Abstract
Global acetylation of histone H4 is a mark of gene transcriptional activation. The c-Myc transcription factor binds to specific DNA sites in cellular chromatin and induces the acetylation of histone H4. In this study, hypoxia (1% Oxygen) induced a decrease in both global acetylated histone H4 (AcH4) and c-Myc in human lung carcinoma A549 cells and in human bronchial epithelial Beas-2B cells, The decline was more stiking in A549 cells compared to Beas2-B cells, when cells were exposed to hypoxic stress for 24 hr. Further studies showed that these alterations of global AcH4 can be attributed to the decrease in c-Myc protein levels. While hypoxia-induced gene activation is known to be mediated by Hypoxia Response Elements (HRE), the mechanism for down-regulation of genes by hypoxia is not known. The decrease in c-Myc protein levels induced by hypoxia may contribute to hypoxia-induced gene repression.
Keywords: Hypoxia, c-Myc, Acetylation, Histone H4
INTRODUCTION
Histones, are small, basic proteins consisting of a globular domain, an N-terminal tail, and a C-terminal tail, and are the major protein components of chromatin. One of the keys to the dynamic nature of chromatin is post-translational modification of these NH2-terminal flexible tails. Such modifications include acetylation, and methylation of lysines (K) and arginines (R), citrullination of arginines, phosphorylation of serines (S) and threonines (T), sumoylation and ubiquitination of lysines, and ADP-ribosylation [1–3]. These modifications play critical roles in chromatin organization and gene transcription. In general, an increased acetylation level of histone H3 or H4 contributes to the formation of an ‘open’ chromatin state and gene transcription, whereas decreased histone acetylation contributes to a ‘closed’ chromatin and transcriptional repression [4, 5].
c-Myc belongs to the Myc family of transcription factors, which also includes N-Myc, L-Myc, S-Myc, and B-Myc. Studies have already shown that the transcription factor c-Myc binds to specific DNA sites in cellular chromatin, induces the acetylation of histone H3 and H4, and recruits other co-factors to activate gene expression [6, 7]. Recent studies demonstrated that c-Myc inactivation induced global changes in chromatin structure associated with a marked reduction of histone H4 acetylation and increased histone H3 K9 methylation [8]; this indicated that c-Myc is required for maintenance of normal histone acetylation and gene activated chromatin [9].
Hypoxia often occurs in a solid tumor, resulting from inadequate and disordered neo-vasculature [10]. Hypoxia-activated gene expression, which is often mediated through hypoxia-inducible factor (HIF), has been well characterized [11]. However, the mechanism for gene repression or aberrant gene silencing during hypoxia has not been delineated. In this study, we report that exposure to hypoxia (1% O2) is able to decrease global acetylated histone H4 (AcH4) levels in both tumorigenic A549 and non-tumorigenic Beas-2B cells. Further study showed that these alterations of global AcH4 can be attributed to a decreased c-Myc protein level. These results indicate that the repression of some genes, such as c-Myc target genes, may due to the reduction and subsequent loss of activity of c-Myc during hypoxia.
MATERIALS and METHODS
Cell lines and culture conditions
Human lung carcinoma A549 cells were cultured in Ham’s F-12 K medium (Invitrogen, Frederick, MD). Human bronchial epithelial Beas-2B cells, Rat-1 cell lines (wild-type Rat-1 cells [c-Myc+/+] and c-Myc knockout Rat-1 cells [c-Myc−/−] [12]) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen). All media were supplemented with 10% fetal bovine serum (FBS, ATLAS Biological, Fort Collins, CO), and 1% penicillin/streptomycin (Grand Island, NY). Cells were maintained at 37°C as monolayers in a humidified atmosphere containing 5% CO2. When cell density reached approximately 70–80% confluence, they were cultured under hypoxic (1% O2) condition.
Whole Cell Lysates and Histone Extraction
Cells were washed with ice-cold 1 × PBS (Phosphate Buffered Saline: 0.13 M NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, and 0.27 mM KCl, [pH 7.4]) twice and lysed with ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, [pH 7.4], 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, and 1 mM EDTA) supplemented with a protease inhibitor mixture (Roche Applied Sciences, Indianapolis, IN) for 10 min on ice. The cell lysates were then transferred to an Eppendorf tube and kept on ice for another 30 min followed by centrifugation at 10000 × g for 10 min. The resultant supernatants were collected as whole-cell lysates (WCL). The pellet was washed once in 10 mM Tris/13 mM EDTA buffer (pH 7.4) and spun down at maximum speed for 5 min. The pellet was then resuspended in 100 μl 0.4 N H2SO4. After at least 1.5 hr of incubation on ice, the sample was centrifuged at ≥ 14,000 × g for 15 min. The resulting supernatant was recovered, then mixed with 1 ml of cold acetone, and kept at −20°C overnight. The histones were then collected by centrifugation at ≥ 14,000 × g for 15 min. After one wash with acetone, the histones were air-dried and re-dissolved in 4 M urea.
