Abstract
Like all human cancers, colorectal cancer is a complicated disease. While a mature body of research involving colorectal cancer has implicated the putative sequence of genetic alterations that trigger the disease and sustain its progression, there is a surprising paucity of well-validated, clinically useful diagnostic markers of this disease. For prognosis or guiding therapy, single gene-based markers of colorectal cancer often have limited specificity and sensitivity. Genome-wide analyses (microarrays) have been used to propose candidate patterns of gene expression that are prognostic of outcome or predict the tumor’s response to a therapy regimen; however, these patterns frequently do not overlap, and this has raised questions concerning their use as biomarkers. The limitation of gene-expression approaches to marker discovery occurs because the change in mRNA expression across tumors is highly variable and, alone, accounts for a limited variability of the phenotype, such as with cancer. More robust and accurate markers of cancer will result from integrating all the information we have about the cell: genomics, proteomics and interactomics. This article will discuss traditional markers in colorectal cancer, both genomic and proteomic, including their respective approaches and limitations, then conclude with examples of systems biology-based approaches for candidate marker discovery, and discuss how this approach is reshaping our view of a biomarker.
Keywords: biomarker, cancer, colon, genomic, interaction, interactomic, networks, protein, proteomic
Epidemiology & genetics of colorectal cancer
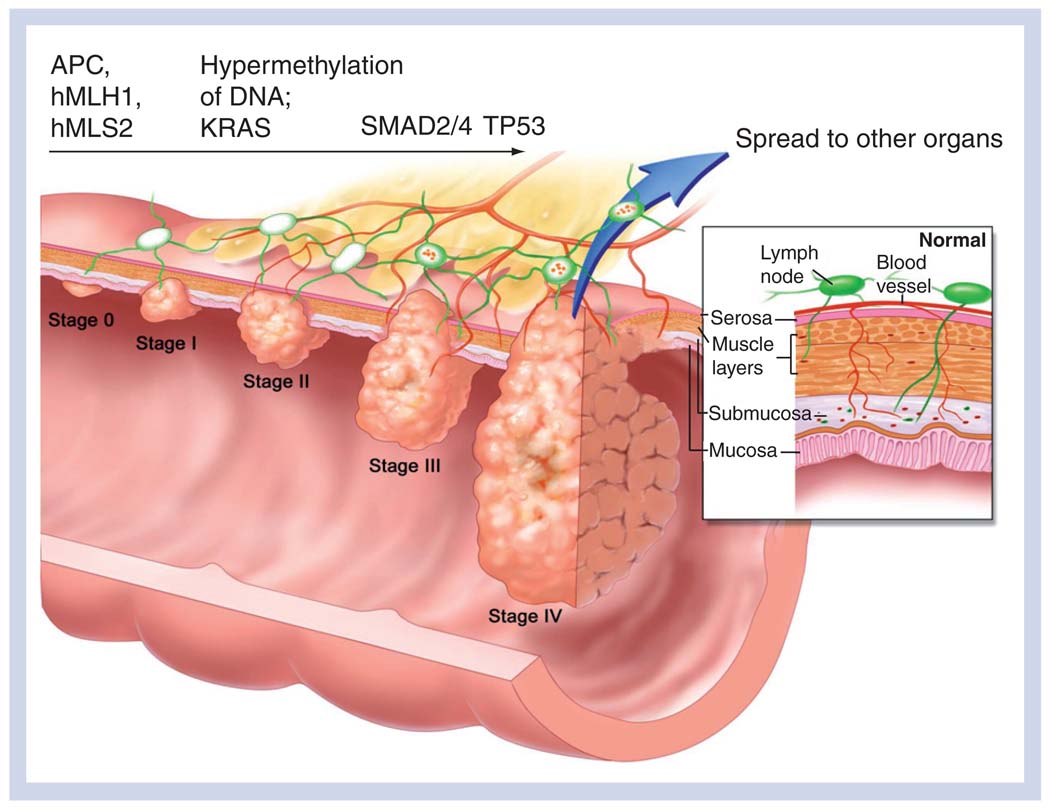
Colorectal cancer (CRC) is the second leading cause of cancer death in the USA and the UK [1]. The disease can be classified into two groups, inherited and sporadic. The former is an inherited predisposition to CRC that may be broadly classified into two forms, familial adenomatous polyposis coli and hereditary nonpolyposis colon cancer, although other syndromes are known. Familial adenomatous polyposis coli is an autosomal-dominant disease that results from a germline mutation in the APC gene, often an N-terminal truncation of the APC protein [2], which inevitably results, before the age of 50 years, in the development of hundreds or more polyps on the colonic wall, one or more of which will unavoidably progress to an established cancer. Often, however, a somatic mutation of the other APC allele is associated with adenoma formation [3] and considered to be the determining event initiating CRC [4]. Hereditary nonpolyposis colon cancer arises from a germline mutation in one or more of the DNA mismatch-repair genes, commonly hMLH1 or hMSH2, and microsatellite instability [5]. Approximately 80% of people with one or more of these mutations will develop CRC, usually by the age of 45 years [101]. By contrast, sporadic CRC, which affects approximately 5–6% of the American population [102], is a progressive disease arising from the accumulation of somatic mutations in colonic epithelial cells. The mutations and epigenetic alterations commonly implicated in the progression are shown in Figure 1, overlaid on the stage (0–IV) of the cancer, during formation of an early adenoma (0), to invasion of the mucosa (I), followed by increasing angiogenesis and lymph node involvement (II–III) and, finally, a breach of the colonic wall and metastasis (IV).
Figure 1. Stage-wise progression of colorectal cancer.
Genetic alterations commonly associated with the progession to subsequent stages are indicated above the arrow.
Reproduced with permission from [104].
Biomarkers: old & new
In a clinical sense, and depending on the context of its application, the optimum biomarker of human cancer may have one or more properties. It would be a single molecule whose level of expression or activity would mark the onset of cancer (diagnostic), indicate the specific treatment regimen for a patient with established cancer (predictive), or indicate the fate of the cancer; for example, good versus poor outcome (prognostic) [6]. The biomarker would be easily assayable by a single test, present in both the disease state and the normal state, and capable of both high specificity and sensitivity. Ideally, it would be readily detectable in body fluid (e.g., serum or urine). Unfortunately, human cancer is a complicated disease [7]. At the molecular level, it is quite unlikely we will find a single optimum marker of CRC, or any cancer, in the traditional sense. Unquestionably, researchers need to take advantage of the tremendous insights resulting from decades of research, as well as clinical and preclinical trials in human cancer (legacy data), but at the same time we need to understand the limitations of any one approach, and guard against bias in favoring the results of one approach over others. The unprecedented volume of data coming from high-throughput experiments in genomics and proteomics is rapidly advancing our understanding of cancer. However, these results need to be placed in the context of specific molecular functions. A more complete understanding of the functional implications of differentially expressed genes or proteins in cancer promises to deliver improved biological markers of the disease [8], markers that are more sensitive to the known heterogeneity of patient tumors and offer improved specificity and sensitivity in classification compared with existing approaches that do not account for function. In turn, these markers will provide an improved focus for follow-up experiments to verify mechanisms, which in turn will help inform the development of novel drug targets.
