Abstract
The active form of vitamin D (1α,25(OH)2D3) is known to have antiproliferative effects and has been implicated in cancers of the colon, breast, and prostate. These cancers occur more frequently among African Americans than Caucasians, and individuals with African ancestry are known to have approximately two-fold lower levels of serum vitamin D (25(OH)D) compared with individuals of European ancestry. However, epidemiological studies of the vitamin D receptor (VDR) have shown inconsistent associations with cancer risk, suggesting that differences in other genes in the pathway may be important. We sought to identify functionally significant polymorphic variants in CYP24A1, a gene that is highly inducible by 1α,25(OH)2D3 and that encodes the primary catabolic enzyme in the pathway. Here we report the identification of six novel SNPs in the human CYP24A1 promoter, including one at nucleotide -279 occurring within the distal vitamin D response element (VDRE2). Our experiments demonstrate that the VDRE2 variant results in decreased protein binding and transactivation in vitro, and reduced expression of CYP24A1 in cultured primary human lymphocytes provides evidence for an effect in vivo. This variant was only observed in our African American population, and represents a first step toward understanding differences in disease risk among racial/ethnic groups.
Keywords: CYP24A1; 24-hydroxylase; promoter; 1α,25(OH)2D3; vitamin D receptor; single nucleotide polymorphism; VDRE
1. Introduction
The biologically active form of vitamin D (1α,25(OH)2D3) has multiple roles in the body, including regulation of intestinal calcium absorption, maintenance of bone mineral density, and modulation of cell growth and apoptosis [1]. Genomic actions of 1α,25(OH)2D3 are mediated through ligand-binding to the vitamin D receptor (VDR), which forms a heterodimer with retinoid x receptor alpha (RXRα) and subsequently binds to vitamin D response elements (VDRE) to either enhance or repress transcription of various genes. These response elements are typically comprised of two conserved hexameric half-sites separated by a three nucleotide spacer, referred to as a DR3 type element. Although it is known that the sequence of a VDRE can have a strong influence on the degree of protein binding, particularly at the fifth position in the half-site [2], previous studies have focused on synthetic variations of response elements and not naturally occurring sequences [3].
It has been known for some time that there are significant racial/ethnic differences in serum vitamin D status, with individuals of African ancestry having approximately two-fold lower levels than those with European ancestry [4-6]. Low levels of serum vitamin D (25(OH)D) have been associated with cancers of the colon, breast, and prostate, as well as an increased risk of cardiovascular disease [7-10]. Compared with whites, African Americans are more frequently diagnosed with these cancers and are at a greater risk for cardiovascular disease [11-13], indicating the possibility that polymorphic variants in the vitamin D pathway could be influencing these disparities. There are several well-known variants in the vitamin D receptor (VDR), but epidemiological studies have shown inconsistent associations with disease outcomes, particularly among different racial/ethnic groups [14-19]. Taken together, this suggests that other variants or combinations of variants within the pathway may underlie the differences in serum vitamin D and disease risk.
The CYP24A1 gene has very low basal expression, but is strongly upregulated by 1α,25(OH)2D3 through two VDRE in the proximal promoter region [20, 21]. The resulting 24-hydroxylase enzyme catalyzes the first step in the catabolic pathway, converting 1α,25(OH)2D3 into the less active intermediate 1,24,25(OH)3D3 [22]. Through activation of this and other negative feedback loops, 1α,25(OH)2D3 can regulate its own metabolism. We hypothesized that there were unidentified SNPs in the promoter of the CYP24A1 gene that could alter expression of the 24-hydroxylase enzyme and impact the rate at which an individual can metabolize 1α,25(OH)2D3. Population studies to date have sequenced only a small number of individuals, particularly individuals of African ancestry, so it is possible that significant polymorphic variants in this region have yet to be identified.
In the work presented here, we report the identification of six novel SNPs in the human CYP24A1 promoter, including one that occurs in the fifth position of the distal vitamin D response element (VDRE2). We demonstrated that this VDRE2 polymorphism results in impaired receptor protein binding and decreased transactivation in vitro, and more importantly, we provide evidence that the variant can lead to decreased expression of the CYP24A1 gene in a heterozygous polymorphic individual.
2. Materials and methods
2.1. Subjects
Healthy individuals who classified themselves as at least 50% Caucasian or at least 50% African American were recruited at Pennsylvania State University General Clinical Research Center during the winter and spring months. The 100 total participants were matched on age (within 2 years), sex, and opposite self-reported race. Each participant donated a venous blood sample that was used to prepare dried blood spot cards and for Ficoll separation with Ficoll-Paque PLUS (GE Healthcare) to obtain lymphocytes.
2.2. DNA isolation and sequencing to screen for CYP24A1 promoter polymorphisms
Genomic DNA for SNP discovery was isolated from dried blood spot cards from 20 randomly selected African American participants in our study using the QIAamp DNA Micro Kit (QIAGEN). A 677 bp region of the proximal CYP24A1 promoter (-618 to +59) including two known VDRE was PCR-amplified using Phusion DNA polymerase (New England Biolabs) and the following primers: 5′-GTGTCAAGGAGGGTAGATGAGATG-3′ (forward) and 5′-TTGCTCAAGTTAAGAAAGTCTCCTC-3′ (reverse). The desired PCR products were gel-isolated using the QIAquick Gel Extraction Kit (QIAGEN), run on agarose gels to verify DNA recovery, and sequenced using the forward PCR primer at the University of Pennsylvania School of Medicine DNA Sequencing Facility by automated cycle sequencing. Polymorphisms were identified by BLAST alignment with the wild-type sequence for human chromosome 20 (GenBank accession number NT_011362) and by visual inspection of printed chromatograms. Some SNPs were also confirmed by restriction fragment length polymorphism analysis. Resulting sequence variants were compared to data from NCBI (build 36, dbSNP b126) and the HapMap project (www.hapmap.org, release 22).
