Abstract
Racemic-metal complexes were used to determine identity, enantiomeric excess (ee) and concentration of chiral diamines using MLCT bands in circular dichroism spectroscopy. It takes under just two minutes per sample to determine [G]t and %R with tolerable errors (19% and 4% respectively). The simplicity of the achiral receptors employed confers to this technique great potential for high-throughput screening.
In recent years high-throughput screening (HTS) assays have been developed for the discovery of efficient catalysts for asymmetric reactions.1 The classic methods employed for enantiomeric excess (ee) determination are HPLC or GC.2 These methods are relatively laborious and slow. Moreover, usually these methods do not permit simultaneous concentration determination. Therefore, it is desirable to create standard protocols which accelerate the quantification of yield and enantiomeric excess (ee) of chiral products in a high-throughput fashion.3 Toward these goals, the development of optical spectroscopic assays eases the instrumentation dependence and increases the speed of HTS assays.4
Progress in the development of indicator-displacement assays (IDA) have been made in recent years by our group,5 with either one receptor, or an array of receptors, combined with pattern recognition protocols for the creation of fast optical techniques that quantitate both ee and concentration. Only a few other techniques have been shown to have this capacity.6
Recently, we reported an even simpler optical technique, which is based on changes in metal-to-ligand charge transfer (MLCT) bands in the circular dichroism (CD) spectra of an inorganic complex. This method allowed us to determine the identity, ee, and concentration of chiral diamines.7 We focused the previous study on the use of one chiral receptor (1) for the ultimate goal of differentiating achiral guests from chiral guests. Here we report a simplification, where we concentrate on the use of racemic receptors. The use of racemic commercial receptors have clear economic and synthetic advantages over using pure enantiomers.8 Herein we describe the use of a small library of racemic complexes and show they can be used to quantitate ee and concentration, and differentiate the analytes. We employ pattern recognition protocols to analyze not only the behavior of one selected analyte, but also to analyze the fingerprint obtained for several analytes from the simultaneously combination of receptors.
CD spectroscopy has become a versatile tool for configuration assignment and the structural evaluation of molecules.9 Because CD active MLCT bands arise from achiral complexes upon chiral analyte binding,7,10 our protocols use simple inorganic complexes. In this study, our analytes are chiral vicinal 1,2-diamines (See Figure 1A).11,12
Figure 1.
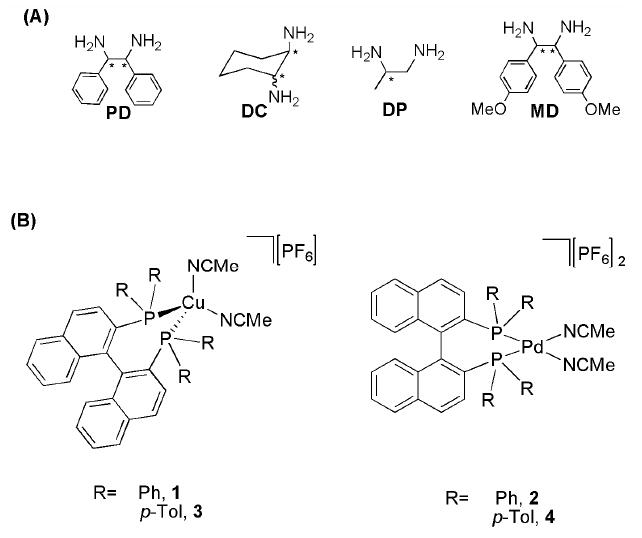
(A) Diamines employed: 1,2-phenylethylenediamine (PD), 1,2-diaminocyclohexane (DC), 1,2-diaminopropane (DP), bis-(4-methoxyphenyl)-1,2-diaminoethane (MD). (B) Racemic-metal complexes employed.
For the creation of racemic metal complexes that have CD active MLCT bands when a chiral analyte is added, we turned our attention to Cu(I) and Pd(II) as metals, and rac-Binap (2,2’-bis (diphenylphosphino) 1,1’-dinaphthyl) and rac-p-TolBinap (2,2’-bis(di-p-tolyl-phosphino)-1,1’-dinaphthyl) as ligands. The compounds [Cu(P-P)(NCMe)2]PF6 (P-P= Binap 1,7 Tol-Binap 313) and [Pd(P-P)(NCMe)2][PF6]2 (P-P= Binap 2, Tol-Binap 4) were prepared (see Supporting Information). The racemic receptors 1-4 do not show any CD signals, as expected. Also, there are no CD signals above 320 nm’s for the chiral diamine analytes, nor for copper or palladium alone with the diamines. Therefore, any signals that arise above this wavelength after the addition of the chiral guests to the receptors would be indicative of the chirality, identity, and concentration of the analyte.
