SUMMARY
Kinetochores couple chromosomes to the assembling and disassembling tips of microtubules, a dynamic behavior that is fundamental to mitosis in all eukaryotes but poorly understood. Genetic, biochemical and structural studies implicate the Ndc80 complex as a direct point of contact between kinetochores and microtubules, but these approaches provide only a static view. Here, using techniques for manipulating and tracking individual molecules in vitro, we demonstrate that the Ndc80 complex is capable of forming the dynamic, load-bearing attachments to assembling and disassembling tips required for coupling in vivo. We also establish that Ndc80-based coupling likely occurs through a biased diffusion mechanism, and that this activity is conserved from yeast to humans. Our findings demonstrate how an ensemble of Ndc80 complexes may provide a ‘slip clutch’ that allows the kinetochore to maintain a load-bearing tip attachment during both microtubule assembly and disassembly.
INTRODUCTION
Chromosomes are organized and separated during cell division by a molecular machine, the mitotic spindle. A central component of this machine is the kinetochore, a specialized site on each chromosome that forms attachments to spindle microtubules. Besides mechanically coupling the chromosomes to microtubules, kinetochores are also regulatory hubs. They control chromosome movement by altering the growth and shortening of attached microtubules, they sense and correct errors in attachment, and they emit diffusible ‘wait’ signals until they are properly attached. These regulatory functions are all vital for accurate mitosis, and they are all related to microtubule attachment. To understand kinetochore function, the basis for its most fundamental behavior—microtubule attachment—must be uncovered.
As the spindle forms, kinetochores make initial attachments to the sides of spindle microtubules. These lateral (‘side-on’) attachments convert quickly into (‘end-on’) tip attachments and thereafter, chromosome movement is coupled to filament assembly and disassembly (Rieder and Salmon, 1998). Kinetochore-microtubule coupling requires a puzzling combination of strength and dynamicity. Molecules at each kinetochore-microtubule interface must maintain a persistent, load-bearing attachment to the filament tip even while thousands of tubulin subunits are added and removed. This strong yet dynamic coupling, often called a ‘slip clutch’ (Maddox et al., 2003), allows kinetochores to harness microtubule shortening to drive poleward chromosome movement and also to track with growing tips during anti-poleward movement (or during microtubule ‘flux’). While the behavior is conserved from yeast to humans (McAinsh et al., 2003; Rieder and Salmon, 1998), its molecular basis remains uncertain.
Models for kinetochore-microtubule coupling fall into several classes. In one class, coupling is based on ATP-powered motor enzymes. Motors likely contribute to coupling in many organisms (Sharp et al., 2000; Yang et al., 2007), and some motors can mediate attachment to disassembling tips in vitro (Lombillo et al., 1995). However, kinetochore-bound motors are largely dispensable in yeast (Grishchuk and McIntosh, 2006; McAinsh et al., 2003) and their depletion in higher eukaryotes does not completely disrupt tip attachment (Kapoor et al., 2006; Weaver et al., 2003; Yang et al., 2007). In a second class of models, peeling protofilaments at disassembling tips drive kinetochore movement either by pushing against a sliding ring (Koshland et al., 1988) or by tugging on tightly-bound kinetochore fibrils (McIntosh et al., 2008). Enthusiasm for the ring model was generated recently by the discovery that the Dam1 kinetochore complex forms rings around microtubules in vitro (Miranda et al., 2005). However, while the Dam1 complex is very likely to contribute to tip coupling, microtubule-based force production, and regulation of kinetochore-attached microtubules in yeast (Asbury et al., 2006; Franck et al., 2007; Tanaka et al., 2007; Westermann et al., 2006), no clear homologues have been identified outside fungi. Moreover, ring formation is not required for microtubule-driven movement of the Dam1 complex in vitro (Gestaut et al., 2008). The fibrils model offers an attractive, ring-independent way for peeling protofilaments to exert force on the kinetochore (McIntosh et al., 2008). However, processive disassembly-driven movement by this mechanism requires tight binding of the fibrils to the microtubule, which would prevent assembly-coupled movement. Thus neither motor-, ring-, nor tight fibril-based mechanisms appear to fully account for tip coupling in all eukaryotes.
In a third class of models, kinetochore-microtubule coupling depends on biased thermal diffusion. This idea was first proposed by Hill (1985), who demonstrated theoretically that an array of individually weak microtubule binders will track with disassembling microtubule tips provided that its diffusion along the filament is sufficiently fast and that its total binding energy is large enough. Importantly, a biased diffusion-based coupler acts exactly like a slip clutch: It supports tension continuously while remaining tip-attached during both assembly and disassembly. This behavior distinguishes it from models based on ATP-powered motor proteins or peeling protofilaments, which are fundamentally unidirectional. Although Hill imagined a sleeve-like arrangement of binding elements completely encircling the filament, the physical underpinnings of his model can apply generally to any array of microtubule binders that are linked together, even a disordered array.
Extensive genetic, biochemical and structural studies have established that the Ndc80 complex is crucial for maintaining a robust attachment between kinetochores and microtubule tips across eukaryotes (reviewed in Tanaka and Desai, 2008). However, the ability of the complex to support the strong yet dynamic attachment required for kinetochore-microtubule coupling has never been demonstrated. Here we show that ensembles of Ndc80 complexes can form load-bearing attachments to assembling and disassembling tips. The properties of this attachment can be fully described by a biased diffusion mechanism. Based on these findings, we propose that the Ndc80 complex forms a slip clutch at the eukaryotic kinetochore.
RESULTS
The Ndc80 complex couples cargo to dynamic microtubules
To study Ndc80-based coupling we adapted a newly developed bead motility assay (Asbury et al., 2006; Franck et al., 2007). All four subunits of the S. cerevisiae Ndc80 complex, Ndc80, Nuf2, Spc24, and Spc25, were co-expressed in E. coli and purified by affinity chromatography followed by gel filtration (Gestaut et al., 2008; Wei et al., 2005) (Figure S1). Purification and binding of the complex to beads were facilitated by a His6-tag on the C-terminus of Spc24, a location near the kinetochore-facing end of the complex (~20 nm away) and farther (~40 nm) from the N-terminal regions of Ndc80 and Nuf2 shown previously to mediate binding to taxol-stabilized microtubules (Cheeseman et al., 2006; Ciferri et al., 2008; Wei et al., 2006; Wei et al., 2005). Beads decorated with the complex were introduced into a chamber containing dynamic microtubules growing from stable, coverslip-anchored seeds. The filaments exhibited dynamic instability typical for microtubules in vitro, switching stochastically between periods of slow growth (at ~10 nm s−1) and rapid shortening (~100 nm s−1). To test for binding, candidate beads diffusing freely in solution were captured with a laser trap and held near the tip of a growing filament. Binding was evident from a sharp reduction in the Brownian motion of the bead, which was monitored with high spatiotemporal resolution using back focal plane interferometry (Asbury et al., 2006; Franck et al., 2007). Beads coated with full-length Ndc80 complex almost always bound microtubules (82%, 42 of 51 beads tested). Control beads coated with a ‘headless’ version of the complex, 2NCC:2S, lacking the N-terminal regions of Ndc80 and Nuf2 (Wei et al., 2005), failed to bind (116 tested). Thus binding of Ndc80-coated beads to assembling microtubule tips in our assay requires the same molecular domains that mediate binding of the free complex to taxol-stabilized microtubules.