Genomic markers in colorectal cancer
There are a variety of indicators evident in nuclear DNA or its transcribed product, mRNA, which may be said to provide biomarkers of cancer. These markers may play a physiologic role in initiating the cancer or regulating its progression, or in mediating its response to drug treatment. CRC patients may have their tumors genotyped for one or more these mutations, or epigenetic modifications, either or both of which may be useful as prognostic or predictive markers. As previously indicated, the clinical utility of these markers is presently limited, neverthe-less, many alterations at the genomic level do play an important role in the disease, and for the purpose of review, these merit discussion.
Gene mutation
It is widely agreed that sporadic CRC is caused by the accumulation of somatic gene mutations evident in colonic epithelial cells [9]. A land-mark study, recently published in the journal Science, revealed the protein-coding genes most frequently mutated in breast and CRC (candidate driver genes), obtained by a genome-wide screen on a cohort of human tumor biopsies [10]. Not surprisingly, APC, TP53, SMAD4 and KRAS were among the 69 driver genes identified in CRC, which mapped to no less than seven distinct gene ontological processes. APC is the ‘gatekeeper’ gene in CRC and was found to be mutated in more than 90% of the 35 tumors used in the discovery and validation screens. Likewise, TP53 and KRAS were mutated in 51 and 44% of tumors, respectively. Three isoforms (2, 3 and 4) of the SMAD tumor-suppressor gene were mutated in more than 5% of the tumors. While the results with respect to these four genes confirmed their known role in CRC, except for APC, up to half of the tumors did not contain mutations in one or more of these genes. Furthermore, the authors noted that in no case did any single cancer specimen have more than six candidate driver genes in common with another sample; overall, each specimen had its own ‘signature’ pattern of somatic mutation. This observation underscores the wide variability of expression patterns in individual tumors, and the limitation of markers in CRC that are exclusively based on changes in the transcriptome. Indeed, to date, it has been suggested that only certain mutations in KRAS are clinically useful as predictive markers for estimating the success of certain chemotherapy treatments in CRC [11], which is later discussed in this article.
Transcriptome changes
Evidence of the ability to quantify genome-wide expression of mRNA by microarrays in cancer was reported over 12 years ago [12]. Since then, thousands of microarray experiments have been conducted with the goal of discovering gene patterns or signatures that change significantly between treated or diseased samples and controls. Encouraged by a call for standards in reporting the results of microarray experiments, owing to their inherent technical variability, a number of public databases were established where the raw data could be deposited along with the relevant annotations and details of sample preparation. Indeed, many journals now require authors who report the results of a microarray experiment to deposit these data in a public database as a condition for publication of their manuscript. One such database is the Gene Expression Omnibus (GEO) hosted at the National Center for Biotechnology Information website [13]. A recent search of this database with the keyword ‘cancer’ returned over 2600 experiments. Refining the search with respect to CRC, 246 experiments were returned, 203 of which had been conducted on human tissue or derived cell lines. Many of these gene-expression profiles have been mined to find signatures that characterize the early stages of CRC tumorigenesis [14], regulate its progression [15] or predict the tumor’s response to a particular therapy [16]. In addition, the high-dimensional nature of these data has proved to be a rich substrate for increasingly sophisticated bioinformatic methods that attempt to overcome the problem obtained when the number of predictor variables (genes) greatly exceeds the number of samples [17]. Despite these advances, however, evidence from studies in other human cancers counsel caution with respect to gene-expression signatures of CRC. For instance, the evaluation of candidate signatures from two landmark studies of breast cancer metastasis revealed strikingly little overlap, although a number of the pathways involving these genes were in common to both studies [18,19]. While technical variation may explain some of the variability, these observations otherwise suggest that the way forward in marker discovery is an integrative ‘omics approach, one that leverages all the relevant information we have regarding the disease, not merely by changes in the transcriptome.
Single-nucleotide polymorphisms & copy-number variation
Single-nucleotide polymorphisms (SNPs) are alterations of one or more nucleotides that occur with an allelic frequency of greater than 1% in members of a species. They are frequently the basis for genome-wide association studies that target susceptibility markers for cancer. These alterations may occur within or outside the protein-coding region of the gene. Copy-number variations (CNVs) are chromosomal aberrations that result from the loss of a gene, its duplication or translocation causing an aberrant number of transcripts to be produced in the cell. As with mutations, genome-wide sequencing may target certain SNPs and CNVs known to predispose an individual to certain cancers, including CRC. For a recent review of the relevance of SNPs and CNVs in CRC, and their relationship to gene expression and chromosomal aberrations, refer to Tsafrir et al. [20].
Epigenetic status
Gene expression in eukaryotes may be regulated by chromatin remodeling, through post-translational modification of histone proteins or by direct modification of a nucleotide; for example, methylation of cytosine, which acts to silence gene transcription. The VIM gene codes for a type III filament protein highly abundant in many tissues, and is transcriptionally silent in both normal and tumor colon epithelial tissue. However, the methylation status of this gene detected in colonic cells that were isolated from the stool of 94 CRC patients predicted the presence of a tumor with 46% sensitivity. In 198 cancer-free controls, it achieved a specificity of 90%. It is worth noting that the same test was 43% sensitive at detecting early cancer, Duke’s stage I and II [21]. Screening for this marker is often recommended in addition to the fecal occult blood test. Recent improvements in the test have raised its prediction rate to 77%, and it has gained the recommendation of the American Cancer Society as a screening tool for CRC.
miRNA: the new ‘ome
Not all RNAs code for protein. miRNAs are small (<22 nucleotides) RNAs known to regulate a variety of cellular processes [22], especially translation and mRNA stability. This has launched a bioinformatic hunt for the precursors in the human genome, and informed a variety of experiments evaluating their role in disease, including CRC [23]. It is too early to say whether the expression of these molecules alone will become useful markers of disease. More likely, as with protein-coding genes, their differential expression in cancer will most likely be one contributor to an integrated molecular phenotype, and not independently diagnostic.