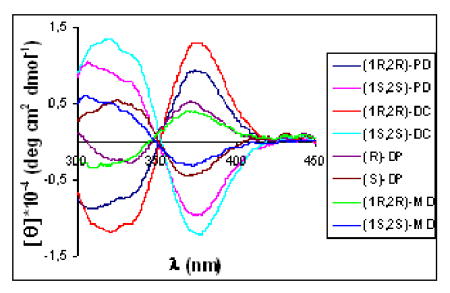
We first focused on the identification of diamines. Therefore, we analyzed complexes 1-4 with two equivalents of the enantiomers of each diamine. The CD showed that the MLCT bands of these complexes were characteristic to the analyte. Every diamine leads to a different CD spectrum, and the enantiomeric analytes gave identical and opposite spectra (see representative examples in Figure 2). These CD spectra allow for enantiomer identification based on the sign of the Cotton effect, as well as the identity of the diamines as judged by the intensity of the signals at selected wavelenghts.
Figure 2.
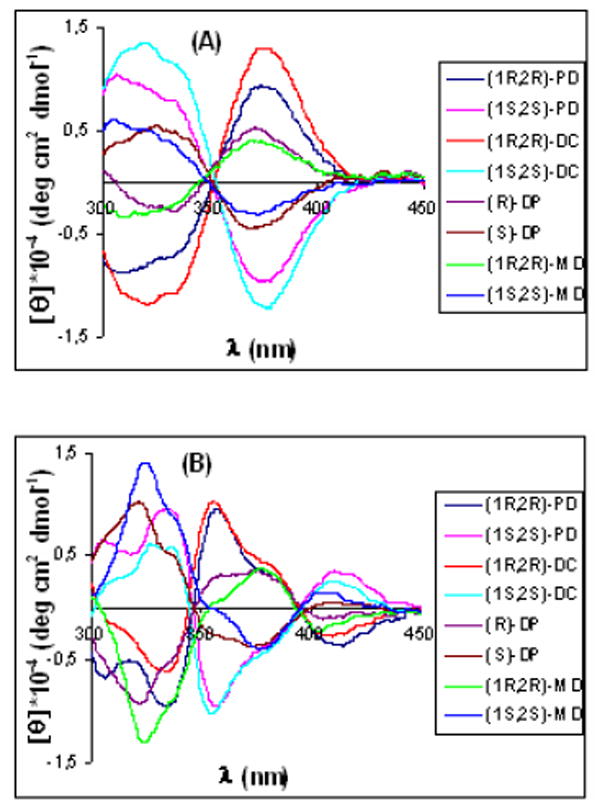
CD spectra of receptors [0.4 mM] with the guests [0.8 mM]: (A) (Rac)-1 and (B) (Rac)-2.
Therefore, to construct an identification pattern the CD spectra of all four receptors at 0.4 mM were recorded at 0.8 mM concentration of each analyte. To ensure reproducibility each experiment was performed 5 times. The data were analyzed at four selected wavelengths (330, 345, 375, 395 nm) for 1 and 3 and at four different wavelengths (320, 335, 355, 410 nm) for 2 and 4, and subjected to linear discriminate analysis (LDA) to identify diagnostic patterns.14
The LDA plots for receptors 1, 2, 3, and 4 show discrimination of all the guests. As an example, Figure 3A shows the results from receptor 1, revealing tight clustering of data relative to spatial separation of diamines and their enantiomers; but only along the F1 axis. This shows that one linear discriminate equation is enough to correlate the differences between ellipticities at the chosen wavelengths to chemical identity and chirality. The same result was found for receptor 3 (see Supporting Information). However, in the case of the palladium complexes 2 (Figure 3B) and 4 (Figure S4B), the clusters are spread through the four quadrants of the LDA plot. This is indicative of the fact that the differences in ellipticities at the four chosen wavelengths for each individual analyte cannot be expressed by one simple linear equation, thereby revealing differential response of the CD curves to each analyte at the wavelengths analyzed. One can readily see this by comparing Figures 2A and 2B, where in 2A the CD spectra have little crossing points while in 2B there is extensive crossing of the spectra.
Figure 3.