In our initial experiments we shut off the laser trap when the bead bound the microtubule tip to record movement in the absence of external load. Without load, the beads exhibited longitudinal diffusion along the filament as the tip continued to grow (65%, 26 of 40 beads tested; see Figure 1) or remained stationary on the microtubule lattice (35%, 14 of 40). Most attachments persisted until the filament tip underwent a spontaneous ‘catastrophe’, disassembling rapidly back to the bead and then carrying it in the direction of shortening (73%, 29 of 40), usually all the way to the coverslip-anchored seed (52%, 21 of 40). Most of the remaining events were terminated artificially (40%, 16 of 40), often because the bead stuck irreversibly to the coverslip. Spontaneous detachment from the microtubules was rare (8%, 3 of 40).
Figure 1. The Ndc80 complex couples cargo to dynamic microtubules.
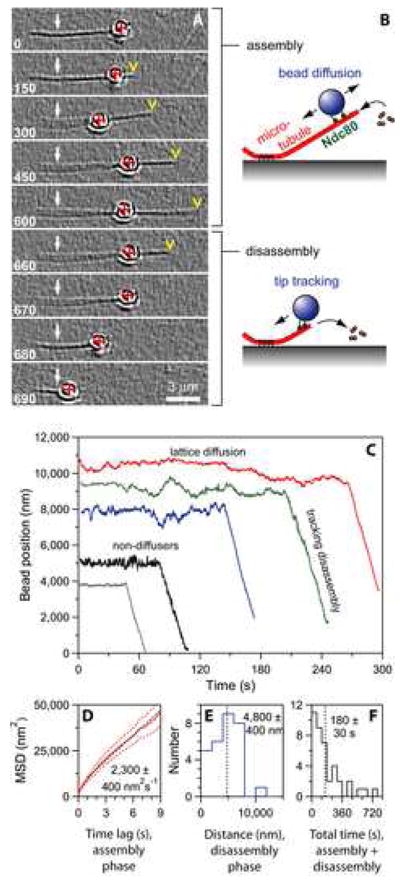
(A) An Ndc80-coated bead diffuses back-and-forth over the microtubule lattice and fails to track with the tip during filament growth (0 – 600 s). During filament shortening, it tracks with the disassembling tip (600 – 690 s). Selected frames from Movie S1 are shown. White arrows indicate the coverslip-anchored portion of the microtubule seed, red crosses indicate the bead center, and yellow arrowheads indicate the microtubule tip. Elapsed times are in seconds.
(B) Schematic diagram of the Ndc80 bead motility assay. In the absence of tension, Ndc80-based linkages usually exhibit lattice diffusion as the filament grows by addition of tubulin subunits to the tip, and they remain tip-bound even as subunits are rapidly lost from the tip during shortening.
(C) Records of bead position versus time, measured without applied force. Increasing position represents movement away from the anchored portion of the microtubule. For clarity, the records are offset vertically by an arbitrary amount.
(D) Mean squared displacement plotted against time for the subset of beads that underwent lattice diffusion. Data are mean (solid red curve) ± s.e.m. (dotted red curves), computed from N = 26 records. Black line shows linear fit used to determine diffusion coefficient.
(E and F) Histograms of bead displacement during microtubule shortening (N = 29 records) and total duration (including growth and shortening phases) of microtubule attachment (N = 40) for a population of beads. Dotted vertical lines indicate mean values. The data in (D – F) are pooled from experiments using beads prepared with 0.6 to 15 nM Ndc80 complex, corresponding to molar ratios of 110 to 2,700 complexes per bead.
We used automated centroid tracking from video recordings to quantify bead position over time (Figure 1C). The Ndc80-based attachments remained bound to the filaments for a mean duration of 180 ± 30 s (Figure 1F) and underwent disassembly-driven movement over 4,800 ± 400 nm on average (Figure 1E). These values underestimate the capacity of the linkages to remain bound and track with disassembly, because most events were terminated by the microtubule shortening completely back to the seed, not by detachment. For the subset of beads that diffused along the filaments, mean squared displacement increased linearly with time and indicated a diffusion constant of 2,300 ± 400 nm2 s−1 (Figure 1D). The diffusion was unbiased. Diffusing and non-diffusing beads alike were able to track with disassembly (Figure 1C). These observations demonstrate that the Ndc80 complex forms persistent dynamic attachments to microtubules, allowing filament disassembly to drive movement.
Ndc80-based couplers harness microtubule disassembly to produce force
Attachments between kinetochores and microtubule tips in vivo withstand tensile forces arising during congression and segregation, but kinetochores lacking the Ndc80 complex fail to establish such attachments (DeLuca et al., 2005; He et al., 2001). To apply force to Ndc80-based linkages in vitro we used a laser trapping-based force clamp. Constant tension was applied in the direction of microtubule growth with a computer controlled piezoelectric stage programmed to maintain a fixed offset between the bead and the trap center (Asbury et al., 2006; Franck et al., 2007). Even with 0.5 – 2.5 pN of continuous tension, the bead-bound Ndc80 complexes maintained an attachment to the filament that persisted through phases of assembly and disassembly and often lasted several minutes (Figure 2; Table S1). During microtubule growth the linkages behaved differently under tension than in its absence. Instead of diffusing along the filament or remaining stationary (as in Figure 1), they stayed associated with the growing tip, traveling an average of 670 ± 70 nm before the tip switched spontaneously into rapid shortening (20%, 64 of 314 events) or until movement was interrupted by bead detachment (72%, 226 of 314) or some other event (e.g., the bead sticking to the coverslip or colliding with another bead; 8%, 24 of 314). Many beads remained attached during disassembly (142 events, including 78 that were not preceded by discernable assembly-coupled movement; see Table S1) and were carried, on average, 1080 ± 140 nm in the direction of filament shortening, against the force of the laser trap (Figure 2E). A few reversed direction when the filament underwent a ‘rescue’, spontaneously resuming growth (3%, 4 of 142). The remaining beads detached during disassembly (58%, 83 of 142) or were otherwise interrupted (39%, 55 of 142). These results establish that Ndc80 complexes can form load-bearing tip attachments that remain bound during microtubule growth, shortening, catastrophe and rescue, and that the attachments are capable of harnessing energy released during filament disassembly to produce mechanical work.