Predictive markers in colorectal cancer therapy: pharmacogenomics
Colorectal cancer is commonly treated with fluoropyrimidines, such as 5-flourouracil (5-FU) or capecitabine, platinum-based drugs, such as oxaliplatin, topisomerase I inhibitors, such as irinotecan, or, particularly in late-stage CRC, drugs that inhibit the EGF receptor (EGFR), such as Eribtux®. Recently, inhibitors of two isoforms of the VEGF receptor have demonstrated efficacy in CRC as well. Certain mutations or polymorphisms of the enzymes involved in the metabolism of these drugs have been investigated as biomarkers for predicting the response to the drug or the disease prognosis. For instance, thymidylate synthase is the target enzyme of the active metabolite of 5-FU. Several studies have evaluated the expression level of this enzyme in patients administered with 5-FU, but reached conflicting conclusions as to whether a high or low level confers a favorable response to 5-FU, or improves prognosis [24–26]. Genetic alterations in certain DNA excision-repair genes may confer differential efficacy in patients treated with oxaliplatin. A particular polymorphism in the protein involved in the glucuronidation (inactivation) of irinotecan (UGT1A1) confers reduced metabolism of the drug, and increases the patient’s chance of mylosupression and diarrhea [27]. Screening for this polymorphism in CRC patients has been approved by the US FDA. KRAS is an important protein involved in the EGFR pathway, often amplified in CRC as well other cancers. Inhibitors of EGFR, such as Erbitux, have differential efficacy depending on the mutation status of KRAS, or certain polymorphisms upstream of the coding region of the gene. Screening for KRAS mutations is now common in the clinic to better inform the oncologist’s decision to treat with these drugs [28]. BRAF is also involved in the EGFR pathway, and several studies have indicated its mutation status is important in predicting the patient’s response to the drug but, unlike KRAS, it has not been widely used in the clinic as a predictive marker for informing therapy [29]. It is beyond the scope of this article to cover, in detail, all the genetic mutations and polymorphisms that have been demonstrated to confer a differential response to a variety of the CRC drugs mentioned. For a thorough review of this topic, refer to Strimpakos et al. [29]. It is suffice to say that only KRAS has gained wide acceptance at the clinical level as genetic biomarker in CRC relevant for predicting the response to EGFR inhibitors.
Limitations & challenges
Setting aside the concern for technical variance, not unique to microarray experiments, gene-expression signatures have limitations as direct markers of biological significance. For instance, many of the so-called driver genes of cancer are not differentially expressed at the level of mRNA or the cancer progression may not be regulated at the level of expression [30]. In addition, the significant, differentially expressed genes in a signature may not resolve to only one or two distinct gene ontological processes, or the pathways they map to are unknown altogether, and this limits their usefulness as guides for mechanistic experiments. Furthermore, the expression of the mRNA does not always correlate with the expression level of the protein [31], which is the immediate effecter of cellular phenotype. In these cases, the level of gene transcription does not necessarily play an important role in the disease. These limitations should not be misunderstood to mean that genome-wide measures of protein-coding mRNAs are no longer useful as indicators of dysregulation, which are possibly important in disease. Rather, it bears repeating that these data are likely to be most useful when integrated into a comprehensive analysis that factors in all the relevant information we have regarding the cell.
Traditional proteomic approaches to markers
Unlike the human genome, estimates of the size of the human proteome are widely variable. The Human Protein Initiative estimates that 20,500 genes could code for over 1 million proteins [103]. Compounded with the myriad post-translational modifications, such as phosphorylation, ubiquination and glycosylation, to name only a few, and recognizing that a modification may occur on one or more protein residues, the full annotation of the human proteome presents an extraordinary challenge. A significant change in the expression or activity (e.g., a kinase) of one or more of these proteomic species between cancer and control indicates dysregulation in the cell, and may be a candidate biomarker. However, at present there is no high-dimensional equivalent to the microarray in proteomics. Even for that portion of the proteome, which is well annotated, it cannot be comprehensively surveyed for expression changes between tumor and control, be the sample tissue, cells or biofluids. However, significant technological improvements have been made. Cox and Mann recently demonstrated that high-resolution mass spectrometry (MS), the workhorse of many proteomic approaches, paired with statistical rigor, can quantify the differential expression change of over 4000 proteins between control and treated mammalian cells in a high-throughput manner [32]. Much progress has been made and many technical hurdles have been cleared so that, now, perhaps only money and cooperation stand in the way of high-throughput, proteome-wide profiling [33]. Furthermore, the empirical evidence that the expression of most protein species does not significantly change between cancer and control is encouraging for biomarker discovery. Therefore, it is not necessary that the entire human proteome be annotated or completely assayable to identify candidate markers of cancer. Indeed, as we discuss in a subsequent section, significant targets found by proteomic profiling are useful inputs to bioinformatic approaches that implicate other proteins with a role in cancer, proteins that lack direct experimental evidence and are unlikely to be found significant by proteomic or genomic profiling alone.
Protein markers from biofluids
At present, the only potentially useful clinical biomarker of CRC is the serum protein CEA, and its value as a predictive marker of disease recurrence has been questioned [11]. As with various genomic markers, certain circulating proteins have demonstrated high sensitivity and specificity as diagnostic, prognostic or predictive markers in cohorts of limited size. Kaaks et al. proposed a model of how chronically high circulating levels of IGFs in serum associated with a higher risk of CRC in women leading a Western lifestyle [34]. Surface-enhanced laser desorption ionization – time of flight (SELDI–TOF) MS has been used to find distinct protein species in biofluids that were able to discriminate the sera of CRC patients from controls [35,36]. When paired with a novel bioinformatic method, a similar approach was apparently able to distinguish adenoma from carcinoma using sera obtained from a large cohort of patients with mixed-stage (Duke’s A–D) CRC [37]. Various quantitative proteomic methods exist that involve covalent modification of the proteins in sample [38]. The proteins in each sample are differentially labeled with moieties of distinct mass, then mixed and digested and the peptides analyzed by liquid chromatography (LC)–MS/MS to determine the relative abundance of parent proteins present in each sample. The relative abundance of a protein between samples may also be measured by label-free strategies using mass spectrometers capable of high mass accuracy [39]. Mass spectrometers capable of high sensitivity and mass accuracy, paired with nano-chromatography are also capable of detecting and quantifying post-translationally modified proteins [40], which are increasingly recognized as playing an important role in cancer [41]. There are two challenges particular to marker discovery in biofluids. One involves the fact that biofluids are frequently highly concentrated in highly abundant proteins. The digestion products of these proteins can overwhelm the mass spectrometer’s detector and mask the detection of less abundant proteins. One strategy to overcome this problem involves depleting the sample of these proteins on columns designed specifically for this purpose. The elute is then digested in the usual manner and the peptides submitted to LC–MS/MS for sequencing. Perhaps the biggest challenge to marker discovery in fluids is the fact that a heterogeneous mix of proteins is secreted from a variety of tissues in the body, making it difficult to attribute a candidate marker to a tissue-specific disease.
