Given the increasing prevalence of patients with congestive heart failure, there is a strong interest in identifying and isolating cardiac progenitor cells capable of differentiating into cardiomyocytes that could be used to repair a damaged, failing heart. Recent work in the zebrafish model system has suggested that the epicardium, the non-myocyte epithelial layer of cells covering the heart, may play a critical role in the regeneration of injured myocardium.1 The epicardium has a distinct embryological origin from both the myocardium and endocardium. It arises from a mass of mesothelial cells termed the proepicardium, located on the wall of the embryonic pericardial cavity just dorsal and caudal to the developing heart.2 During early embryonic development, cells of the proepicardium migrate onto the heart at the atrioventricular junction and then spread over the surface of the heart to form a primitive epicardium. Some cells of the epicardium then undergo a mesenchymal transformation and invade into the sub-epicardial space. There these epicardial-derived cells further differentiate into cells that form the coronary vasculature as well as cardiac fibroblasts. Through this multi-step differentiation pathway, the proepicardium serves as an important source for the non-cardiomyocyte cellular component of the heart.
Over the last few years, the pathways that regulate the development and differentiation of the proepicardium have begun to be revealed. First, the formation of the proepicardium is critically dependent on the transcription factor GATA4 and inductive signals from the developing liver bud.3, 4 Once formed, cells of the proepicardium express the transcription factors Tbx18 and Wt-1. They may also become specified to a particular cell fate (i.e. fibroblast, smooth muscle cell, endothelial cell) before beginning their migration to the developing heart as suggested by proepicardial retroviral labeling studies in chick.5 The proepicardium has been receiving increasing attention as recent work has suggested that these cells have the potential to differentiate into cardiomyocytes as well as cells of the epicardium. Indeed, lineage-tracing experiments utilizing cre-lox technology with cre expressed under the control of the Tbx18 or WT-1 gene promoters have suggested that a fraction of these proepicardial cells become cardiomyocytes in the developing heart.6, 7 However, this conclusion is somewhat controversial as some cardiomyocytes may express low levels of these markers later in development, complicating the interpretation of these lineage tracing experiments.8
Additional support for the ability of proepicardial cells to differentiate into cardiomyocytes has come from in vitro culture experiments of proepicardial explants. Both BMP-2 and BMP-4 were found to induce cardiomyocyte formation from proepicardial explant cultures.9, 10 In contrast, FGF-2 was found to promote the differentiation of proepicardial cells along non-cardiomyocyte cardiac lineages. Further, FGF-2 also blocks the ability of BMP-2 to induce cardiomyocyte differentiation in these cultures, suggesting an interaction between these two signaling pathways during proepicardial differentiation.
In work reported in this issue of Circulation Research, Van Wijk and colleagues provide further evidence for the potential of proepicardial cells to differentiate into cardiomyocytes.11 Using vital cell labeling with DiI, they show that cells within a field of the chick proepicardium can become cardiomyocytes in the inflow tract myocardium, supporting the notion that proepicardial cells have cardiomyocyte potential. However, these experiments are unable to determine whether cells within this field are truly bi-potential (i.e., having both myocardial and epicardial potential), or if this field of cells contains two distinct populations, one with solely myocardial potential and the other with epicardial potential. Further work will be necessary to resolve this question.
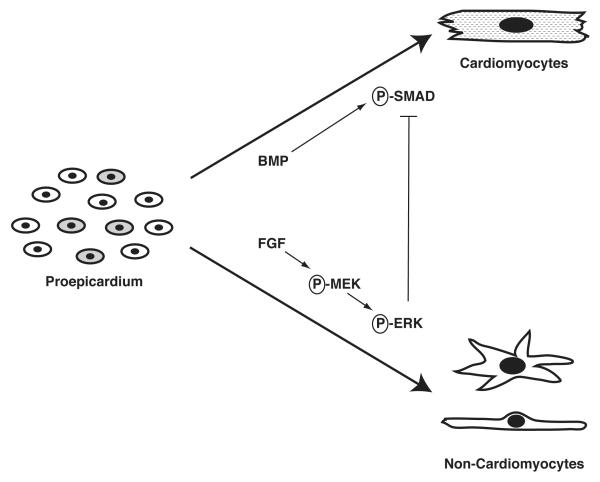
Van Wijk and colleagues also take an important step forward in our understanding of the interplay of signaling pathways regulating the differentiation of cells within the proepicardium (see Figure 1). Consistent with previous work, they show that BMP-2 can induce cardiomyocyte differentiation in proepicardial cultures and FGF-2 can block it, but in this report they extend these observations by defining the downstream signaling pathways activated by each of these ligands. They show that FGF-2 activates a kinase cascade that results in the phosphorylation of ERK, while BMP-2 signaling is mediated through phosphorylation of SMAD1/5/8. Interestingly, immunohistochemistry of the proepicardium using anti-phospho-ERK or anti-phospho-SMAD antibodies reveals significant heterogeneity within this structure, as some proepicardial cells express phospho-SMADs, others express phospho-ERK, and still others express both. This observation is consistent with the idea that the proepicardium is not a homogenous field of cells and thus may contain cells with distinct developmental potentials.