Figure 2. Ndc80-based couplers support tension.
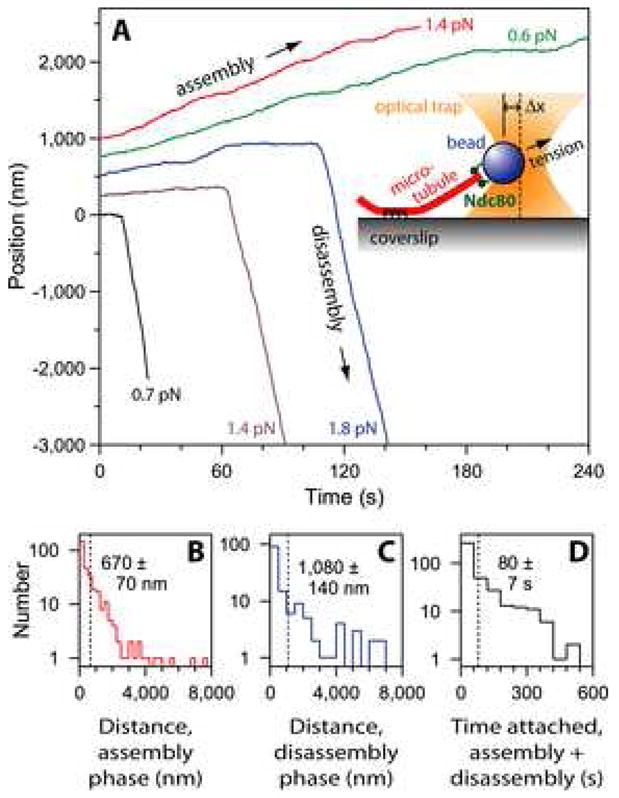
(A) Records of bead position versus time during continuous application of tensile load. Increasing position represents assembly-coupled movement in the direction of applied force, away from the anchored portion of the microtubule (e.g., blue trace < 120 s). Decreasing position represents disassembly-driven movement against the applied force (e.g., blue trace > 120 s). These data were recorded using beads prepared with ≤ 2 nM Ndc80 complex, corresponding to ≤ 360 complexes per bead. For clarity, the records are offset vertically by an arbitrary amount. The inset shows a schematic diagram of the force-clamp experiment. The bead is held by an optical trap (orange). As the microtubule grows and shortens, the coverslip is moved by computer to keep a fixed separation (Δx) between the bead and the trap, thereby keeping a constant level of tension on the Ndc80-based linkage.
(B – D) Histograms of distance moved during assembly (N = 314 records), distance moved during disassembly (N = 142), and total attached time (including both assembly and disassembly phases; N = 392) for a population of tip-attached beads moving under 0.5 – 2.5 pN of tension. Dotted vertical lines indicate the average for each distribution. These data are pooled from experiments using beads prepared with 0.6 to 15 nM Ndc80 complex, corresponding to molar ratios of 110 to 2,700 complexes per bead.
Coupling and force production are conserved properties of the Ndc80 complex
To determine whether the ability to form dynamic, load-bearing tip attachments is conserved in the human Ndc80/Hec1 complex, all four subunits were co-expressed in E. coli and purified by affinity chromatography followed by gel filtration (Figure S1). As above, a His6-tag on the C-terminus of Spc24 was used both for purification and bead binding. Beads coated with the full-length Ndc80/Hec1 complex behaved similarly in force clamp experiments to those coated with the yeast complex. After attachment to a growing tip and placement under 0.5 to 2.5 pN of tension, the beads often remained tip-bound for many minutes through phases of filament growth and shortening (Table S1; Figure S2A). The mean displacements during filament assembly, 1,900 ± 300 nm (N = 46), and disassembly, 970 ± 270 nm (N = 25), and the average duration of attachment, 280 ± 50 s (N = 50), were comparable to those measured for the yeast complex (see Figure S2B – D). These results show that the capacity of the Ndc80 complex to couple force to dynamic microtubule tips is conserved from yeast to humans.
Coupling and force production persist at low surface densities
Kinetochores in vivo contain 6 to 30 copies of the Ndc80 complex per attached microtubule (Emanuele et al., 2005; Joglekar et al., 2008). To estimate the number of complexes required in vitro for a load-bearing coupler we measured coupling performance as a function of the surface density of Ndc80 on the beads. Anti-His beads (at 5.6 pM) were incubated with the His6-tagged yeast Ndc80 complex at molar ratios ranging from 110 to 2,700 complexes per bead. At each molar ratio, we measured displacements for a population of tip-attached beads subject to 1 pN of continuous tension during microtubule growth and, if possible, during shortening. We also calculated detachment rates for each condition by counting the number of detachment events and dividing by the total observation time. At molar ratios of ~360 complexes per bead and higher (i.e., ≥ 2 nM Ndc80 complex), we routinely observed tip attachments that persisted for minutes and moved 400 to 1,000 nm during growth and shortening (Figure 3A). Below ~360 complexes per bead, the detachment rate from growing tips increased (Figure 3B), an effect that reduced the distance moved during growth and made measurements during shortening more difficult (because fewer beads remained bound long enough for the filament to undergo catastrophe). Below ~110 complexes per bead (< 0.6 nM Ndc80 complex), microtubule binding still occurred but the beads detached within a few seconds when placed under tension. These observations suggest a lower bound for the formation of a load-bearing coupler of ~360 complexes per bead.
Figure 3. Low surface densities of Ndc80 are sufficient to form load-bearing couplers.