Protein markers from tissue biopsy
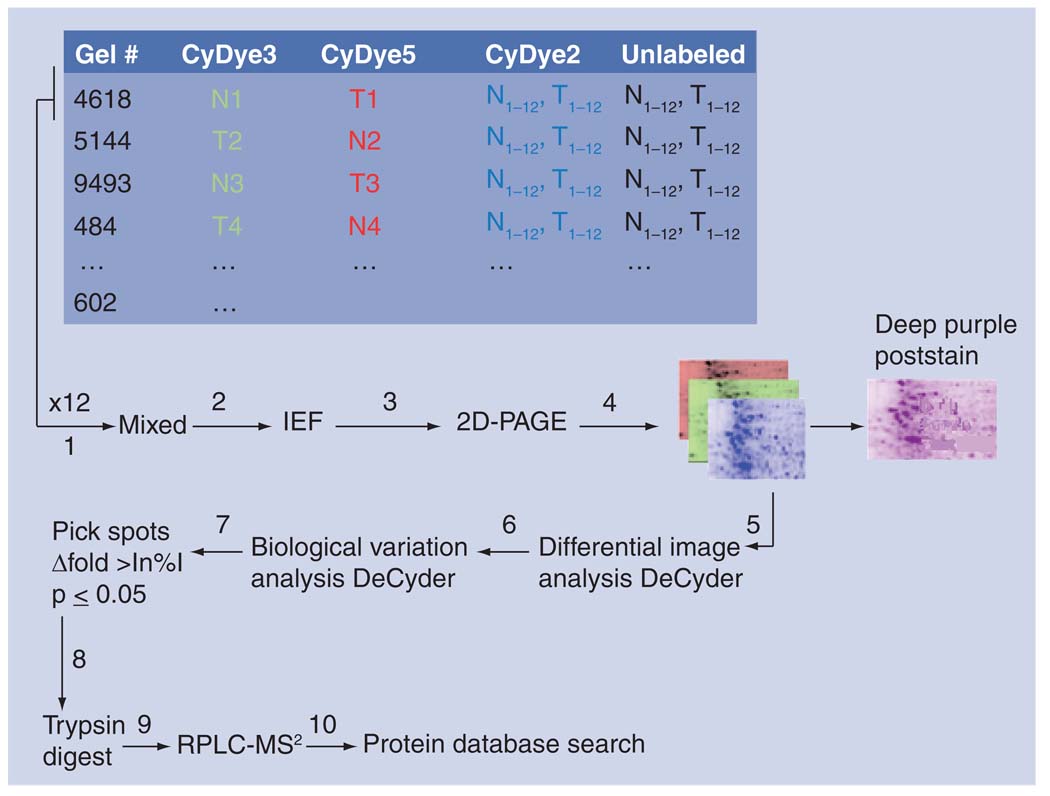
Profiling for changes in oncogenic proteins between tumor and control in tissue has three distinct advantages over biofluids: a putative marker protein may not be secreted to biofluids, the ambiguity of source tissue is eliminated and the sample is enriched in changes for tumor versus control. A common method for separating proteins collected from tissue is 2D-difference gel electrophoresis, a variant of the 2D-polyacrylimide gel electrophoresis method that allows the multiplexing of up to three samples in a single gel, typically normal, tumor and an internal standard. Each sample is labeled by a distinct flurophore and then the samples are mixed, loaded onto a poly-acrylimide gel and separated by isoelectric value and molecular weight. Follow-up image analysis allows for the identification of spots significant for the tumor phenotype. The spots are excised from the gel, digested by trypsin and the peptides submitted for sequencing by LC–MS/MS. The identification of proteins in the samples is performed subsequently by database search (Figure 2). A number of studies have used this approach to identify significantly changing proteins between matched normal and tumor tissues obtained from CRC patients [42–44]. Verification of select findings is commonly carried out by western blot or appropriate MS-based methods, optimally with samples not used in the discovery phase. The method has an ascertainment bias for detecting highly expressed proteins, but is considered suitable for identifying post-translationally modified proteins or isoforms. The bias may be overcome by prefractioning the samples to focus on proteins differentially expressed in a particular subcellular compartment (e.g., mitochondria). An alternative approach involves mapping the proteins to protein-interaction networks and, subsequently, analyzing the activity of a suite of protein interactions (a network) between tumor and control. The end point of this analysis is the inference of a functional role of a well-connected set of proteins in disease, one that is readily testable in an in vivo model. An example of this approach to functional marker discovery is discussed in the final section.
Figure 2. Proteomic profiling: 2D-differential gel electrophoresis.
Tripartite samples of N, T and pooled control were mixed (1); samples were separated by isoelectric focusing (2); then by molecular weight (3); and each fluorophore imaged independently (4). Using the DeCyder software, spots were matched on an intragel basis with differential image analysis DeCyder (5), and on an intergel basis with biological variation analysis Decyder to assess biological variation (6). Significant spots were selected for robotic excision (7), digested by trypsin (8) and the peptides separated by reverse-phase chromatography and detected by tandem mass spectrometry (9). MS2 spectra were searched against the Sequest database (10). 2D-PAGE: 2D polyacrylimide gel electrophoresis; IEF: Isoelectric focusing; MS2: Tandem mass spectrometry; N: Normal; RPLC-MS: Reverse-phase liquid chromatography-mass spectrometry; T: Tumor.
Protein markers from cell culture
Cell culture continues to be one of the most expedient experimental models for testing biological hypotheses. At the level of the genome, modern tools of cell and molecular biology allow for genes to be knocked in, knocked out, systematically mutated or modified in a myriad of ways, and the phenotype analyzed by a wide variety of methods capable of impressive precision and specificity. Similarly, at the level of the proteome, many perturbation experiments can be conducted (e.g., interference of protein expression, ectopic overexpression, pharmacological inhibition or constitutive activation) and the ensuing phenotype analyzed. As differential expression of protein continues to be viewed as a quality indicator of cellular dysregulation in cancer, methods have been developed that have enabled quantitative protein analysis between treated and control cells by sensitive MS [45]. In addition, proteins differentially modified in disease may also be found in cell models of CRC. Kim et al. surveyed the phosphoproteome of HT-29 cells and found 238 unique phosphorylation sites that the authors suggested may be used as surrogate markers implicating the differential activity of a suite of kinases [46]. Cell culture is an equally useful model for verification of mechanistic hypothesis involving markers found in tissue or biofluids. If one has a cell line with a similar genetic background and pathologic stage to a cohort of clinical samples used to screen for candidate markers, mechanistic hypotheses involving that marker may be tested in a cell model. In addition, the conditioned media in which the cells grow can be quantitatively assayed for proteins in the secretome, and in this way used to verify the candidate markers found in a screen of biofluids [47]. The limitation of cell culture as a model of human disease lies in the fact that cells grown in (2D) culture are known to have altered metabolism, and the microenvironment is often strikingly different, lacking features of angiogenesis, for example; xenografts or other animal models of disease may then be more appropriate.
