000074680. [DOI] [PubMed] [Google Scholar]
- Maule AG, Shaw C, Halton DW, Curry WJ, Thim L. RYIRFamide: A turbellarian FMRFamide-related peptide (FaRP) Regul Pept. 1994;50:37–43. doi: 10.1016/0167-0115(94)90189-9. [DOI] [PubMed] [Google Scholar]
- McVeigh P, Alexander-Bowman S, Kidd E, Mousley A, Marks NJ, Maule AG. Neuropeptide-like protein diversity in phylum nematoda. Int J Parasitol. 2008;38:1493–1503. doi: 10.1016/j.ijpara.2008.05.006. [DOI] [PubMed] [Google Scholar]
- McVeigh P, Leech S, Mair GR, Marks NJ, Geary TG, Maule AG. Analysis of FMRFamide-like peptide (FLP) diversity in phylum nematoda. Int J Parasitol. 2005;35:1043–1060. doi: 10.1016/j.ijpara.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Minault M, Lecron JC, Labrouche S, Simonnet G, Gombert J. Characterization of binding sites for neuropeptide FF on T lymphocytes of the jurkat cell line. Peptides. 1995;16:105–111. doi: 10.1016/0196-9781(94)00159-4. [DOI] [PubMed] [Google Scholar]
- Moneypenny CG, Kreshchenko N, Moffett CL, Halton DW, Day TA, Maule AG. Physiological effects of FMRFamide-related peptides and classical transmitters on dispersed muscle fibres of the turbellarian, Procerodes littoralis. Parasitology. 2001;122:447–455. doi: 10.1017/s0031182001007508. [DOI] [PubMed] [Google Scholar]
- Mousley A, Marks NJ, Maule AG. Neuropeptide signalling: A repository of targets for novel endectocides? Trends Parasitol. 2004;20:482–487. doi: 10.1016/j.pt.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci U S A. 2001;98:14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LS, Kim K, Memmott JE, Li C. FMRFamide-related gene family in the nematode, Caenorhabditis elegans. Brain Res Mol Brain Res. 1998;58:103–111. doi: 10.1016/s0169-328x(98)00106-5. [DOI] [PubMed] [Google Scholar]
- Omar HH, Humphries JE, Larsen MJ, Kubiak TM, Geary TG, Maule AG, Kimber MJ, Day TA. Identification of a platyhelminth neuropeptide receptor. Int J Parasitol. 2007;37:725–733. doi: 10.1016/j.ijpara.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Osugi T, Ukena K, Sower SA, Kawauchi H, Tsutsui K. Evolutionary origin and divergence of PQRFamide peptides and LPXRFamide peptides in the RFamide peptide family. insights from novel lamprey RFamide peptides. FEBS J. 2006;273:1731–1743. doi: 10.1111/j.1742-4658.2006.05187.x. [DOI] [PubMed] [Google Scholar]
- Pfister D, De Mulder K, Hartenstein V, Kuales G, Borgonie G, Marx F, Morris J, Ladurner P. Flatworm stem cells and the germ line: Developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev Biol. 2008;319:146–159. doi: 10.1016/j.ydbio.2008.02.045. [DOI] [PubMed] [Google Scholar]
- Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of ecdysozoa, lophotrochozoa, and protostomia. Mol Biol Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science. 1977;197:670–671. doi: 10.1126/science.877582. [DOI] [PubMed] [Google Scholar]
- Robb SM, Ross E, Sanchez Alvarado A. SmedGD: The Schmidtea mediterranea genome database. Nucleic Acids Res. 2008;36:D599–606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Riutort M, Littlewood DT, Herniou EA, Baguna J. Acoel flatworms: Earliest extant bilaterian metazoans, not members of platyhelminthes. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci U S A. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sithigorngul P, Pupuem J, Krungkasem C, Longyant S, Panchan N, Chaivisuthangkura P, Sithigorngul W, Petsom A. Four novel PYFs: Members of NPY/PP peptide superfamily from the eyestalk of the giant tiger prawn Penaeus monodon. Peptides. 2002;23:1895–1906. doi: 10.1016/s0196-9781(02)00176-6. [DOI] [PubMed] [Google Scholar]
- Sundstrom G, Larsson TA, Brenner S, Venkatesh B, Larhammar D. Evolution of the neuropeptide Y family: New genes by chromosome duplications in early vertebrates and in teleost fishes. Gen Comp Endocrinol. 2008;155:705–716. doi: 10.1016/j.ygcen.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Matsushima O, Morishita F, Fujimoto M, Ikeda T, Minakata H, Nomoto K. A myomodulin-CARP-related peptide isolated from a polychaete annelid, Perinereis vancaurica. Zoolog Sci. 1994;11:33–38. [PubMed] [Google Scholar]
- Varshavsky A. ‘Spalog’ and ‘sequelog’: Neutral terms for spatial and sequence similarity. Curr Biol. 2004;14:R181–3. doi: 10.1016/j.cub.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Venancio TM, DeMarco R, Almeida GT, Oliveira KC, Setubal JC, Verjovski-Almeida S. Analysis of Schistosoma mansoni genes shared with deuterostomia and with possible roles in host interactions. BMC Genomics. 2007;8:407. doi: 10.1186/1471-2164-8-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Price DA, Sahley CL. Identification and characterization of a myomodulin-like peptide in the leech. Peptides. 1998;19:487–493. doi: 10.1016/s0196-9781(97)00419-1. [DOI] [PubMed] [Google Scholar]
- Weaver RJ, Audsley N. Neuropeptides of the beetle, Tenebrio molitor identified using MALDI-TOF mass spectrometry and deduced sequences from the Tribolium castaneum genome. Peptides. 2008;29:168–178. doi: 10.1016/j.peptides.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Yang HY, Tao T, Iadarola MJ. Modulatory role of neuropeptide FF system in nociception and opiate analgesia. Neuropeptides. 2008;42:1–18. doi: 10.1016/j.npep.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