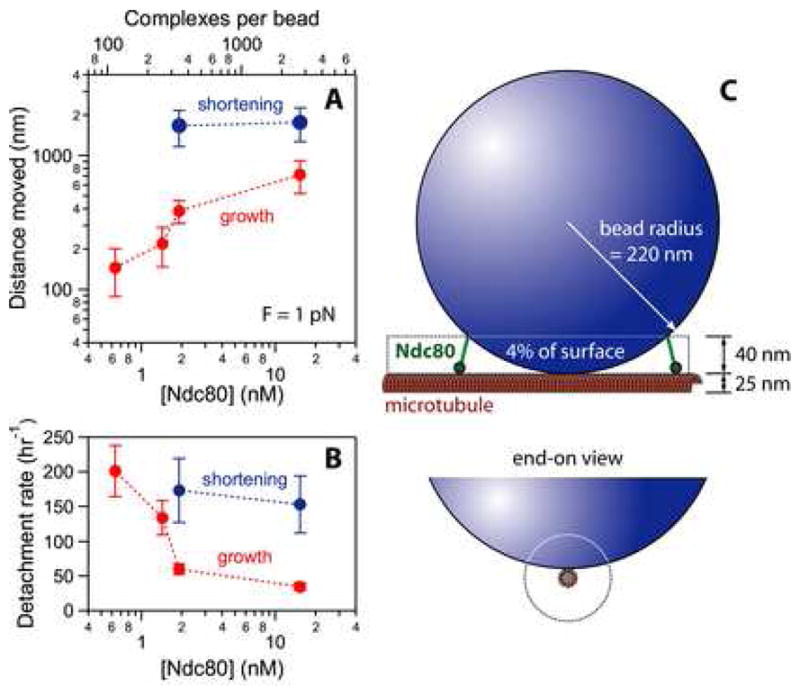
(A) Mean displacement as a function of Ndc80 density for tip-attached beads under 1 pN of tension. The ratio of Ndc80 complexes to beads and the concentration of Ndc80 during incubation (with 5.6 pM beads) are shown on the top and bottom axes, respectively. At molar ratios of 360 complexes per bead and higher (i.e. ≥ 2 nM Ndc80 complex), tip attachments persisted for minutes and moved 400 to 1,000 nm during growth and shortening. Each red point represents the average of at least 30 events. Blue points are averages of at least 17 events. Uncertainties represent s.e.m.
(B) Rate of bead detachment from growing and shortening tips, estimated by counting the number of detachment events and dividing by the total observation time in each phase. Each red point was calculated from at least 29 detachments during growth. Each blue point was calculated from at least 14 detachments during shortening. Uncertainties represent counting error.
(C) Schematic diagram, drawn approximately to scale, showing that Ndc80 complexes on a small fraction of the bead surface, ≤ 4%, are capable of simultaneously binding to the microtubule. The configuration shown, where the bead rests against the side of a filament whose tip extends well past the point of contact, maximizes the amount of bead surface in close proximity to the microtubule. Thus it provides an upper limit for the fraction of bead surface within 40 nm. The bead may instead adopt a more end-on configuration, which would reduce the number of complexes capable of binding.
Simple geometric considerations indicate that Ndc80 complexes on only a small fraction of the bead surface, ≤ 4%, can simultaneously interact with the microtubule (Figure 3C). Assuming a random distribution of complexes on the bead surface, 360 complexes per bead corresponds to 14 ± 5 interacting with the filament. This calculation may overestimate the number of interactions, since it assumes (1) that all Ndc80 complexes are functional, (2) that they bind very efficiently to the bead surface, (3) that every bead-bound Ndc80 complex within 40 nm can attach to the filament, and (4) that the bead rests directly against the side of a filament whose tip extends > 120 nm past the point of contact. Relaxing any of these assumptions would lead to a lower estimate. We also note that our estimate includes a relatively large uncertainty (± 33%) arising from the inherent uncertainty in our measurements of bead and protein concentration, and from counting statistics (see Materials and methods). Nevertheless, we can conclude that the number of microtubule-interacting Ndc80 complexes required to form a load-bearing coupler in our bead assay is close to the 6 to 30 found at kinetochores in vivo.
Individual Ndc80 complexes exhibit diffusion along microtubules
To study interactions between Ndc80 complexes and microtubules at the level of single molecules, we applied total internal reflection fluorescence (TIRF) microscopy (Figure 4). A fluorescent-tagged version of the yeast complex was created by fusing green fluorescent protein (GFP) to the C-terminus of Nuf2, a modification which causes no detectable phenotype in vivo (Shimogawa et al., 2006). At the low concentrations of free Ndc80 necessary for recording single molecules (i.e., < 10 nM), we observed very little binding in a standard microtubule growth buffer. However, in buffers with reduced ionic strength and at a concentration of 20 pM, individual fluorescent particles of Ndc80 complex bound transiently to taxol-stabilized microtubules and underwent one-dimensional diffusion along the filaments (Figure 4B). The brightness of the particles suggested they were composed of one or two complexes. Most disappeared in a single step, which probably represents detachment from the microtubule (Figure 4C). However, a small fraction showed a stepwise loss of half their intensity while they remained attached to the filament, consistent with photobleaching of one GFP molecule within a particle containing two. We fit the initial brightness distribution for all the particles by the sum of two Gaussian functions (Figure 4D), corresponding to a large population (89%) with a unitary brightness of 16,200 ± 5,500 arbitrary units and a small population (11%) with twice the brightness (32,400 ± 5,500 a.u.). The unitary brightness was similar to that of individual GFP molecules, 17,000 ± 11,000 a.u., measured in control experiments where GFP-MCAK was bound tightly to microtubules under otherwise identical conditions (data not shown).
Figure 4. Ndc80 complexes exhibit one-dimensional diffusion along microtubules.
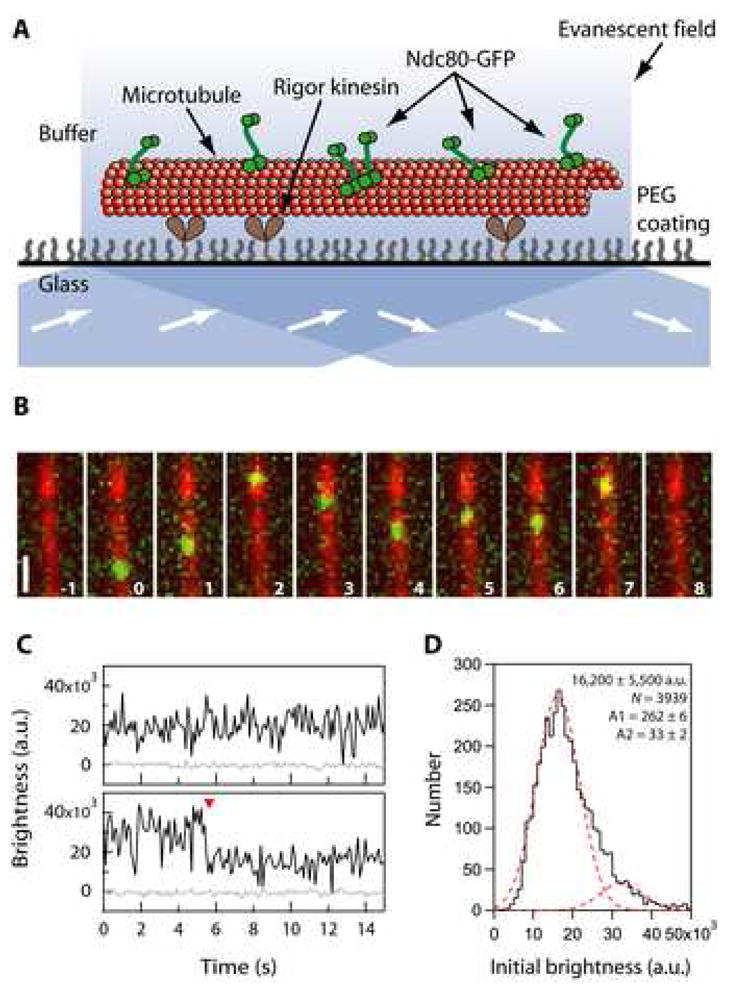
(A) Schematic of the TIRF assay for observing GFP-tagged Ndc80 complexes (green rods) interacting with individual microtubules (red). Excitation by total internal reflection, in combination with a surface treatment that blocks non-specific adsorption, allowed movement of single Ndc80 complexes to be recorded in the evanescent field.