Interactome
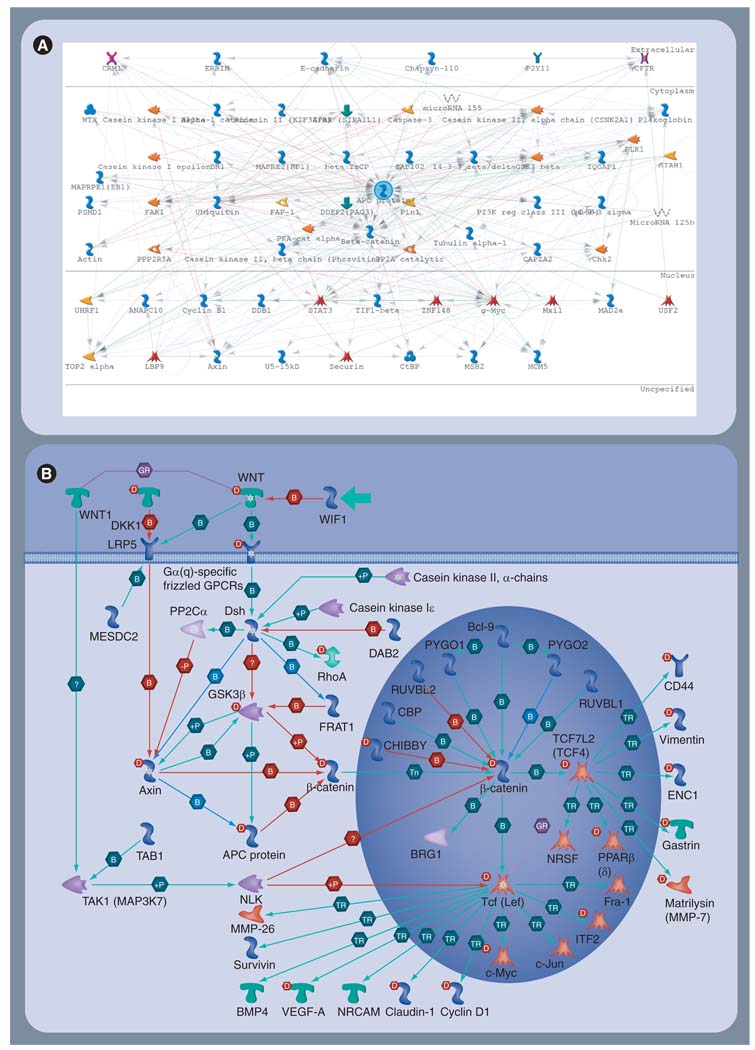
Proteins in the cell do not function independently. Cellular phenotype is the result of proteins interacting with each other and with other molecules in the cell (e.g., lipids, RNA, DNA, hormones and drugs). How these interactions are coordinated and regulated is, to a large extent, unclear. There is, however, wide agreement that cancer is caused and sustained by dysregulated pathways (networks) driven by mutant proteins. Many efforts are underway to annotate and catalog individual interactions between proteins (and other molecules) in computer databases, and these databases may be used to build large network graphs of interactions. These graphs reveal the daunting complexity we must deal with if we are to understand the functional implications of differentially expressed genes or proteins in disease. For example, Figure 3A is an interaction graph that depicts the proteins (70) known to interact with APC, the so-called ‘gatekeeper’ gene, mutated in over 90% of CRC tumors [10]. By contrast, Figure 3B shows the relatively few interactions involving APC in the well-studied WNT-signaling pathway, known to be dysregulated in CRC. Clearly, Figure 3A indicates there are many more interactions on the APC axis that may play an important role in causing, sustaining or suppressing the cancer phenotype. Furthermore, it is certainly conceivable that the differences in the activity of these various interactions may account for the heterogeneity of tumors in patients, the difference in the aggressiveness of their tumors and, ultimately, may be the source of the differential response to treatment frequently observed in the clinic [48]. The implication is clear; improved markers of CRC will need to account for the coordinated, differential expression of many genes or proteins that synergistically accelerate or retard the activity of networks responsible for disease [49].
Figure 3. Relevant colorectal cancer subnetworks and pathways.
(A) Protein interaction graph of the APC (center) axis. Nodes (n = 70) are indicated as glyphs (e.g., enzymes, transcription factors and receptors) and interactions with APC are indicated by lines. Evidence of the interactions is based on literature curation. The nominal cellular compartment of each protein is indicated by annotation on the right. (B) Map of the canonical WNT-signaling pathway, including APC (left, center). MetaCore©.
It is beyond the scope of this article to delve into the details of all the interactomic databases presently available, both public and commercial. It is sufficient to note that many are species specific; the first attempt to build a comprehensive protein–protein interaction (PPI) database was carried out in yeast [50]. Human PPIs have also been constructed based on a variety of evidence; from pull-down experiments (e.g., yeast two-hybrid or coimmunoprecipitation), inference from homology, computational predictions from binding motifs, evidence found in the literature or a combination of these [51]. For a review of publicly available human PPIs, refer to Mathivanan et al. [52]. Human PPIs mark a milestone for biomarker discovery in cancer because they provide a functional context in which to analyze the mechanistic role of genes or proteins found to be significantly differentially expressed by traditional screens.
Network-based markers in colorectal cancer
Traditional genomic and proteomic approaches to biomarker discovery in cancer, by themselves, have limitations. Integrating all the information regarding the cell into a functional model may improve the robustness and reliability of markers in cancer. Indeed, Chuang et al. recently demonstrated a network-modeling approach, which integrated gene-expression profiles with interaction networks to more reliably and robustly predict breast cancer metastasis [53]. Their approach mapped microarray data to a human PPI and then searched for small subnetworks within the PPI that could distinguish metastatic and nonmetastatic patients. A classifier constructed from these subnetworks was more accurate at predicting metastasis when compared with single gene markers. In a similar approach, Jonsson et al. mapped a consensus of 346 cancer genes to a carefully constructed human PPI (based on homology), and found that these genes had, on average, twice as many connections as noncancer genes [54]. In a related approach, Segal et al. used a series of gene-expression signatures and human curated annotations to identify cancer modules, some of which generalized to tumorigenesis while others were found to be stage or tissue specific [55]. These studies, in addition to others, provide compelling evidence that coexpressed genes in cancer concentrate nonrandomly in ‘hotspots’, and when mapped to the interactome revealed well-connected sets of proteins (modules). In an evolutionary sense, it is these modules (or subnetworks) that are seen as being selected for the growth advantage they convey in cancer.