2.3. DNA isolation and SNP Genotyping for the VDRE2 polymorphism
Genomic DNA was isolated from frozen lymphocytes of all 100 participants with the QIAamp DNA Mini Kit (QIAGEN). A custom TaqMan SNP Genotyping assay (Applied Biosystems) was designed for the newly identified VDRE2 polymorphism and subsequently tested on samples of known genotype to conduct an internal validation of the assay. Quantitative Real-Time PCR and allelic discrimination were conducted at the Functional Genomics Core Facility at the Penn State College of Medicine using 10 ng genomic DNA per assay. Genotypes were successfully determined for 99% of the samples with a reliability rate of 100% when a random 10% sample was again genotyped in a separate experiment. Samples were also genotyped for the presence or absence of the M1T polymorphism in VDR (rs10735810) using a predesigned TaqMan assay (Applied Biosystems, assay ID# C_12060045_20) with similar success.
2.4. Plasmid construction and site-directed mutagenesis
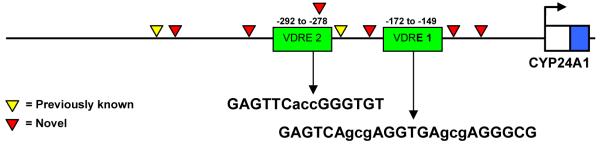
The hCYP24p-Luc construct was generated by first PCR-amplifying and gel-isolating a 677 bp region of the proximal CYP24A1 promoter (-618 to +59) as described above. The purified product was then subjected to a second round of amplification using two nested primers designed to introduce Xho I and Hind III restriction sites for subcloning: 5′-TGCTCgAGTTAAGAAAGTCTCCTCTTC-3′ (forward) and 5′-GGACCAaGCtTTTATGGAGACAGA-3′ (reverse). The new PCR product of 603 bp (-617 to -15) was digested with the above restriction endonucleases to expose the cohesive ends and was gel-purified as before. The pGL3-Promoter vector (Promega) was similarly digested and gel-purified, then ligated overnight with the CYP24A1 promoter fragment and transformed into chemically competent E. coli. Sequencing was done to confirm the orientation and integrity of the newly created hCYP24p-Luc construct, which contains the proximal CYP24A1 promoter (-611 to -25) driving expression of firefly luciferase under the control of two VDRE: GAGTCAgcgAGGTGAgcgAGGGCG at -169 to -145 (VDRE 1) and GAGTTCaccGGGTGT at -289 to -274 (VDRE2).
To create the hCYP24pV2SNP-Luc construct, we used the hCYP24p-Luc plasmid as template for site-directed mutagenesis with the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) as described by the manufacturer. Briefly, the primers 5′-CGAAGCACACCCGGTGGACTCCGGGCTT-3′ (sense) and 5′-AAGCCCGGAGTCCACCGGGTGTGCTTCG-3′ (antisense) were annealed to the hCYP24p-Luc plasmid to allow synthesis of the mutated promoter with Pfu DNA polymerase, followed by digestion of the parental plasmid by Dpn I, transformation into XL1-Blue chemically competent cells, and screening of the resulting colonies by restriction endonuclease digestion. Positive clones were verified by sequencing, and the new construct was identical to the wild-type promoter with the exception of a single base pair change in VDRE2 (GAGTTCaccGGGTGT was mutated to GAGTCCaccGGGTGT) to reflect the polymorphism identified at -279 during sequencing of our human subjects.
The r24OHase-Luc construct, a generous gift from Dr. H. DeLuca at the University of Wisconsin-Madison, contains the proximal 942 bp of rat 24-hydroxylase (CYP24A1) promoter with two VDRE cloned into pMAMneo-Luc vector upstream of firefly luciferase. This was used as a positive control for induction by 1α,25(OH)2D3 in transfection experiments.
Expression constructs for VDR and RXRA were generated from an existing human cDNA pool made by reverse-transcription of human liver RNA, a gift from Dr. Philip Lazarus at Penn State University. Coding sequences for both VDR and RXRA were specifically amplified with the following primers: 5′-GGTCTGAAGTGTCTGTGAGACCTC-3′ (VDR-forward), 5′-ACAAACAGCAACTCCTCATGGCTG-3′ (VDR-reverse), 5′-GGGCATGAGTTAGTCGCAGA-3′ (RXRA-forward), and 5′-AAACAGGCCAGGCAGAGAAG-3′ (RXRA-reverse). Amplified cDNAs were gel-isolated and sequenced to verify that they were wild-type, then TA-cloned separately into the pcDNA3.1/V5-His-TOPO vector (Invitrogen) and transformed into chemically competent E. coli. Multiple transformed colonies were screened for the presence and orientation of the cDNA inserts, and positives were confirmed by bidirectional DNA sequencing. The resulting constructs (pcDNA3.1-VDR and pcDNA3.1-RXRA) contain the wild-type coding sequences with native stop codons, and expression is driven by the CMV promoter.
2.5. Cell culture, transient transfection, and luciferase assays
Human breast cancer cells (MCF-7) and human lung carcinoma cells (H1299) were maintained at 5% CO2 in MEM supplemented with 10% fetal bovine serum, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen).
H1299 cells were seeded in 6-well plates and allowed to attach for at least 18 hours. Prior to transfection, cells were rinsed once with PBS to remove traces of serum and replaced with serum-free, antibiotic free MEM. Lipofectamine 2000 reagent (Invitrogen) was used as described by the manufacturer to form DNA complexes containing 0.6 μg luciferase reporter construct (r24OHase-Luc, hCYP24p-Luc, or hCYP24pV2SNP-Luc), 0.6 μg of each expression vector (pcDNA3.1-VDR and pcDNA3.1-RXRA), and 0.2 μg pRL-SV40 vector (Promega) to control for transfection efficiency. Whenever necessary, pUC19 (carrier DNA) was added to bring the total amount of DNA per well to 2 μg. Four hours post-transfection, complexes were removed by aspiration and one rinse with PBS prior to the addition of complete MEM and 100 nM 1α,25(OH)2D3 (Alexis Biochemicals) or vehicle (EtOH). After 16 hours of treatment, cells were harvested by trypsinization, centrifuged for 5 min. at 100 × g to pellet, resuspended thoroughly in passive lysis buffer (Promega), and stored at -70°C. MCF-7 cells were transfected in a similar manner using a total of 4 μg DNA per well, keeping the same plasmid ratios.