(B) Selected frames from Movie S3 showing one-dimensional diffusion of the Ndc80 complex (green) along a microtubule (red). Elapsed times are in seconds. Scale bar, 1μm.
(C) Records of brightness versus time for most diffusing particles were roughly constant over time (as in the upper trace). However, some particles showed a stepwise loss of half their intensity while they remained attached to the filament (red arrowhead, lower trace), consistent with photobleaching of one GFP molecule within a particle containing two GFPs.
(D) Distribution of initial brightness values for particles of Ndc80-GFP diffusing on taxol-stabilized microtubules. Data are fit by the sum of two Gaussians (dashed red curves), corresponding to a large population (89%) with a unitary brightness of 16,200 ± 5,500 a.u. (mean ± s.d.) plus a small population (11%) with twice the brightness, 32,400 ± 5,500 a.u. (N = 3,939 events on 172 microtubules in 34 recordings).
Sensitivity to ionic strength is a hallmark of protein-protein interactions that rely on electrostatic attraction. To quantify the sensitivity of Ndc80-microtubule interactions to ionic strength, we recorded particle binding and diffusion on taxol-stabilized microtubules across a range of total potassium ion concentrations from 30 to 90 mM (Figure 5). At each potassium ion concentration, we measured association and dissociation rates, kon and koff, and used these values to estimate an equilibrium dissociation constant, Kd = koff·kon−1. Diffusion coefficients were estimated for each condition from linear fits to plots of mean squared displacement versus time. Raising the ionic strength lowered the association rate (i.e., fewer particles bound) and also increased the dissociation rate (residence times became shorter). As a result the apparent Kd increased 4-fold, from 0.23 ± 0.02 μM at 30 mM potassium to 0.84 ± 0.12 μM at 90 mM potassium (Figure 5D). These Kd values fall within the range of previous estimates, which vary from 0.04 μM to 3 μM depending on the Ndc80 construct and the buffer composition (Cheeseman et al., 2006; Ciferri et al., 2008; Wei et al., 2007). Raising the ionic strength also caused a modest (50%) increase in the apparent diffusion coefficient, which went from 0.117 ± 0.004 μm2 s−1 at 30 mM potassium to 0.17 ± 0.01 μm2 s−1 at 90 mM potassium (Figure 5E). We note that these changes are unlikely to reflect differences in the oligomeric state of the complex, because particle brightness values were indistinguishable at the three ionic strengths (Figure S3). Collectively, these results show that particles consisting of one or two Ndc80 complexes form transient, electrostatically based interactions with the microtubule that allow rapid diffusion along the filament lattice. Lattice diffusion implies that the energetic barrier for complete detachment of the particles from the microtubule is higher than for movement between neighboring sites on the filament lattice. This confirms a key property required for effective tip coupling via Hill’s biased diffusion mechanism.
Figure 5. Binding of individual Ndc80 complexes to microtubules is sensitive to ionic strength.
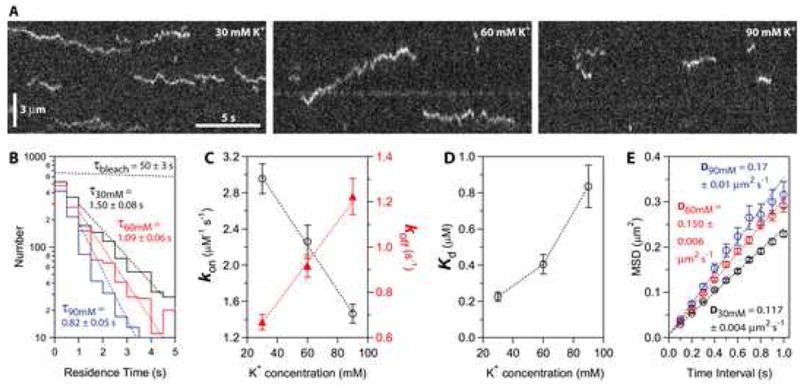
(A) Kymographs showing binding and diffusion of Ndc80-GFP on taxol-stabilized microtubules in buffers with varying ionic strength (30, 60 and 90 mM K+). Position along the microtubule is depicted on the vertical axis while time changes along the horizontal axis. Long-duration events (> 3 s) were more common at lower ionic strength. Concentration of Ndc80 complex, 20 pM.
(B) Residence time distributions for Ndc80 complexes in 30 mM K+ (black histogram, N = 1,729 binding events on 61 microtubules in 12 recordings totaling 40 min), 60 mM K+ (red histogram, N = 1,333 events, 57 microtubules, 11 movies, totaling 37 min) and 90 mM K+ (blue histogram, N = 877 events, 54 microtubules, 11 movies, totaling 37 min). Dotted lines show weighted exponential fits used to determine mean residence times. The exponential distribution of bleach times for single GFP molecules in control experiments is depicted by the uppermost dotted black line, corresponding to a mean bleach time of τbleach = 50 ± 3 s.
(C and D) Raising the ionic strength lowered the association rate, kon (left axis in C, open black circles), and increased the dissociation rate, koff (right axis in C, filled red triangles). As a result, the apparent equilibrium dissociation constant, Kd = koff·kon−1, increased 4-fold (D). On-rates were calculated as the number of observed binding events per tubulin dimer per second, divided by the Ndc80 concentration. Off-rates were taken from τ−1 in (B). Uncertainties represent s.e.m.
(E) Raising the ionic strength speeds lattice diffusion of Ndc80 complexes. Mean squared displacement (MSD) is plotted against time for each ionic strength. Dotted lines show linear fits used to determine diffusion coefficients. Data are mean ± s.e.m., computed from N = 2,002 events for 30 mM K+, N = 1,302 events for 60 mM K+, and N = 664 events for 90 mM K+.