Western blot
The protein concentration was determined using Bio-Rad DC protein assay (Bio-Rad, Hercules, CA). Five μg of purified histones or 50 μg whole cell lysates (WCL) were separated by SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Antibodies against acetylated histone H4 (AcH4) were purchased from Upstate (Lake Placid, NY). c-Myc antibody (N-262) was from Santa Cruz Biotechnology (Santa Cruz, CA), and α-tubulin antibodies were from Sigma (St. Louis, MO). HRP-conjugated anti-rabbit or mouse secondary antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). The detection was accomplished by chemical fluorescence following an ECL Western blotting protocol (Amersham, Piscataway, NJ). After transfer to PVDF membranes, the gels were stained with Bio-safe Coomassie stain (Bio-Rad) to assess the loading of histones.
RESULTS
Hypoxia decreases global acetylated histone H4 (AcH4) levels
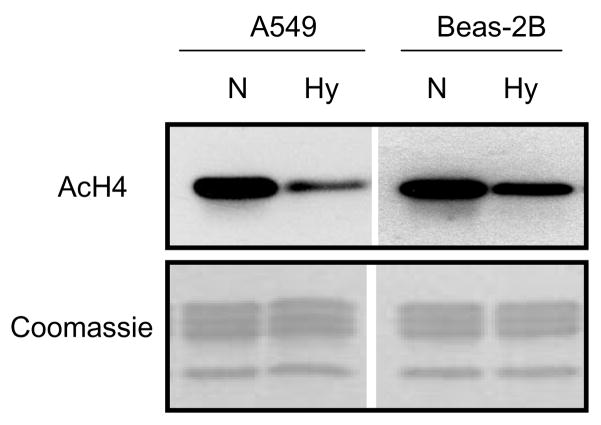
Previous studies in our laboratory have shown that hypoxia induced trimethylation of H3K9 (H3K9m3) and decreased the acetylation of histone H3 (AcH3) in A549 cells [13–17]. Here, we show that exposure to hypoxia for 24 hr decreased the levels of acetylated histone H4 (AcH4) in both A549 and Beas-2B cells (Figure 1). theeffect was more evident in tumorigenic A549 cells than in non-tumorigenic Beas-2B cells.
Figure 1.
Hypoxia (Hy) decreased the global levels of acetylated histone H4 (AcH4) in both non-tumorigenic Beas-2B and tumorigenic A549 cell lines. Cells were cultured under normoxic (N) or hypoxic (Hy: 1% O2) conditions for 24 hr. Total histones (5 μg per lane) were separated on 15% SDS-PAGE gels and then subjected to Western blotting with antibodies against AcH4. The lower panels show the gels stained with Coomassie Blue (after being transferred) in order to monitor for the loading of the histones.
Hypoxic stress decreases c-Myc protein levels
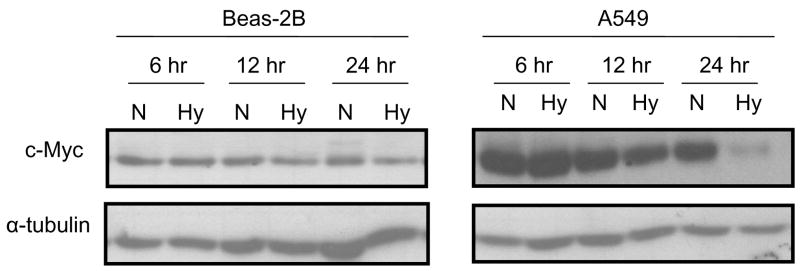
Studies have been shown that c-Myc up-regulates global acetylation of histone H4. To determine whether c-Myc plays a role in hypoxia-induced alterations in AcH4, c-Myc protein levels were measured after A549 and Beas-2B cells were exposed to hypoxic (1% O2) stress for 6, 12 or 24 hr. The results showed that hypoxia decreased c-Myc protein levels in both A549 and Beas-2B cells (Figure 2), especially after 24. However, the decreased c-Myc was more striking in tumorigenic A549 cells than in non-tumorigenic Beas-2B cells, which was consistent with the effect of hypoxia on global AcH4 (Figure 1).