To enhance cell lysis prior to the luciferase assay, samples were thawed for 3 minutes in a room temperature water bath following removal from storage at -70°C. Thawed lysates were centrifuged at 4°C for one minute at high speed in a microcentrifuge to pellet cellular debris and the supernatant was transferred to a new microcentrifuge tube on ice. The Dual Luciferase Assay Kit (Promega) was used according to the manufacturer’s instructions and samples were measured using a dual-injector luminometer (Pharmingen). Data were expressed as a ratio of firefly to Renilla luciferase units.
2.6. Electrophoretic mobility shift assays (EMSA)
To prepare the probes for gelshift assays, the following sets of complementary oligonucleotides containing the desired response element were synthesized with a 5′ biotin label as such (sequences given for the plus strand only, and VDRE is in uppercase): wild-type VDRE2 from hCYP24A1 promoter, 5′-cgaagcACACCCggtGAACTCcgggctt-3′; and polymorphic VDRE2 from hCYP24A1 promoter, 5′-cgaagcACACCCggtGGACTCcgggctt-3′. (Identical oligos were also synthesized without the 5′ biotin label for competitive binding reactions.) Double-stranded DNA probes were made by boiling 150 pmoles of each complementary oligo in the presence of 50 mM NaCl for 5 minutes, then slowly cooling to room temperature overnight to allow annealing. Binding reactions were set up using 100 ng recombinant human VDR protein (BIOMOL) and/or 100 ng recombinant human RXRα (OriGene) in binding buffer (10 mM Tris, 50 mM KCl, 1 mM DTT; pH 7.5) with 1 mM EDTA, 5% glycerol, 1 μg BSA, and 0.1 μg poly (dI-dC) in the presence or absence of 1 μM 1α,25(OH)2D3. In some cases, binding reactions were set up with additional salt for a total of 150 mM KCl. After a 30 min preincubation, 25 fmol biotinylated dsDNA probe was added to each reaction and incubated for an additional 20 min at room temperature. For competitive binding studies, 5 pmol unlabeled competitor DNA was added at the preincubation step to be in 200-fold molar excess of the labeled probe. Reactions were separated on 6% native polyacrylamide gels, transferred to Biodyne B membranes (Pierce), and crosslinked using a UV Stratalinker (Stratagene). Detection was performed using the LightShift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer’s instructions.
2.7. Quantitative Real-Time PCR of CYP24A1 and VDR
Primary lymphocytes from one subject with the CYP24A1 VDRE2 polymorphism and one subject wild-type for VDRE2 were matched on age (+/- 3 years), male sex, and race. The cells were rapidly thawed and placed in culture medium (RPMI, 10% heat-inactivated fetal bovine serum, L-glutamine, and pen/strep, all from Invitrogen) and allowed to recover overnight in an incubator (37°C, 5% CO2). The cells were then counted by use of trypan blue exclusion dye and a hemacytometer and each subject’s cells were divided equally into two wells of a 6-well plate in culture medium supplemented with 2 μg/ml PHA-M and 20 ng/ml TPA (both from Sigma) for activation. After 14 hours, either 100 nM 1α,25(OH)2D3 (Alexis Biochemicals) or vehicle (ethanol) was added to each set of lymphocytes and allowed to incubate an additional 6 hours. Total RNA was isolated from 5×105 cells using the Absolutely RNA Microprep Kit (Stratagene) as described by the manufacturer. The quality of RNA yield was determined at the Functional Genomics Core Facility at Penn State College of Medicine on an Agilent BioAnalyzer with the Total RNA Nano assay (Agilent Technologies, Inc.). Approximately 250 ng of total RNA from each sample were used as template for cDNA synthesis with SuperScript II Reverse Transcriptase and oligo(dT)12-18 (Invitrogen) as described by the manufacturer. The resulting cDNA was template for Real-Time PCR using prevalidated TaqMan Gene Expression Assays for human CYP24A1, VDR, and GAPDH (Applied Biosystems assay ID#s Hs00167999-m1, Hs01045840_m1, and Hs99998805_m1, respectively), and the relative quantification of treated versus control lymphocytes was determined using SDS software and the ΔΔCT method.
3. Results
3.1. Identification and genotyping of polymorphisms in the CYP24A1 promoter
Direct sequencing results yielded 8 SNPs in the 500 nucleotide segment of proximal CYP24A1 promoter among African Americans. Of these, 6 were novel low-prevalence polymorphisms currently not listed on NCBI or HapMap (Table 1 and Figure 1); all were identified only in heterozygotes. The SNP at -279 relative to the transcription start site occurs in the fifth position of the hexameric repeat in VDRE2 in three of the twenty samples sequenced. After genotyping all 100 participants from our study (African Americans and Caucasians) for the VDRE2 polymorphism with the custom SNP genotyping assay, we were able to identify one more heterozygous individual in our African American population and no homozygous polymorphic individuals (4/50 heterozygotes, or 4% allelic prevalence). The SNP was not observed in any of the 49 individuals successfully genotyped in the Caucasian population.