Ensembles are required for tracking with disassembling tips
Another critical assumption of Hill’s model is that when the coupler encounters a disassembling tip its otherwise random diffusion must become biased in favor of movement back toward the filament lattice. To test this we modified the TIRF assay to study interactions with dynamic rather than stabilized microtubules. Short, GMPCPP-stabilized microtubule seeds were anchored to the coverslip and dynamic extensions of several micrometers were grown from them by the addition of free tubulin and GTP. Fluorescent Ndc80 complex was then introduced by buffer exchange, which also lowered the level of free tubulin to trigger filament disassembly. We used 1 nM Ndc80 complex, a concentration at which the microtubules were crowded with diffusing particles, so that each tip would encounter many particles as it disassembled. No clear tip tracking was evident. However, the density of particles sometimes (15%, 20 of 132 tips recorded) became noticeably higher at a disassembling tip as compared to the lattice. When individual particles could be discerned as they encountered a disassembling tip, they sometimes appeared to ‘bounce’ repeatedly off the tip, diffusing back onto the lattice rather than dissociating (Figure 6A, top). These observations confirm that the diffusion of Ndc80 particles becomes biased at disassembling tips, as required for the biased diffusion mechanism.
Figure 6. Small ensembles of Ndc80 complexes track with disassembling microtubule tips.
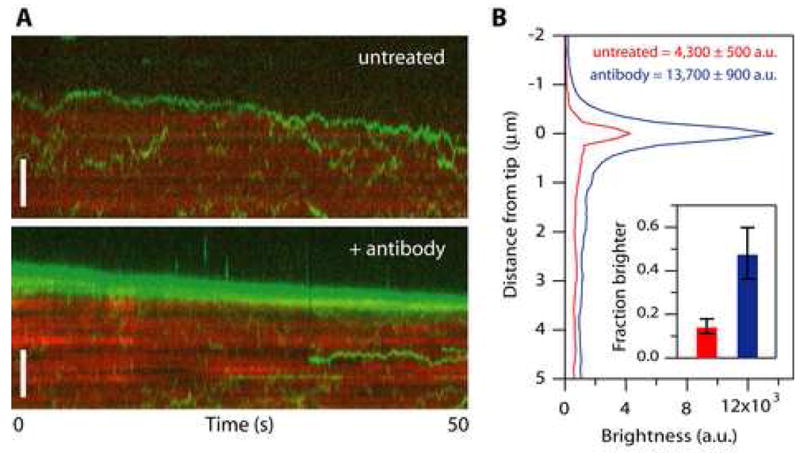
(A) Kymographs showing movement of particles composed of Ndc80 complexes (green) on disassembling microtubules (red). At 1 nM Ndc80 complex the microtubules were crowded with diffusing particles, many of which encountered the disassembling tip. No clear tip tracking was evident. However, in some cases the density of particles became noticeably higher at a disassembling tip relative to the lattice. When individual particles could be discerned as they encountered a disassembling tip, they sometimes appeared to ‘bounce’ repeatedly off the tip (upper kymograph). After pre-treating the Ndc80 complex with anti-His antibody to drive oligomerization, more disassembling tips accumulated fluorescent particles, and some long episodes of processive tip tracking were observed (lower kymograph). Scale bars, 3 μm.
(B) Profiles of average fluorescence versus position along the microtubule for a population of disassembling filaments decorated with Ndc80 complexes. Only intervals when the tip appeared to accumulate fluorescent particles were included in the averages. During these events, the disassembling tips became brighter if the Ndc80 complex had been pre-treated with antibody (blue curve, N = 21 events from 17 microtubules in 10 recordings) than without antibody pre-treatment (red curve, N = 24 events from 20 microtubules in 27 recordings). The fraction of disassembling tips that accumulated fluorescence was also increased after antibody pre-treatment (inset). Uncertainties in peak brightness represent s.d., and in the inset they represent counting error.
Given the results from our bead assays, we reasoned that tip tracking might occur in our TIRF assay if larger particles containing more complexes could be generated. To test this we recorded the behavior of Ndc80 complexes that had been pre-incubated with antibodies directed against the His6-tag on Spc24, to drive oligomerization. After antibody treatment, the fraction of disassembling tips that accumulated fluorescent particles was higher (46%, 17 of 35 tips) and they were 3-fold brighter than those in control experiments without antibody treatment (Figure 6B). In addition, some brighter particles in the antibody-treated samples exhibited clear episodes of processive tip tracking (Figure 6A, bottom). Together these observations indicate that tip tracking is not a property exhibited by individual Ndc80 complexes. Assembly of an array of complexes is required.
Biased diffusion fully describes the behavior of Ndc80-based couplers
To test whether the biased diffusion mechanism can account for the performance of Ndc80-based couplers, we used computational (Monte Carlo) methods to simulate Hill’s model (1985) with parameters chosen to match the properties of individual Ndc80 complexes. The simulations recapitulate our experimental observations very closely (Figure 7). When bound to growing microtubules in the absence of external load, the simulated couplers have no tendency to follow the tip. Instead, they remain on the microtubule lattice where they exhibit unbiased diffusion at a rate that slows as the number of binders in the ensemble increases (Figure 7B). At disassembling tips their random movement becomes biased: Small, highly diffusive couplers (e.g., with M = 2 binding elements) tend to stay ahead of the disassembling tip and only occasionally ‘bounce’ off the tip (Figure 7A). In contrast, couplers with more elements track unidirectionally with disassembly (Figure 7B). Interestingly, couplers with very high numbers of binders (e.g., M = 100) diffuse at undetectably slow rates, yet they still track with disassembling tips (see Supplemental Discussion). When tensile loads are applied, couplers with very few binding elements detach quickly but larger couplers can sustain substantial forces indefinitely. Couplers with M = 14 elements, for example, support 2.5 pN while remaining persistently attached to the filament tip over many micrometers of growth and shortening (Figure 7C). These observations show that the behavior of Ndc80-based couplers is well described by a biased diffusion mechanism.
Figure 7. Simulated behavior of a diffusion-based coupler.