Figure 2.
Hypoxia (Hy) decreased c-Myc protein levels in both non-tumorigenic Beas-2B and tumorigenic A549 cell lines in a time-dependent manner. Beas-2B and A549 cells were cultured under normoxic (N) or hypoxic (Hy: 1% O2) conditions for 6, 12 and 24 hr. After treatment, cells were lysed and 50 μg of total protein was separated on 7.5% SDS-PAGE gels, followed by Western blotting using antibodies against c-Myc or α-tubulin (to control for loading). Similar data were obtained in at least two other independent experiments; a representative Western blot is shown.
c-Myc contributes to a decrease of acetylated histone H4 (AcH4) during hypoxic stress
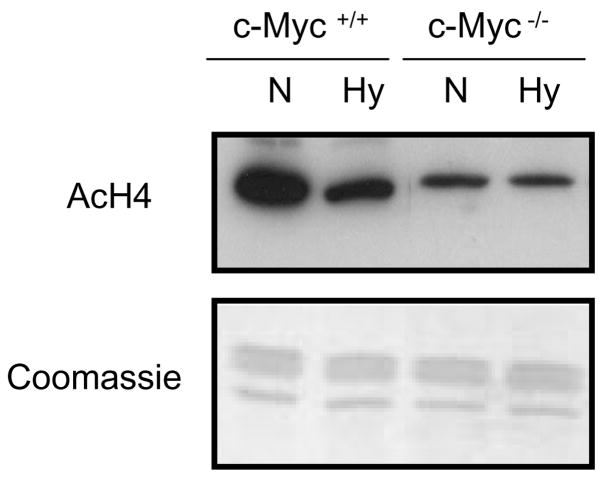
To further assess whether hypoxia-induced alterations in global AcH4 levels were dependent upon c-Myc inactivation, the Rat-1 cell lines (wild-type Rat-1 cells [c-Myc+/+] and c-Myc knockout Rat-1 cells [c-Myc−/−]) were exposed to hypoxic stresss for 24 hr. These studies showed that hypoxia decreased AcH4 levels in the wild-type Rat-1 cells, but this decline was attenuated in the c-Myc−/− cells (Figure 3). These results indicated that the reduction of c-Myc levels due to hypoxia was the factor that most likely resulted in the decrease of global AcH4 levels.
Figure 3.
The effect of c-Myc on the hypoxia (Hy) mediated decrease in the global levels of acetylated histone H4 (AcH4) Wild-type (c-Myc+/+) and c-Myc knockout (c-Myc−/−) Rat-1 cells were cultured under normoxic (N) or hypoxic (Hy: 1% O2) conditions for 24 hr. Total histones (5 μg per lane) were separated on 15% SDS-PAGE gels and then subjected to Western blotting with antibodies against AcH4. The lower panels show the gels stained with Coomassie Blue (after being transferred) in order to monitor for loading of the histones.
DISCUSSION
Rapidly growing tumors generally develop hypoxic regions. Tumor cells adapt to their hypoxic microenvironment through angiogenesis, enhanced glucose metabolism and decreased mitochondrial respiration. These adaptive metabolic alterations are carried out by up- or down-regulating gene expression. The up-regulation of gene expression during hypoxia are often mediated through hypoxia-inducible factor (HIF) [11]. Hypoxia also decreases the expression of several genes, such as cellular adhesion proteins and DNA repair proteins, and leads to genetic instability and metastasis of tumor cells [18–20]. However, the mechanism by which hypoxic stress represses gene expression remains unclear. Here, hypoxia was found to induce a reduction in global AcH4 levels, an important active epigenetic mark. This reduction is likely linked to the repression of several genes during hypoxic stress.