Table 1. CYP24A1 Promoter SNPs Identified From Direct Sequencing.
| Position Relative to Start Site |
SNP reference # |
Base Pair Change |
Genotypea |
SNP Allelic Frequency |
||
|---|---|---|---|---|---|---|
| -103 | novel | C > G |
CC 19 |
CG 1 |
GG 0 |
2.5% |
| -127 | novel | G > C |
GG 19 |
GC 1 |
CC 0 |
2.5% |
| -226 | novel | C > T |
CC 19 |
CT 1 |
TT 0 |
2.5% |
| -261 | rs2762943 | G > T |
GG 0 |
GT 0 |
TT 20 |
100% |
| -279b | novel | T > C |
TT 17 |
TC 3 |
CC 0 |
7.5% |
| -320 | novel | G > C |
GG 17 |
GC 3 |
CC 0 |
7.5% |
| -437 | novel | C > T |
CC 19 |
CT 1 |
TT 0 |
2.5% |
| -451 | rs2585427 | G > C |
GG 3 |
GC 7 |
CC 10 |
67.5% |
Sequencing was done using DNA from 20 African Americans.
This polymorphism occurs at the 5th position of the 5′ half element in VDRE 2.
Figure 1.
Polymorphisms observed in sequencing the human CYP24A1 promoter. The positions of both VDRE and all eight SNPs are shown relative to the transcription start site. Sequences indicated for each VDRE are on the opposite strand from the one that is transcribed.
3.2. Differential transactivation of wild-type and polymorphic CYP24A1 promoter constructs
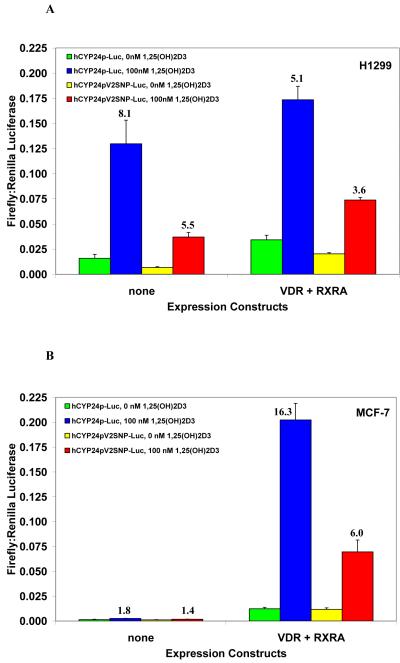
It is known that the 24-hydroxylase gene has very low basal expression and that strong induction by 1α,25(OH)2D3 is mediated through two vitamin D response elements (VDRE) in the proximal promoter region. [20, 21] To test for differences in inducibility of the wild-type and polymorphic variant VDRE2 by 1α,25(OH)2D3, we transiently transfected H1299 cells with three different luciferase reporter constructs: 1) r24OHase-Luc [23], which contains the rat 24-hydroxylase promoter and was used to test for induction by 1α,25(OH)2D3 (data not shown), 2) hCYP24p-Luc, which contains approximately 600 bp of the wild-type proximal hCYP24 promoter, and 3) hCYP24pV2SNP-Luc, which is identical to the previous construct with the exception of a single base pair change to create the SNP identified at -279 in VDRE2. Each reporter plasmid was transfected either alone or co-transfected with expression constructs for VDR and RXRA. The wild-type human 24-hydroxylase promoter was induced 8-fold by 1α,25(OH)2D3 even in the absence of coexpressed VDR and RXRα, and the polymorphic human construct was induced about 5.5-fold (Figure 2A). The same experiment was performed in MCF-7 cells, and while little to no induction was seen without the coexpression of VDR and RXRα, the activation was still markedly higher with the wild-type human construct than the polymorphic variant at 16-fold and 6-fold, respectively (Figure 2B).
Figure 2.
Transactivation of wild-type and polymorphic CYP24A1 promoter constructs by 1α,25(OH)2D3. (A) H1299 and (B) MCF-7 cells were transfected with either wild-type or polymorphic CYP24A1 promoter luciferase constructs alone or in combination with expression plasmids for VDR and RXRα as shown and as described in Section 2. Following 16 h treatment with 100 nM 1α,25(OH)2D3 or vehicle, firefly luciferase activity was measured and normalized to Renilla luciferase. Fold inductions by 1α,25(OH)2D3 for each of the reporter constructs are shown above their respective bars. All conditions were done in duplicate, and values are the mean of three independent experiments with standard deviations as shown.
3.3. Impaired VDR/RXRα binding to the polymorphic VDRE
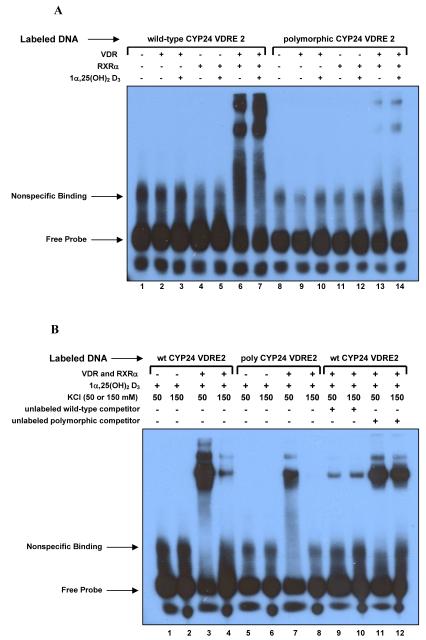
It has been shown previously that VDR and RXRα heterodimerize and bind to both VDRE in the CYP24A1 promoter [20, 21, 24-26]. The nucleotide at the fifth position in the hexameric repeat of the VDRE can have a dramatic effect on VDR/RXRα binding [2]. To determine whether the polymorphism we observed in the African American population would have an effect on protein binding to the response element, we utilized EMSA to test binding to both the wild-type and polymorphic sequences for VDRE2. The DNA probes used were comprised of the wild-type or polymorphic VDRE2 and surrounding sequence (-295 to -268) and were incubated with proteins and 1α,25(OH)2D3 as indicated (Figure 3A). As predicted, VDR and RXRα bound the wild-type VDRE as a heterodimer, but binding to the polymorphic VDRE was markedly reduced. The presence of 1α,25(OH)2D3 enhanced these interactions, but was not required for binding. To confirm the results seen with our polymorphic sequence, we ran a second EMSA using competitive binding reactions with either unlabeled wild-type or unlabeled polymorphic VDRE2 probes in a 200-fold molar excess of the labeled DNA probe. Reactions were also run using both 50 and 150 mM KCl, as others have shown a difference in binding relative to salt content [27],[28]. The results (Figure 3B) demonstrate that high salt conditions decreased binding, as the wild-type DNA-protein complex formation was greatly reduced and the polymorphic DNA-protein complex did not form under high salt conditions. More importantly, the degree of binding to the polymorphic probe was much less than that of the wild-type probe under any conditions. When included, unlabeled wild-type DNA was able to effectively out-compete the labeled wild-type DNA for binding under both low and high salt conditions, while the unlabeled polymorphic DNA had little to no impact on the ability of VDR and RXRα to bind the wild-type sequence. Taken together, the results from Figures 2 and 3 provide strong evidence that the polymorphic variant identified in VDRE2 may impact the regulation of CYP24A1 expression in vivo.