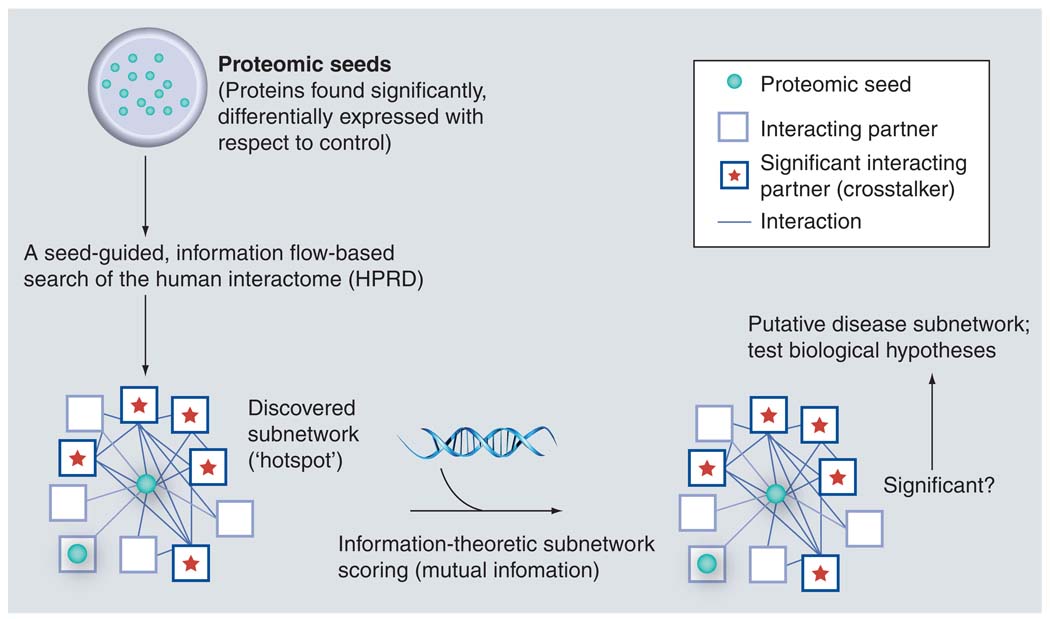
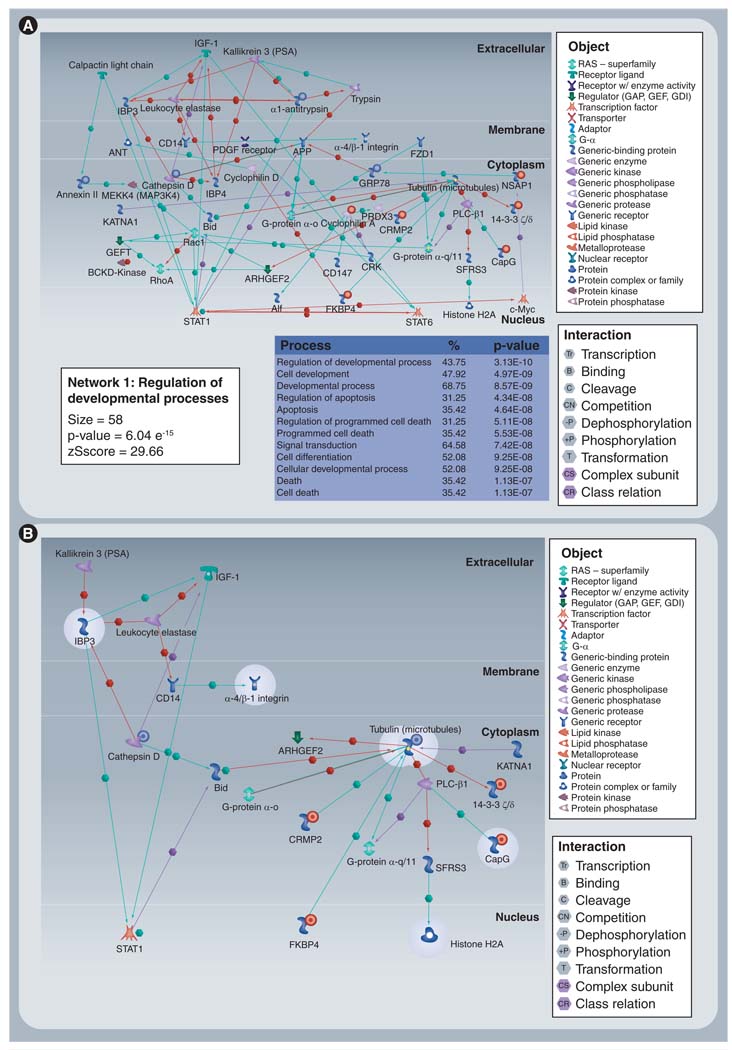
Building on this evidence, Nibbe et al. hypothesized that the highly significant targets (n = 67) found by a proteomics screen in tissue for a CRC phenotype, would be an optimal dataset to ‘seed’ a search for subnetworks significant for the phenotype (Figure 4) [56]. Instead of searching the entire network, their approach was guided by the hypothesis that these targets would lie on or near hotspots in the interactome. Using a well-annotated PPI constructed of high-confidence interactions, they were able to discover significant subnetworks variably saturated with their proteomic targets. Adapting the scoring method used by Chuang et al., they then systematically pruned these subnetworks to find well-connected sets of proteins that were highly discriminative of the cancer phenotype (Figure 5). In a separate cohort of clinical samples, they verified the protein expression change of these targets. The result indicated coregulation of the targets both at the level of translation and transcription (as measured by microarray).
Figure 4. Proposed bioinformatic flow.
The proteomic first approach begins with a seed of targets that are found to be significant for disease. These are used to search for hotspots in the interactome – subnetworks of well-connected proteins that reveal significant proximity and interactivity (crosstalk) to the targets in the seed. Using differential mRNA expression between normal and tumor (microarray) as a surrogate for network activity, each subnetwork is scored and pruned for its ability to discriminate normal from tumor. The pruned subnetworks are evaluated for their role in the disease state, and may be used to inform mechanistic verification experiments. HPRD: Human Protein Reference Database.