c-Myc is a very strong proto-oncogene and it is very often found to be overexpressed in many human tumours [21]. However, c-Myc mRNA and protein are generally expressed at low levels in normal proliferating cells [22]. In agreement with these reports, c-Myc protein levels were found much higher in tumorigenic A549 cells than in non-tumorigenic Beas-2B cells. In this study, we found that hypoxic stress also decreased c-Myc protein levels in both tumorigenic A549 and non-tumorigenic Beas-2B cells. The decreased c-Myc induced by hypoxic stress was also reported in other cell lines, such as RCC4 and HCT116 cells [23–25]. In addition the decrease in both AcH4 and c-Myc protein levels were more evident in A549 than in Beas-2B cells. Since c-Myc protein level is different in tumorigenic and non-tumorigenic cells, we surmise that c-Myc is regulated by different mechanism in A549 and Beas-2B cells. In non-tumorigenic cells, c-Myc protein level is tightly controlled at a low level and hypoxia-induced reduction in c-Myc protein level protects hypoxic cells from c-Myc-mediated apoptosis. In fact, we observed that Beas-2B cells have lower apoptotic rate under hypoxia than normoxia (data not shown), which reinforced this hypothesis. However, in cancer cells, c-Myc is frequently overexpressed and stabilized by an unknown mechanism for promoting cell proliferation and mitochondrial respiration through increasing mitochondrial biogenesis [26]. In contrast, HIF have already been shown to inhibit mitochondrial biogenesis [23]. The opposite effects of HIF and c-Myc on mitochondrial biogenesis may adjust c-Myc expression and degradation to ensure efficient mitochondrial biogenesis, which may result in a larger decrease in c-Myc protein levels in tumorigenic A549 cells.
It has been shown that c-Myc itself can directly induce acetylation of histone H4 (AcH4); this has been proposed to play an essential permissive role allowing c-Myc target gene activation by additional signals normally triggered by mitogens [6]. Further studies showed that the decrease in AcH4 levels due to hypoxia was dependent on c-Myc. These results indicate that the repression of some genes, such as c-Myc target genes, is due to the decrease and subsequent loss of activity of c-Myc protein during hypoxia. Since c-Myc can control up to 15% of all of our genes resulting in both increased and decreased expression, the results shown here may help explain how hypoxia up-regulates gene expression independent of HIF-1α DNA-binding and transactivation domains (such as CDKN1A) [27], as well as how hypoxia down-regulates gene expression.
Acknowledgments
The authors would like to thank Dr. John M. Sedivy from Brown University for providing us with Rat-1 cell lines (wild-type Rat-1 cells [c-Myc+/+] and c-Myc knockout Rat-1 cells [c-Myc−/−]) used in this study. This work was supported by grants ES014454, ES005512, and ES000260 from the National Institutes of Environmental Health Sciences, and grant CA16087 from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 2.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM, Probert L, Casaccia-Bonnefil P, Moscarello MA. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci. 2006;26:11387–11396. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 5.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 6.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. Embo J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 11.Semenza GL. O2-regulated gene expression: transcriptional control of cardiorespiratory physiology by HIF-1. J Appl Physiol. 2004;96:1173–1177. doi: 10.1152/japplphysiol.00770.2003. discussion 1170–1172. [DOI] [PubMed] [Google Scholar]
- 12.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke Q, Li Q, Ellen TP, Sun H, Costa M. Nickel compounds induce phosphorylation of histone H3 at serine 10 by activating JNK-MAPK pathway. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Kent JR, Placek B, Whelan KA, Hollow CM, Zeng PY, Fraser NW, Berger SL. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J Virol. 2006;80:5740–5746. doi: 10.1128/JVI.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016. doi: 10.1158/0008-5472.CAN-06-0101. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Ke Q, Kluz T, Yan Y, Costa M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol Cell Biol. 2006;26:3728–3737. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sook Kim M, Hyen Baek J, Bae MK, Kim KW. Human rad21 gene, hHR21(SP), is downregulated by hypoxia in human tumor cells. Biochem Biophys Res Commun. 2001;281:1106–1112. doi: 10.1006/bbrc.2001.4488. [DOI] [PubMed] [Google Scholar]
- 20.Hasan NM, Adams GE, Joiner MC, Marshall JF, Hart IR. Hypoxia facilitates tumour cell detachment by reducing expression of surface adhesion molecules and adhesion to extracellular matrices without loss of cell viability. Br J Cancer. 1998;77:1799–1805. doi: 10.1038/bjc.1998.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hann SR, Eisenman RN. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudolph C, Adam G, Simm A. Determination of copy number of c-Myc protein per cell by quantitative Western blotting. Anal Biochem. 1999;269:66–71. doi: 10.1006/abio.1999.3095. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. Embo J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corn PG, Ricci MS, Scata KA, Arsham AM, Simon MC, Dicker DT, El-Deiry WS. Mxi1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer Biol Ther. 2005;4:1285–1294. doi: 10.4161/cbt.4.11.2299. [DOI] [PubMed] [Google Scholar]
- 26.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack FA, Patel JH, Biju MP, Haase VH, Simon MC. Decreased growth of Vhl−/− fibrosarcomas is associated with elevated levels of cyclin kinase inhibitors p21 and p27. Mol Cell Biol. 2005;25:4565–4578. doi: 10.1128/MCB.25.11.4565-4578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]