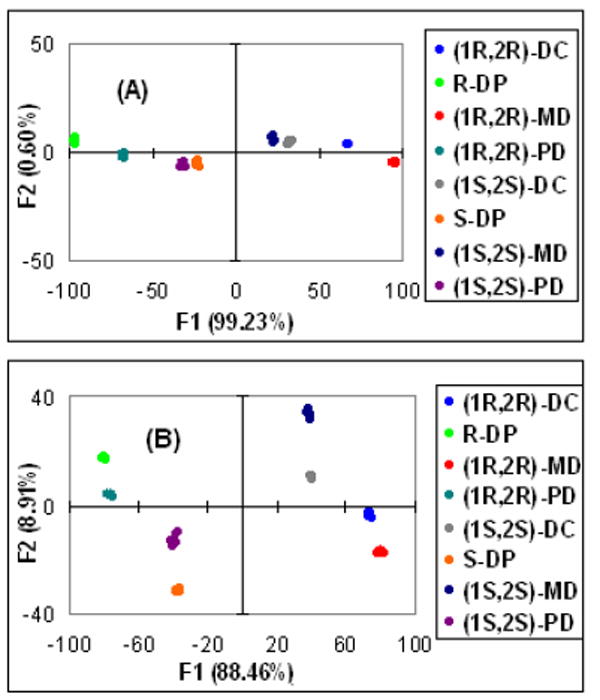
LDA plots obtained with the: (A) receptor 1, (B) receptor 2.
To test the ability of a combination of all the receptors to discriminate the analytes, we created an LDA plot using the CD data of all the receptors, employing the same wavelengths previously cited (See Figure 4). The LDA shows the analytes clearly discriminated with the greatest cross-reactivity reflected in the axes. The precision of the LDA plots was examinated using a jackknife analysis15 and 100% of accuracy in chemo- and enantioselectivity was established.
Figure 4.
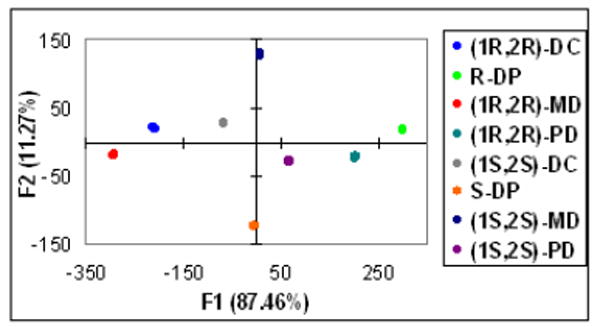
Linear discriminate analysis for a combination of all receptors.
After the determination of chirality and identity, we decided to investigate the capacity to determine both concentration and ee for a known analyte. We employed receptor 2 and DP combined with a robotically controlled 96-well plate CD reader (JASCO ASU-605). The wells were loaded with 0.4 mM of receptor 2 in acetonitrile, then charged with four concentrations of DP (0.2 mM, 0.4 mM, 0.8 mM, and 1.4 mM) with eleven ee values for each (1, 0.8, 0.6, 0.4, 0.2, 0, -0.2, -0.4, -0.6, -0.8, and -1). The same plate was loaded with five unknown samples created independent of the training set.
We analyzed the trainings sets using Principle Component Analysis (PCA) (See Figure 5A).14 This plot shows that the F1 axis defines chirality with negative scores for the samples enriched in (R)-enantiomer and positive scores for the (S)-enantiomer. The F2 axis defines concentration. From 0.2 mM to 0.8 mM the differences in concentration are considerable; however after 0.8 mM the differences relative to 1.4 mM are clearly smaller. These results correlate with the titrations which show that after approximately 0.8 mM saturation is achieved (Figure 5B).
Figure 5.
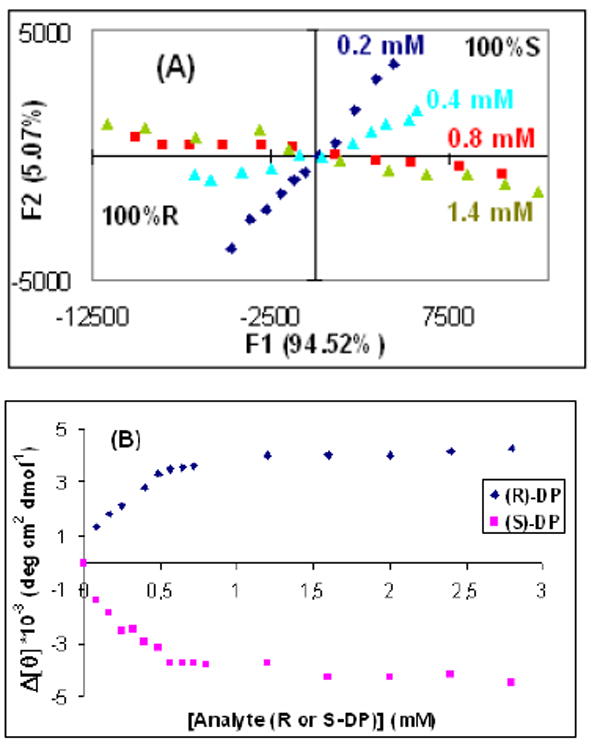
(A) Titration of receptor 2 [0.4 mM] with enantiomers of DP at 381 nm. (B) PCA plot of DP with four ee training sets at four different [G]t values: 0.2, 0.4, 0.8, 1.4 mM.