Figure 5. Discovered, significant subnetworks.
(A) Discovered subnetwork that is annotated as that expressed in late-stage colorectal cancer and (B) the pruned version. Significant targets in pruned version (highlighted in gray) were extended one interaction to infer functional relevance. Proteins for which there was direct proteomic evidence (seed) have adjacent red (upregulated) or blue (downregulated) circles.
Reproduced with permission from [56].
Future perspective
Alterations to individual genes or proteins in CRC provide only one clue to the complex cellular changes driving this cancer. Likewise, candidate patterns of gene expression account for only a limited variability of the disease phenotype, preventing their adoption in the clinic as useful prognostic or predictive markers of CRC. A rich literature now exists for systems biology-based approaches to marker discovery in a variety of human diseases. The challenges of the ‘omics revolution are large, but the early results are encouraging. The integration of high-dimensional results from genomics and proteomics, combined with legacy data underwriting interactomic databases, holds promise to pave the way for improved classifiers of disease.
Executive summary.
Epidemiology & genetics of colorectal cancer
Colorectal cancer (CRC) is the second leading cause of cancer death in adult Americans.
Specific genetic alterations drive inherited forms of CRC (familial adenomatous polyposis coli and hereditary nonpolyposis colon cancer).
A diverse accumulation of somatic mutations cause and propagate sporadic CRC tumors.
Biomarkers: old & new
Diagnostic marker: presage the early onset of cancer.
Prognostic marker: predicts the likely progression of the tumor and patient outcome.
Predictive marker: predicts response of the tumor to a specific therapy regimen.
Ideal markers would be readily measured and useful in the clinic.
Genomic markers of colorectal cancer
Gene mutations: commonly APC, TP53, SMAD4 and KRAS. At present, only KRAS may be clinically useful as a predictive marker.
Transciptomic changes: measured by genotyping or genome-wide analysis (microarray).
Single-nucleotide polymorphisms and copy-number variations: used in genome-wide association studies. Typically, these only account for a limited variability of phenotype.
miRNAs: new and promising, but as a solitary they are measure unlikely to account for complex cancer phenotypes.
Proteomic approaches to markers
Proteome-wide profiling is not presently feasible, however, it looks promising in the fuutre.
Markers from biofluids: serum markers of CRC have been identified by mass spectrometry approaches, but have not been validated in multiple, large cohorts.
Markers from tissue: offers three important advantages over proteomic profiling in biofluids, but markers still require verification and validation in larger cohorts.
Markers in cell culture: ideal basis for verification of markers discovered in fluids or tissue, but the model lacks the features of an in situ tumor.
Interactome
A database of protein–protein interactions that may be mined for small networks of interactions which may be differentially active between tumor and control.
A powerful basis for integrative approaches for finding improved, functional markers of disease.
Commonly species specific; several high-confidence human protein–protein interactions are now available.
Network-based markers in colorectal cancer
Emerging evidence indicates that proteins with a functional role in cancer concentrate in hotspots in the proteome.
Cancer proteins have been shown to be highly connected (modules) in the proteome.
Some modules have been shown to predict tumorigenesis while others were stage specific.
Network-based markers were shown to be more accurate predictors of metastasis in breast cancer, compared with traditional approaches based on gene-expression analysis alone.
A proteomics-first approach provides a novel way to search for functional subnetworks of proteins discriminative of CRC.
Systems biology-based approaches integrate all the information about the cell, and promise to deliver more robust and accurate classifiers of disease, compared with traditional approaches.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was supported, in whole or in part, by NIH grants UL1-RR024989 from the National Center for Research Resources (Clinical and Translational Science Awards), P30-CA043703, CWRU Cancer Research Center Proteomics core and T32-GM008803 from the National Institute of General Medical Sciences (NIGMS) (Institutional National Research Service Award). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪ ▪ of considerable interest
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Powell SM, Petersen GM, Krush AJ, et al. Molecular diagnosis of familial adenomatous polyposis. N. Engl. J. Med. 1993;329:1982–1987. doi: 10.1056/NEJM199312303292702. [DOI] [PubMed] [Google Scholar]
- 3.Markowitz SD, Dawson DM, Willis J, Willson JK. Focus on colon cancer. Cancer Cell. 2002;3:233–236. doi: 10.1016/s1535-6108(02)00053-3. [DOI] [PubMed] [Google Scholar]
- 4.Kinzler KW, Vogelstein B. Lessons from hereditary colon cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 5.Green RC, Parfrey PS, Woods MO, Younghusband HB. Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J. Natl Cancer Inst. 2009;101:331–340. doi: 10.1093/jnci/djn499. [DOI] [PubMed] [Google Scholar]
- 6.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 7.Hornberg JJ, Bruggeman FJ, Westerhoff HV, Lankelma J. Cancer: a systems biology disease. Biosystems. 2006;83:81–90. doi: 10.1016/j.biosystems.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 8. Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. ▪ Good review of the genetic basis for human colorectal cancer, including the relevant downstream signaling pathways, and where the opportunities lie for further research.
- 9.Calvert PM, Frucht H. The genetics of colorectal cancer. Ann. Intern. Med. 2002;137:603–612. doi: 10.7326/0003-4819-137-7-200210010-00012. [DOI] [PubMed] [Google Scholar]
- 10. Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. ▪▪ Landmark paper that identified the driver genes (and passenger genes) implicated in the initiation and/or progression of colorectal cancer (CRC).
- 11. Saltz LB. Biomarkers in colorectal cancer: added value or just added expense? Expert Rev. Mol. Diagn. 2008;8:231–233. doi: 10.1586/14737159.8.3.231. ▪ Concise and critical review of the state of clinically useful biomarkers in CRC.