Figure 3.
Binding of VDR and RXRα to the wild-type and polymorphic vitamin D response elements. (A) EMSA showing impaired binding of VDR-RXRα heterodimers to the polymorphic VDRE probe (lanes 13 and 14) compared with wild-type (lanes 6 and 7). Where indicated, 100 ng recombinant human VDR and/or RXRα proteins were added in the presence or absence of 1 μM 1α,25(OH)2D3. (B) Second EMSA testing the impact of 50 vs. 150 mM salt and competitive binding. Binding to wild-type (lanes 1-4) and polymorphic (lanes 5-8) VDRE probe was assayed under both low and high salt conditions with proteins and 1α,25(OH)2D3 added as indicated. Competitive binding reactions were also done with both salt conditions using a 200-fold molar excess of unlabeled competitor probes for the wild-type (lanes 9-10) or polymorphic elements (lanes 11-12).
3.4. Decreased induction of CYP24A1 expression by 1α,25(OH)2D3 in a heterozygous polymorphic individual
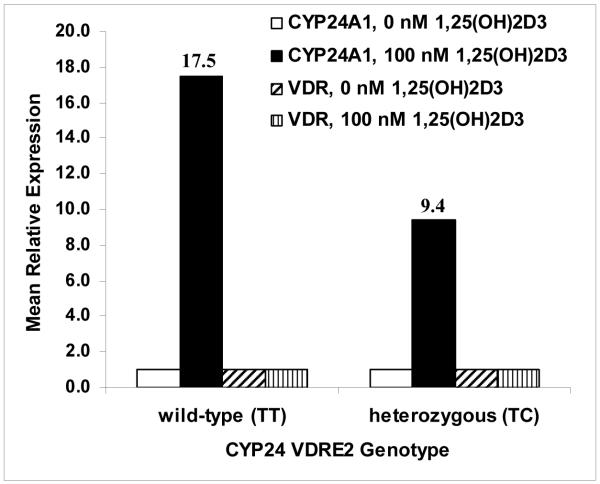
To determine whether the presence of a single copy of the VDRE2 polymorphism would be enough to impact CYP24A1 gene expression, we treated primary lymphocytes from two individuals (one heterozygous polymorphic and one wild-type) with 0 or 100 nM 1α,25(OH)2D3 for 6 hours and isolated RNA to synthesize cDNA for Real-Time PCR. TaqMan gene expression assays were performed to determine the relative quantification of both CYP24A1 and VDR in comparison to the endogenous control GAPDH. The basal levels of CYP24A1 expression in untreated samples were similar for both wild-type and polymorphic (data not shown). The fold induction by 1α,25(OH)2D3 in the heterozygous polymorphic individual was approximately half that of the wild-type individual with mean relative expression values of 9.4 and 17.5, respectively (Figure 4). Expression of VDR was essentially unchanged with treatment for either sample. These preliminary results suggest that the presence of a single copy of the VDRE2 allelic variant may be sufficient to impair induction of CYP24A1 gene expression in heterozygous individuals, and that a homozygous polymorphic individual would likely demonstrate a more dramatic reduction in inducibility.
Figure 4.
Quantitative PCR of CYP24A1 and VDR gene expression in cultured primary lymphocytes. As described in the Materials and Methods, RNA was isolated from treated and untreated primary lymphocytes from subjects who were either wild-type or heterozygous polymorphic for the CYP24A1 VDRE2 variant. Following reverse transcription, equal amounts of cDNA from each were used for qRT-PCR of CYP24A1 and VDR. Mean relative expression was determined using the SDS program by normalizing to GAPDH and using the ΔΔCt method to calculate the relative quantification of treated vs. untreated cells from the same person. Fold induction of CYP24A1 expression by 1α,25(OH)2D3 is shown for each subject above their respective bars. All reactions were done in triplicate and are the mean of two separate qRT-PCR runs.
4. Discussion
There is limited epidemiological research on polymorphic variants in CYP24A1. Studies of selected SNPs report no association with prostate cancer [29] or postmenopausal breast cancer risk in Caucasians [19]. However, a major drawback of most current SNP research is the lack of accompanying functional data, as the significance of the variants being tested is often unknown. It is also widely assumed that most important genetic variants have already been identified, and while somewhat true, it is likely that many low-prevalence polymorphisms have yet to be discovered. The proximal promoter region of CYP24A1 is highly GC-rich (approximately 71% in the first 550 nucleotides) and is challenging to sequence, which could explain the relative lack of known polymorphic variants in that region. The NCBI dbSNP database lists 31 variants in the 5 kb immediately upstream of the transcriptional start site, but only two of these have been genotyped and validated by the HapMap project [30]. In this study, we have identified six novel SNPs in the CYP24A1 proximal promoter, including a naturally-occurring polymorphism in a vitamin D response element. This polymorphic variant appears to occur with low frequency (∼4%) among healthy African Americans, although it is possible that the polymorphism is also present in the Caucasian population at a lower prevalence and was not represented in our sample set. Here we have demonstrated that the change in VDRE2 sequence has functional impacts on protein binding, transactivation, and gene expression.