Figure 1. Crosstalk between FGF and BMP Signaling Pathways during Proepicardial Differentiation.
Proepicardial cells, likely a heterogeneous population (as indicated by shading), differentiate down the cardiomyocyte lineage in response to BMP signaling or down a non-cardiomyocyte cardiac lineage in response to FGF signaling. Further, the FGF signaling pathway is dominant over the BMP pathway as FGF signaling through ERK can block BMP-induced cardiomyocyte differentiation.
The ability of FGF-2 to block BMP-induced cardiomyocyte differentiation of proepicardial cells is also explained in this report through the demonstration that activation of FGF signaling in proepicardial cells attenuates BMP-induced SMAD phosphorylation and nuclear localization. This presumably results in a failure to transcriptionally activate SMAD-dependent target genes important for the induction of the cardiomyocyte cell fate. Further, the ability of FGF-2 to block BMP-induced differentiation is mediated through a MEK/ERK-dependent pathway as the MEK inhibitor U0126 blocked these effects. Consistent with their in vitro results, Van Wijk et al. show that treatment of whole chick embryos in ovo with BMP2+U0126 blocked the migration of the proepicardium onto the developing heart tube and enhanced myocardial formation in the venous pole of the heart, while treatment with FGF-2 led to enhanced epicardial formation. Together, these results suggest that the there is crosstalk between the FGF and BMP signaling pathways in the differentiation of cells of the proepicardium into myocardial vs. non-myocardial cardiac lineages (see Figure 1).
While this work is a step forward in our understanding of the pathways regulating proepicardial and epicardial development, important questions remain. First, what are the downstream transcriptional targets of FGF and BMP signaling that modulate proepicardial differentiation? Second, what signaling pathways modulate proepicardial lineage decisions leading to a cardiac fibroblast, smooth muscle, or endothelial cell fate? Third, are cells of the proepicardium truly committed to a particular lineage before leaving the proepicardium, or do they receive additional inductive cues upon their arrival to the epicardium or sub-epicardial space? And finally, can mature epicardium be re-programmed into a proepicardial—like state for potential use in myocardial regeneration strategies? It would not be surprising to find that a number of “conversations” are occurring in the proepicardium to direct these lineage decisions. The challenge for the field will be to identify who’s talking to whom.
Acknowledgments
Sources of Funding E.C.S. is supported by NIH grant # HL71063.
Footnotes
Disclosures None.
References
- 1.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127(3):607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 2.Olivey HE, Compton LA, Barnett JV. Coronary vessel development: the epicardium delivers. Trends in Cardiovascular Medicine. 2004;14(6):247–251. doi: 10.1016/j.tcm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Ishii Y, Langberg JD, Hurtado R, Lee S, Mikawa T. Induction of proepicardial marker gene expression by the liver bud. Development. 2007;134(20):3627–3637. doi: 10.1242/dev.005280. [DOI] [PubMed] [Google Scholar]
- 4.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci USA. 2004;101(34):12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci USA. 1992;89(20):9504–9508. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008 doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008 doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christoffels VM, Grieskamp T, Norden J, Mommersteeg MTM, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458(7240):E8–9. doi: 10.1038/nature07916. discussion E9-10. [DOI] [PubMed] [Google Scholar]
- 9.Kruithof BP, van Wijk B, Somi S, Kruithof-de Julio M, Pomares JM Pérez, Weesie F, Wessels A, Moorman AF, van den Hoff MJ. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Developmental Biology. 2006;295(2):507–522. doi: 10.1016/j.ydbio.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 10.Schlueter J, Männer J, Brand T. BMP is an important regulator of proepicardial identity in the chick embryo. Developmental Biology. 2006;295(2):546–558. doi: 10.1016/j.ydbio.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 11.van Wijk B, van den Berg G, Abu-Issa R, Barnett P, van der Velden S, Schmidt M, Ruijter JM, Kirby ML, Moorman AF, van den Hoff MJ. Epicardium and myocardium separate from a common precursor pool by crosstalk between BMP- and FGF-signaling pathways. Circulation Research. 2009 doi: 10.1161/CIRCRESAHA.109.203083. [DOI] [PMC free article] [PubMed] [Google Scholar]