- 12.DeRisi J, Penland L, Brown PO, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 13.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;337(Database issue):D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabates-Bellver J, Van der Flier LG, de Palo M, et al. Transcriptome profile of human colorectal adenomas. Mol. Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X, Tan J, Li J, et al. DACT3 is an epigenetic regulator of Wnt/β-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529–541. doi: 10.1016/j.ccr.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer J, Allen WL, McLean EG, et al. Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res. 2006;66:2765–2777. doi: 10.1158/0008-5472.CAN-05-2693. [DOI] [PubMed] [Google Scholar]
- 17.Huang DS, Zheng CH. Independent component analysis-based penalized discriminant method for tumor classification using gene expression data. Bioinformatics. 2006;22:1855–1862. doi: 10.1093/bioinformatics/btl190. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 19.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 20.Tsafrir D, Bacolod M, Selvanayagam Z, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 21.Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J. Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc. Natl Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edler D, Glimelius B, Hallstrom M, et al. Thymidylate synthase expression in colorectal cancer: a prognostic and predictive marker of benefit from adjuvant fluorouracil-based chemotherapy. J. Clin. Oncol. 2002;20:1721–1728. doi: 10.1200/JCO.2002.07.039. [DOI] [PubMed] [Google Scholar]
- 25.Soong R, Shah N, Salto-Tellez M, et al. Prognostic significance of thymidylate synthase, dihydropyrimidine dehydrogenase and thymidine phosphorylase protein expression in colorectal cancer patients treated with or without 5-fluorouracil-based chemotherapy. Ann. Oncol. 2008;19:915–919. doi: 10.1093/annonc/mdm599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prall F, Ostwald C, Schiffmann L, Barten M. Do thymidylate synthase gene promoter polymorphism and the C/G single nucleotide polymorphism predict effectiveness of adjuvant 5-fluorouracil-based chemotherapy in stage III colonic adenocarcinoma? Oncol. Rep. 2007;18:203–209. [PubMed] [Google Scholar]
- 27.Cote JF, Kirzin S, Kramar A, et al. UGT1A1 polymorphism can predict hematologic toxicity in patients treated with irinotecan. Clin. Cancer Res. 2007;13:3269–3275. doi: 10.1158/1078-0432.CCR-06-2290. [DOI] [PubMed] [Google Scholar]
- 28.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 29.Strimpagos AS, Syrigos KN, Saif MW. Pharmacogenetics and biomarkers in colorectal cancer. Pharmacogenomics J. 2009;9:147–160. doi: 10.1038/tpj.2009.8. [DOI] [PubMed] [Google Scholar]
- 30.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 31.Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 33. Service RF. Proteomics ponders prime time. Science. 2008;321:1758–1761. doi: 10.1126/science.321.5897.1758. ▪ Adequately covers the challenges facing the proteomics community in identifying biomarkers in human disease.
- 34.Kaaks R, Toniolo P, Akhmedkhanov A, et al. Serum C-peptide, insulin-like growth factor (IGF)-I, IGF-binding proteins, and colorectal cancer risk in women. J. Natl Cancer Inst. 2000;92:1592–1600. doi: 10.1093/jnci/92.19.1592. [DOI] [PubMed] [Google Scholar]
- 35.Ward DG, Suggett N, Cheng Y, et al. Identification of serum biomarkers for colon cancer by proteomic analysis. Br. J. Cancer. 2006;94:898–905. doi: 10.1038/sj.bjc.6603188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habermann JK, Paulsen U, Roblick UJ, et al. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes Chromosomes Cancer. 2007;46:10–26. doi: 10.1002/gcc.20382. [DOI] [PubMed] [Google Scholar]
- 37.Chen YD, Zheng S, Yu JK, et al. Artificial neural networks analysis of surface-enhanced laser desorption/ionization mass spectra of serum protein pattern distinguishes colorectal cancer from healthy population. Clin. Cancer Res. 2004;10:8380–8385. doi: 10.1158/1078-0432.CCR-1162-03. [DOI] [PubMed] [Google Scholar]
- 38.Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief. Funct. Genomic. Proteomic. 2006;5:112–120. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 39.Asara JM, Christofk HR, Freimark LM, et al. A label-free quantification method by MS/MS TIC compared with SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994–999. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 40.Gafken PR, Lampe PD. Methodologies for characterizing phosphoproteins by mass spectrometry. Cell Commun. Adhes. 2006;13:249–262. doi: 10.1080/15419060601077917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 42.Friedman DB, Hill S, Keller JW, et al. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2004;4:793–811. doi: 10.1002/pmic.200300635. [DOI] [PubMed] [Google Scholar]
- 43.Alfonso P, Núñez A, Madoz-Gurpide J, et al. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics. 2005;5:2602–2611. doi: 10.1002/pmic.200401196. [DOI] [PubMed] [Google Scholar]
- 44.Mazzanti R, Solazzo M, Fantappié O, et al. Differential expression proteomics of human colon cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:1329–1338. doi: 10.1152/ajpgi.00563.2005. [DOI] [PubMed] [Google Scholar]
- 45.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 46.Kim JE, Tannenbaum SR, White FM. Global phosphoproteome of HT-29 human colon adenocarcinoma cells. J. Proteome Res. 2005;4:1339–1346. doi: 10.1021/pr050048h. [DOI] [PubMed] [Google Scholar]
- 47.Volmer MW, Stühler K, Zapatka M, et al. Differential proteome analysis of conditioned media to detect Smad4 regulated secreted biomarkers in colon cancer. Proteomics. 2005;5:2587–2601. doi: 10.1002/pmic.200401188. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Shannon WD, Watson MA, et al. Gene expression profiling of the irinotecan pathway in colorectal cancer. Clin. Cancer Res. 2005;11:2053–2062. doi: 10.1158/1078-0432.CCR-04-1254. [DOI] [PubMed] [Google Scholar]
- 49.McMurray HR, Sampson ER, Compitello G, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2008;8:437–439. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 51.De Las Rivas J, de Luis A. Interactome data and databases: different types of protein interaction. Comp. Funct. Genomics. 2004;5:173–178. doi: 10.1002/cfg.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathivanan S, Periaswamy B, Gandhi TK, et al. An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics. 2006;7(Suppl 5):S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chuang HY, Lee E, Liu YT, et al. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140–150. doi: 10.1038/msb4100180. ▪▪ Widely influential paper that showed network-based markers of breast cancer were more accurate classifiers of metastatic breast cancer, compared with a traditional microarray approach.
- 54.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2007;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segal E, Friedman N, Koller D, et al. A module map showing conditional activity of expression modules in cancer. Nat. Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 56. Nibbe RK, Markowitz S, Myeroff L, et al. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol. Cell. Proteomics. 2009;8:827–845. doi: 10.1074/mcp.M800428-MCP200. ▪▪ Describes a novel ‘proteomics-first’ approach to identify protein subnetworks discriminative of late-stage CRC versus control, which provide mechanistic models for hypothesis testing in vivo.
Websites
- 101. [reviewed 2008];Bethesda (MD): Lynch syndrome; National Library of Medicine (USA), Specialized Information Services Division. 2003 http://chem.sis.nlm.nih.gov/chemidplus/chemidlite.jsp.
- 102.American Cancer Society. 2008 www.cancer.org/docroot/cri/content/cri_2_4_1x_what_are_the_key_statistics_for_colon_and_rectum_cancer.asp.
- 103.Human Proteome Initiative. 2008 http://ca.expasy.org/sprot/hpi.
- 104.National Cancer Institute VisualsOnline. http://visualsonline.cancer.gov/details.cfm?imageid=7181.