Transfection of H1299 cells with CYP24A1 promoter constructs revealed that induction by 1α,25(OH)2D3 was consistently 30% lower using the polymorphic construct compared with the wild-type. In MCF-7 cells the difference was even more striking, with induction of the polymorphic construct being 63% lower than wild-type. Mobility shift assays illustrated reduced protein binding to the polymorphic versus the wild-type response element under multiple conditions, including low and high salt content. Competitive binding reactions also provided compelling evidence for the impact of the polymorphic variant, as the unlabeled polymorphic probe was shown to be largely ineffective at outcompeting the labeled wild-type probe for binding of the VDR-RXRα heterodimer. Prior studies have shown the fifth position of the hexameric repeat to be particularly important in influencing protein:DNA interactions [2]. Consistent with those results, the VDRE polymorphism we observed in the fifth position also has a detrimental impact on binding.
The transfection and binding assay results strongly indicate that the VDRE2 polymorphism may cause functional impairment in individuals that are homozygous polymorphic. Of greater interest is the potential impact in a heterozygous individual and the severity of the effect. Analysis of CYP24A1 gene expression in primary lymphocytes suggest there may be a moderate decrease in the ability of heterozygous individuals to upregulate CYP24A1 expression, and we hypothesize that the difference in a homozygous polymorphic individual would be more pronounced. These results are limited by the small and finite number of samples in our laboratory, and since we did not identify any individuals who were homozygous for the SNP in our study, we were unable to test its impact in vivo.
It is unclear why a lack of induction of the CYP24A1 promoter constructs was observed in MCF-7 cells in the absence of cotransfected expression constructs for VDR and RXRα, as previous reports have shown a response using a different 24-hydroxylase promoter construct without the addition of exogenous protein [31]. We did not explore this possibility, but as our cells are from a higher passage they may have a diminished response to 1α,25(OH)2D3 treatment due to low expression of VDR [32], as the addition of exogenous protein restored sensitivity to 1α,25(OH)2D3.
In our EMSA binding studies we observed that higher salt content was inhibitory towards complex formation, regardless of which DNA probe was used. This is in contrast with a previously published report showing enhanced binding with higher salt [27], although direct comparison to this study is not possible because the binding reactions were dissimilar. It is also difficult to draw definitive conclusions from the previous experiments, as the low salt reactions were done only in the absence of ligand and using less protein. Human physiological sodium concentrations are at or above 140 mM [33], so the reactions done at high salt (150 mM) are likely to be more reflective of the conditions in vivo.
One potentially interesting question not addressed in the current study is the effect of variants in the vitamin D receptor in combination with changes in the vitamin D response element. The VDR has a highly prevalent start codon polymorphism (SNP ID: rs10735810, historically identified by its RFLP as FokI), with the variant allele having a frequency of greater than 55% in each of the four major populations genotyped by HapMap [30]. This particular SNP results in a VDR protein that is three amino acids shorter in length due to a nonsynonymous alteration of the first codon that causes translation to initiate at a second (in-frame) Methionine. There have been conflicting results from studies of the two VDR forms (short and long), with some reporting no difference in transactivation or ligand-binding [16], and another finding that the short form interacts more efficiently with the transcription factor TFIIB [34]. More recently, a third group demonstrated that the short form of VDR is better at activating immune cells and can enhance transcription through some factors, but showed no difference in transactivation through a classical DR3 type VDRE [35]. It is possible that the effects vary from one response element to another; the authors of the latter study used a luciferase reporter construct driven by three copies of the mouse osteopontin VDRE, and thus the results may not necessarily extend to all positively regulated VDRE. As the functional significance of the shorter form of the VDR protein has not been conclusively determined, the CYP24A1 expression assays in this study were conducted using lymphocytes from two individuals with different VDRE2 genotypes but the same VDR genotype (both homozygous for the short form) for ease of comparison. Unpublished observations from our lab suggest that the short form of the VDR may have a greater capacity to induce expression of CYP24A1 in comparison to the long form. Further studies are needed from multiple individuals who are both wild-type and polymorphic for the CYP24A1 VDRE2 variant and possess the three different VDR genotypes (homozygous short-form, heterozygous, and homozygous long-form) in order to determine the true differences in CYP24A1 expression and the effect of the different VDR forms. It also remains to be seen whether the decrease in gene expression translates to a measurable decrease in 24-hydroxylase protein.
The importance of the vitamin D pathway in cancer etiology is an emerging field, and while it has been well-established that 1α,25(OH)2D3 has anti-proliferative properties against many cancer cell lines [26, 36, 37], the exact mechanisms of action are still being determined. Results from several recent reports would suggest that the vitamin D pathway is dysregulated in certain cancers, including changes in CYP24A1. For example, one group has shown by immunohistochemistry that the cellular localization of CYP24A1 protein changes dramatically in malignant colon tissues, with a significant elevation in the amount of cytoplasmic protein compared with normal colonic epithelium [38]. A separate group has also recently demonstrated that certain antineoplastic agents can selectively destabilize CYP24A1 mRNA transcripts [39]. This effectively increased the half-life of 1α,25(OH)2D3 by decreasing the amount of enzyme present for its inactivation. Further studies are needed to see whether the previous in vitro work can be recapitulated in vivo, but show promise in enhancing current strategies in chemotherapy by targeting the vitamin D pathway.
In summary, our study identified a naturally-occurring polymorphism in a vitamin D response element of the human CYP24A1 gene that appears to impair receptor protein binding, decrease transactivation, and decrease expression of the CYP24A1 gene in vivo. Further molecular studies are needed to determine the extent of the effect in humans, both alone and in combination with other defects in vitamin D pathway genes. These results are a first step towards identifying the causes of differential cancer susceptibility among both individuals and racial/ethnic groups. By evaluating the contribution of multiple, small genetic changes, future studies will increase our understanding of how they interact to affect an individual’s lifetime risk of disease and should lead to more informed epidemiological research.