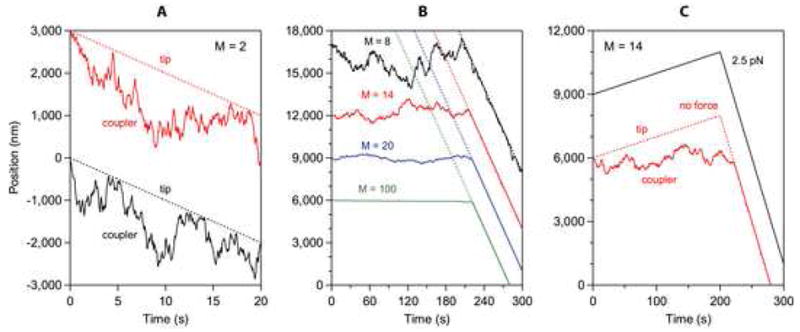
(A – C) Output from Monte Carlo simulations with parameters defined as in Hill (1985) but chosen to match our observations. The number of binding elements in the coupler, M, was varied between 2 and 100, as indicated. In (A and B), no external load was applied. The couplers in (A) were initially located at a disassembling tip. In (B) they were initially located on the lattice, where they exhibited unbiased movement for several minutes until being captured and carried by the disassembling tip (denoted by dotted lines). In (C) the couplers were initially located at an assembling tip, which later began to disassemble (> 200 s). A load of 2.5 pN was applied in one case (black trace), whereas no load was applied in the other case (red trace). The other parameter values were held constant as follows: r ≡e−b/kT = 0.75, s ≡ e−w/kT = 10−6, l = 0.62 nm, κrl2 = 0.170 μm2 s−1, αc = 10 nm s−1, βs = 100 nm s−1. Note that the parameters, s, l, κrl2, αc, and βs are constrained by the binding equilibrium for single Ndc80 complexes, the spacing of tubulin subunits in the microtubule lattice, the diffusion speed for single Ndc80 complexes, and the microtubule growth and shortening speeds, respectively. The only free parameter, r, was chosen to fall in the same range considered by Hill. The records are offset vertically by arbitrary amounts, for clarity.
DISCUSSION
The Ndc80 complex can make a direct and significant contribution to kinetochore-microtubule coupling
We measured the mechanical performance of Ndc80-based tip attachments in experiments that recapitulate the in vivo situation far more closely than previous assays. Even in the absence of other kinetochore components, we find that the Ndc80 complex can form dynamic tip attachments that permit assembly- and disassembly-coupled movement over distances comparable to the excursions of tip-attached kinetochores in vivo, which range from 0.5 to 5 μm depending on the cell type (Pearson et al., 2001; Stumpff et al., 2008). Under continuous tensile load the linkages often persist for many minutes, similar to the 2 to 10 minutes during which kinetochores support tension in yeast (Pearson et al., 2001). Also, the number of Ndc80 complexes required to form a load-bearing coupler in our assay is close to the 6 to 30 complexes found per microtubule at kinetochores in vivo (Emanuele et al., 2005; Joglekar et al., 2008).
The loads we applied here are comparable to the forces supported by kinetochore-microtubule junctions in vivo. Thermodynamic considerations suggest that a disassembling microtubule tip might be capable of producing as much as 20 to 60 pN of tensile force (Desai and Mitchison, 1997). However, based on the amount of stretching between sister kinetochores and the elasticity of the chromatin that links them, the tension transmitted to bi-oriented sister kinetochores during normal chromosome movement is probably closer to 0.4 to 8 pN per attached microtubule (Brower-Toland et al., 2002; Nicklas, 1988; Pearson et al., 2001; see Supplemental discussion). Ndc80-based linkages in our assay support similar forces of 0.5 to 2.5 pN, so the experiments provide a close approximation to the physiological situation.
Ndc80-based coupling likely depends on biased diffusion
At the level of individual complexes, Ndc80 binds microtubules transiently, exhibiting moderate (micromolar) affinity and rapid lattice diffusion. These properties make it ideally suited for efficient tip coupling through a biased diffusion mechanism (Hill, 1985). An alternative view proposed recently, that the complex mediates processive attachment to depolymerizing tips by binding tightly to peeling protofilaments (McIntosh et al., 2008), seems less likely given the mobility and transiency of the Ndc80-microtubule interaction (but the mechanism could apply to other kinetochore components). Instead, a mechanism based purely on biased diffusion is sufficient to describe the ability of Ndc80-based couplers to mediate attachments to both polymerizing and depolymerizing tips. Coupling by this mechanism relies on a gradient in binding energy, which exists at any tip, independent of whether it is assembling or disassembling. Importantly, the mechanism accounts for the ability of Ndc80-based couplers to sustain tensile forces comparable to those found in vivo. It will also provide a molecular friction that resists translocation of the microtubule through the attachment (Hill, 1985). Together these observations suggest that the Ndc80 complex forms a slip clutch at the kinetochore.
General implications for kinetochore-microtubule coupling
The idea that the Ndc80 complex forms a slip clutch at the kinetochore can explain aspects of spindle physiology. For example, when 9G3 antibodies against Ndc80/Hec1 are injected into dividing cells, kinetochore-microtubule attachments are stabilized, the chromosomes stop oscillating, sister kinetochores become hyperstretched, and kinetochore fiber flux slows (DeLuca et al., 2006). These changes are all consistent with an increase in molecular friction at the kinetochore. If an ensemble of Ndc80 complexes provides a kinetochore slip clutch, then antibody treatment could dramatically increase its friction by rafting the complexes together or by otherwise tightening their interaction with the filament (e.g., by blocking phosphorylation). The slip clutch idea can also explain the seemingly incongruous observations that the Ndc80 complex possesses only moderate microtubule affinity, yet its depletion results in severe detachment defects without major loss of other microtubule-binding kinetochore components (DeLuca et al., 2002). Notably, the affinity of the complex for microtubules depends on flexibly tethered positive charges that interact with negative charges on the E-hook of tubulin (Ciferri et al., 2008; Wei et al., 2007). This pliant, electrostatic binding is reminiscent of the charge-charge interactions that allow DNA-scanning enzymes to slide freely over the DNA phosphate backbone (Blainey et al., 2006). If flexibly tethered charges on the Ndc80 complex are the basis for a slip clutch activity at the kinetochore, this would explain why their removal severely compromises kinetochore-microtubule coupling (Guimaraes et al., 2008; Miller et al., 2008).
The fact that kinetochores in all eukaryotes maintain load-bearing attachments to growing and shortening microtubule tips suggests that a common molecular mechanism underlies this behavior. We propose that ensembles of Ndc80 complexes acting through a biased diffusion mechanism supply the combination of plasticity and strength that allows kinetochores to hold on loosely to microtubule tips without letting go.
MATERIALS AND METHODS
Protein expression and purification
The S. cerevisiae Ndc80 complex was expressed from two vectors encoding the Ndc80/Nuf2 and Spc24/Spc25 sub-complexes (Wei et al., 2005). For fluorescent studies, the Nuf2 gene was substituted with Nuf2-GFP from yeast strain MSY107-5d (Shimogawa et al., 2006). For the human Ndc80/Hec1 complex, a polycistronic vector encoding all four subunits was cloned (see Supplemental materials and methods). All complexes included a C-terminal His6-tag on Spc24 for affinity purification and bead binding. Purification was carried out as described previously except buffers more amenable to the Ndc80 complex were used (Asbury et al., 2006; Franck et al., 2007; Gestaut et al., 2008). The buffer for purification over talon resin was 50 mM HEPES (pH 7.6) with 300 mM NaCl, 2.5 U mL−1 benzonase, and protease inhibitors (0.01 mg mL−1 chymostatin, 0.01 mg mL−1 aprotinin, 0.01 mg mL−1 leupeptin, 0.01 mg mL−1 pepstatin, 0.002 mg mL−1 benzamadine and 1 mM phenylmethylsulfonyl fluoride). For gel filtration, 50 mM HEPES (pH 7.6) with 200 mM NaCl was used.