Acknowledgements
This work was dually supported by grants from the Penn State Population Research Institute and by a General Clinical Research Center grant from NIH (M01RR10732) and GCRC Construction Grant (C06RR016499) awarded to the Pennsylvania State University College of Medicine. The authors would like to thank Dr. Philip Lazarus for the gift of normal human liver RNA and Dr. Hector DeLuca for the gift of the r24OHase-Luc plasmid. Drs. Ryan Dellinger, L. Michael Carastro, and Bandana Chatterjee all provided helpful suggestions on experimental design. Excellent technical assistance was provided by Rick Ball at the GCRC and Robert Brucklacher at the Functional Genomics Core Facility of the Section of Research Resources, Penn State College of Medicine. Ms. Diane Pague recruited and scheduled study participants, and Drs. Ryan Dellinger, Carol Ely Hepfer, and Marian Walters kindly assisted in critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007;7(9):684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- [2].Jin CH, Pike JW. Human vitamin D receptor-dependent transactivation in Saccharomyces cerevisiae requires retinoid X receptor. Molecular Endocrinology. 1996;10(2):196–205. doi: 10.1210/mend.10.2.8825559. [DOI] [PubMed] [Google Scholar]
- [3].Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65(7):1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Harris SS, Dawson-Hughes B. Seasonal changes in plasma 25-hydroxyvitamin D concentrations of young American black and white women. Am J Clin Nutr. 1998;67(6):1232–1236. doi: 10.1093/ajcn/67.6.1232. [DOI] [PubMed] [Google Scholar]
- [5].Harris SS, Soteriades E, Coolidge JA, Mudgal S, Dawson-Hughes B. Vitamin D insufficiency and hyperparathyroidism in a low income, multiracial, elderly population. J Clin Endocrinol Metab. 2000;85(11):4125–4130. doi: 10.1210/jcem.85.11.6962. [DOI] [PubMed] [Google Scholar]
- [6].Zadshir A, Tareen N, Pan D, Norris K, Martins D. The prevalence of hypovitaminosis D among US adults: data from the NHANES III. Ethnicity & disease. 2005;15(4 Suppl 5):S5-97–101. [PubMed] [Google Scholar]
- [7].Holick MF. Sunlight and vitamin D for bone health and prevention of autoimmune diseases, cancers, and cardiovascular disease. Am J Clin Nutr. 2004;80(6 Suppl):1678S–1688S. doi: 10.1093/ajcn/80.6.1678S. [DOI] [PubMed] [Google Scholar]
- [8].Lou YR, Qiao S, Talonpoika R, Syvala H, Tuohimaa P. The role of Vitamin D3 metabolism in prostate cancer. J Steroid Biochem Mol Biol. 2004;92(4):317–325. doi: 10.1016/j.jsbmb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- [9].Peehl DM, Feldman D. Interaction of nuclear receptor ligands with the Vitamin D signaling pathway in prostate cancer. J Steroid Biochem Mol Biol. 2004;92(4):307–315. doi: 10.1016/j.jsbmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- [10].Trump DL, Hershberger PA, Bernardi RJ, Ahmed S, et al. Anti-tumor activity of calcitriol: pre-clinical and clinical studies. J Steroid Biochem Mol Biol. 2004;89-90(15):519–526. doi: 10.1016/j.jsbmb.2004.03.068. [DOI] [PubMed] [Google Scholar]
- [11].Jemal A, Clegg LX, Ward E, Ries LA, et al. Annual report to the nation on the status of cancer, 1975-2001, with a special feature regarding survival. Cancer. 2004;101(1):3–27. doi: 10.1002/cncr.20288. [DOI] [PubMed] [Google Scholar]
- [12].Jemal A, Siegel R, Ward E, Murray T, et al. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- [13].Geronimus AT, Bound J, Keene D, Hicken M. Black-white differences in age trajectories of hypertension prevalence among adult women and men, 1999-2002. Ethnicity & disease. 2007;17(1):40–48. [PubMed] [Google Scholar]
- [14].Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338(2):143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- [15].Slattery ML. Vitamin D receptor gene (VDR) associations with cancer. Nutrition reviews. 2007;65(8 Pt 2):S102–104. doi: 10.1111/j.1753-4887.2007.tb00332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gross C, Krishnan AV, Malloy PJ, Eccleshall TR, et al. The vitamin D receptor gene start codon polymorphism: a functional analysis of FokI variants. J Bone Miner Res. 1998;13(11):1691–1699. doi: 10.1359/jbmr.1998.13.11.1691. [DOI] [PubMed] [Google Scholar]
- [17].Gross C, Musiol IM, Eccleshall TR, Malloy PJ, Feldman D. Vitamin D receptor gene polymorphisms: analysis of ligand binding and hormone responsiveness in cultured skin fibroblasts. Biochem Biophys Res Commun. 1998;242(3):467–473. doi: 10.1006/bbrc.1997.7986. [DOI] [PubMed] [Google Scholar]
- [18].Arai H, Miyamoto K, Taketani Y, Yamamoto H, et al. A vitamin D receptor gene polymorphism in the translation initiation codon: effect on protein activity and relation to bone mineral density in Japanese women. J Bone Miner Res. 1997;12(6):915–921. doi: 10.1359/jbmr.1997.12.6.915. [DOI] [PubMed] [Google Scholar]
- [19].McCullough ML, Stevens VL, Diver WR, Feigelson HS, et al. Vitamin D pathway gene polymorphisms, diet, and risk of postmenopausal breast cancer: a nested case-control study. Breast Cancer Res. 2007;9(1):R9. doi: 10.1186/bcr1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen KS, DeLuca HF. Cloning of the human 1 alpha,25-dihydroxyvitamin D-3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim Biophys Acta. 1995;1263(1):1–9. doi: 10.1016/0167-4781(95)00060-t. [DOI] [PubMed] [Google Scholar]
- [21].Armbrecht HJ, Hodam TL, Boltz MA, Partridge NC, et al. Induction of the vitamin D 24-hydroxylase (CYP24) by 1,25-dihydroxyvitamin D3 is regulated by parathyroid hormone in UMR106 osteoblastic cells. Endocrinology. 1998;139(8):3375–3381. doi: 10.1210/endo.139.8.6134. [DOI] [PubMed] [Google Scholar]
- [22].Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, et al. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998;13(3):325–349. doi: 10.1359/jbmr.1998.13.3.325. [DOI] [PubMed] [Google Scholar]
- [23].Arbour NC, Ross TK, Zierold C, Prahl JM, DeLuca HF. A highly sensitive method for large-scale measurements of 1,25-dihydroxyvitamin D. Analytical biochemistry. 1998;255(1):148–154. doi: 10.1006/abio.1997.2439. [DOI] [PubMed] [Google Scholar]
- [24].Zou A, Elgort MG, Allegretto EA. Retinoid X receptor (RXR) ligands activate the human 25-hydroxyvitamin D3-24-hydroxylase promoter via RXR heterodimer binding to two vitamin D-responsive elements and elicit additive effects with 1,25-dihydroxyvitamin D3. J Biol Chem. 1997;272(30):19027–19034. doi: 10.1074/jbc.272.30.19027. [DOI] [PubMed] [Google Scholar]
- [25].Toell A, Polly P, Carlberg C. All natural DR3-type vitamin D response elements show a similar functionality in vitro. Biochem J. 2000;352(Pt 2):301–309. [PMC free article] [PubMed] [Google Scholar]
- [26].Thompson PD, Jurutka PW, Whitfield GK, Myskowski SM, et al. Liganded VDR induces CYP3A4 in small intestinal and colon cancer cells via DR3 and ER6 vitamin D responsive elements. Biochem Biophys Res Commun. 2002;299(5):730–738. doi: 10.1016/s0006-291x(02)02742-0. [DOI] [PubMed] [Google Scholar]
- [27].Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26(17):6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zella LA, Kim S, Shevde NK, Pike JW. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Molecular Endocrinology. 2006;20(6):1231–1247. doi: 10.1210/me.2006-0015. [DOI] [PubMed] [Google Scholar]
- [29].Holick CN, Stanford JL, Kwon EM, Ostrander EA, et al. Comprehensive Association Analysis of the Vitamin D Pathway Genes, VDR, CYP27B1, and CYP24A1, in Prostate Cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(10):1990–1999. doi: 10.1158/1055-9965.EPI-07-0487. [DOI] [PubMed] [Google Scholar]
- [30].The International HapMap Project Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- [31].Narvaez CJ, Byrne BM, Romu S, Valrance M, Welsh J. Induction of apoptosis by 1,25-dihydroxyvitamin D3 in MCF-7 Vitamin D3-resistant variant can be sensitized by TPA. J Steroid Biochem Mol Biol. 2003;84(23):199–209. doi: 10.1016/s0960-0760(03)00029-3. [DOI] [PubMed] [Google Scholar]
- [32].Jensen SS, Madsen MW, Lukas J, Bartek J, Binderup L. Sensitivity to growth suppression by 1alpha,25-dihydroxyvitamin D(3) among MCF-7 clones correlates with Vitamin D receptor protein induction. J Steroid Biochem Mol Biol. 2002;81(2):123–133. doi: 10.1016/s0960-0760(02)00057-2. [DOI] [PubMed] [Google Scholar]
- [33].Frohlich BT, De Bernardez Clark ER, Siber GR, Swartz RW. Improved pertussis toxin production by Bordetella pertussis through adjusting the growth medium’s ionic composition. Journal of Biotechnology. 1995;39(3):205–219. doi: 10.1016/0168-1656(95)00013-g. [DOI] [PubMed] [Google Scholar]
- [34].Jurutka PW, Remus LS, Whitfield GK, Thompson PD, et al. The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Molecular Endocrinology. 2000;14(3):401–420. doi: 10.1210/mend.14.3.0435. [DOI] [PubMed] [Google Scholar]
- [35].van Etten E, Verlinden L, Giulietti A, Ramos-Lopez E, Branisteanu DD, Ferreira GB, Overbergh L, Verstuyf A, Bouillon R, Roep BO, Badenhoop K, Mathieu C. The vitamin D receptor gene FokI polymorphism: Functional impact on the immune system. Eur J Biochem. 2007;37(2):395–405. doi: 10.1002/eji.200636043. [DOI] [PubMed] [Google Scholar]
- [36].Wang Q, Yang W, Uytingco MS, Christakos S, Wieder R. 1,25-Dihydroxyvitamin D3 and all-trans-retinoic acid sensitize breast cancer cells to chemotherapy-induced cell death. Cancer Res. 2000;60(7):2040–2048. [PubMed] [Google Scholar]
- [37].Zhao XY, Peehl DM, Navone NM, Feldman D. 1alpha,25-dihydroxyvitamin D3 inhibits prostate cancer cell growth by androgen-dependent and androgen-independent mechanisms. Endocrinology. 2000;141(7):2548–2556. doi: 10.1210/endo.141.7.7549. [DOI] [PubMed] [Google Scholar]
- [38].Matusiak D, Benya RV. CYP27A1 and CYP24 Expression as a Function of Malignant Transformation in the Colon. J Histo & Cytochem. 2007;55(12):1257–1264. doi: 10.1369/jhc.7A7286.2007. [DOI] [PubMed] [Google Scholar]
- [39].Tan J, Dwivedi PP, Anderson P, Nutchey BK, et al. Antineoplastic agents target the 25-hydroxyvitamin D3 24-hydroxylase messenger RNA for degradation: implications in anticancer activity. Molecular cancer therapeutics. 2007;6(12 Pt 1):3131–3138. doi: 10.1158/1535-7163.MCT-07-0427. [DOI] [PubMed] [Google Scholar]