Bead motility assays
Recombinant His6-tagged Ndc80 complex was linked to 0.44 μm-diameter streptavidin-coated polystyrene beads (Spherotech, Lake Forest, IL) using biotinylated His5-antibody (Qiagen, Valencia, CA) essentially as described in Asbury et al. (2006) and Franck et al. (2007). The amount of complex per bead was adjusted by incubating dilutions of 0.6 – 15 nM Ndc80 complex, prepared in BRB80 (80 mM PIPES, 1mM MgCl2, 1mM EGTA at pH 6.9) plus 8 mg mL−1 bovine serum albumin (BSA) and 1 mM DTT, with a fixed amount of beads (5.6 pM) at 4° C for 90 min. Flow chambers were created by sandwiching two strips of double-sided sticky tape between a KOH-washed coverslip and a standard glass slide, and functionalized by coating with 5 mg mL−1 biotinylated BSA (Vector Labs, Burlingame, CA), washing, and then incubating with 0.3 mg mL−1 avidin DN (Vector Labs). Stable, biotinylated microtubule seeds were introduced and then washed with growth buffer (BRB80 plus 8 mg mL−1 BSA with 1 mM GTP) before introduction of Ndc80-coated beads, which were diluted 8-fold into growth buffer supplemented with 1.5 to 1.9 mg mL−1 tubulin, 1 mM DTT and an oxygen scavenging system consisting of 250 μg mL−1 glucose oxidase, 30 μg mL−1 catalase and 4.5 mg mL−1 glucose. Assays were performed at 22° C.
TIRF microscopy assay
Flow cells for TIRF experiments were prepared as described in Gestaut et al. (2008). After rinsing with ddH2O, the coverslip was incubated with ‘rigor’ kinesin diluted in BRB20 (20 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.9) plus 200 mM KCl and 8 mg mL−1 BSA for 5 min. Taxol-stabilized Alexa-647-labeled microtubules were bound for ~1 min and washed with BRB20 plus 0, 30 or 60 mM KCl (for final concentrations of 30, 60 and 90 mM K+, respectively) and 10 μM taxol. GFP-tagged Ndc80 complex, diluted in BRB20 with 10 μM taxol, an oxygen scavenger system (consisting of 200 μg mL−1 glucose oxidase, 35 μg mL−1 catalase, 25 mM glucose and 5 mM DTT) and the reported amount of K+, was then introduced.
For experiments with dynamic microtubules, Alexa-647-labeled GMPCPP-stabilized seeds were bound and washed with growth buffer (BRB80 plus 8 mg mL−1 BSA and 1 mM GTP). Extensions were assembled in growth buffer plus 2 mg mL−1 tubulin (1% Alexa-647-labeled), and oxygen scavenger. After ~5 min the buffer was exchanged with BRB20 plus 1 mM GTP, 2 mg mL−1 tubulin (1% Alexa-647-labeled), oxygen scavenger. Then 1 nM GFP-labeled Ndc80 complex was introduced in a buffer lacking both free tubulin and GTP (BRB20 plus oxygen scavenger), to induce depolymerization. For antibody-induced oligomerization, 525 nM Ndc80 complex was pre-incubated with 250 nM biotin-conjugated His5-antibody (Qiagen, Valencia, CA) for ~60 min. For all experiments the Ndc80 complex was passed through a 0.22 μm filter after dilution into the final buffer.
Instrumentation, data collection, and analysis
Our optical force clamp has been described previously (Asbury et al., 2006; Franck et al., 2007). Bead-trap separation was sampled at 40 kHz while stage position was updated at 50 Hz to maintain the desired load. Position data were decimated to 200 Hz before storing to disk. Records of bead position versus time were analyzed using custom software written in Igor Pro (Wavemetrics, Lake Oswego, OR). Periods of microtubule growth and shortening were identified in the records by eye. Catastrophes were scored only when a bead moved in the direction of filament shortening by an amount that clearly exceeded the level of random noise (typically 5 – 10 nm rms during force clamping). Rescues were scored when an episode of rapid disassembly was clearly followed by slow growth. In assays without load, mean squared displacements versus time for individual traces were averaged to estimate the diffusion coefficient during microtubule assembly.
TIRF movies were analyzed using software developed in Labview (National Instruments, Austin, TX) for tracking the position and brightness of individual GFP spots. Association rates were estimated by dividing the total number of events by the total microtubule length, the total observation time and the concentration of free complex in solution. Matlab software (Mathworks, Natick, MA) was developed to calculate the average GFP intensity per pixel as a function of distance from disassembling MT tips. Only intervals when the tips appeared to accumulate more fluorescence than the lattice for > 4 s were included in the averages.
Computer simulations of the biased diffusion mechanism
Simulations of the biased diffusion model were performed in Igor Pro using standard iterative Monte Carlo methods. Briefly, for each iteration the rate constants for ‘inward’ and ‘outward’ movement, ki and ko, were first calculated based on the current state of the system, as described by Hill (1985). A random number between 0 and 1 was then generated and compared with these rates to determine whether the coupler moved during the iteration. The time interval corresponding to each iteration was kept very short (typically < 5 μsec) to avoid missing events. Shortening the interval further had no effect on the simulations, confirming that the fraction of missed events was negligible. We also verified that the program recapitulates the steady-state distributions shown in Hill’s paper (i.e., Figure 3) when matching parameters are used.
Supplementary Material
Acknowledgments
We thank Stephen Harrison and Jennifer DeLuca for plasmids, Anthony Azevedo for conducting early experiments with the 2NCC:2S construct, Ryan Lemke for technical assistance, Jason Stumpff for helpful comments on the manuscript, and all members of the Seattle Mitosis Club for valuable discussion. This work was supported by an NIH traineeship (T32 GM07270, awarded to A.F.P.), an NSF IGERT traineeship (DGE-0504573, to A.D.F.), a Searle Scholar Award (06-L-111, to C.L.A.), a Packard Fellowship for Science and Engineering (2006-30521, to C.L.A.), and by grants R01GM40506, R01GM79373 and R01GM69429 from the National Institute of General Medical Sciences (to T.N.D., C.L.A. and L.W., respectively).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell. 2005;16:4882–4892. doi: 10.1091/mbc.E05-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. Embo J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci U S A. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol. 2003;162:377–382. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, LGE, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. Kinetochore Attachments Require an Interaction between Unstructured Tails on Microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, Molk JN, Ruse C, Niessen S, Yates JR, 3rd , et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