Furthermore, we analyzed the same data using multilayer perceptron (MLP) artificial neural network (ANN)16 due to our previous experience,5d,7,17 and as a means to test the accuracy of this protocol for ee and concentration determination. The architecture of MLP consists of an input, an output, and a hidden layer where the data is processed through a back propagation algorithm. The input layer consists of 51 ellipticities from CD320 to CD420 at 2 nm intervals. The training set data resulted in a MLP-ANN with 12 hidden layers. The five unknowns were computed, and the values of [G]t and %R had average errors of 18.6% and 3.8% (Table 1), which is clearly amenable to a first approximation in a HTS protocol.
Table 1.
Determination of Concentration and ee of DP Samples by ANN.
| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| [G]t (mM) | 0.80 | 1.40 | 0.50 | 1.00 | 1.20 |
| [G]t (mM) (ANN) | 0.92 | 0.94 | 0.66 | 0.97 | 1.36 |
| %R | 90 | 80 | 50 | 30 | 20 |
| %R (ANN) | 85 | 80 | 42 | 26 | 18 |
In summary, we described a technique based on a library of achiral receptors that fingerprints chemical identity, chirality, and concentration. The optical CD signatures can be interpreted by a variety of pattern recognition protocols depending on the goal of the analysis. A CD spectrometer interfaced with a robotic 96-well plate serves to generate data that trains an ANN, and subsequently yields aceptable accuracy in the analysis of true unknowns. The technique allows for the determination of both concentration and ee values in under two minutes per sample. In this specific study we avoided chiral receptor synthesis and/or resolution, analyte derivatization, and pretreatment of the data, as well as the necessity of knowing the analyte concentration in advance. We are currently expanding the technique to a wide variety of chiral organic functional groups.
Supplementary Material
Experimental procedures, and crystallographic data (CIF). This material is avalaible free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by the National Institutes of Health, Grant GM77437 and the Welch Foundation. S.N. thanks the Spanish Ministry of Education and Science for a postdoctoral fellowship.
References
- 1.(a) Wahler D, Reymond J-L. Curr Opin Biotechnol. 2001;12:535–544. doi: 10.1016/s0958-1669(01)00260-9. [DOI] [PubMed] [Google Scholar]; (b) Stambuli JP, Hartwig JF. Curr Opin Chem Biol. 2003;7:420–426. doi: 10.1016/s1367-5931(03)00056-5. [DOI] [PubMed] [Google Scholar]; (c) Gennari C, Piarulli U. Chem Rev. 2003;103:3071–3100. doi: 10.1021/cr020058r. [DOI] [PubMed] [Google Scholar]
- 2.(a) Welch CJ, Szczerba T, Perrin SR. J Chromatogr A. 1997;758:93–98. [Google Scholar]; (b) Welch CJ, Grau B, Moore J, Mathre DJ. J Org Chem. 2001;66:6836–6837. doi: 10.1021/jo015966n. [DOI] [PubMed] [Google Scholar]; (c) Welch CJ, Fleitz F, Antia F, Yehl P, Waters R, Ikemoto N, Armstrong IJD, Mathre DJ. Org Process Res Dev. 2004;8:186–191. [Google Scholar]; (d) Sigman MS, Jacobsen EN. J Am Chem Soc. 1998;120:4901–4902. [Google Scholar]; (e) Wolf C, Hawes PA. J Org Chem. 2002;67:2727–2729. doi: 10.1021/jo025534s. [DOI] [PubMed] [Google Scholar]
- 3.For recently publications see: Bailey DM, Hennig A, Uzunova VD, Nau WM. Chem Eur J. 2008;14:6069–6077. doi: 10.1002/chem.200800463.Truppo MD, Escalettes F, Turner NJ. Angew Chem Int Ed. 2008;47:2639–2641. doi: 10.1002/anie.200705046.Chen Z-H, He Y-B, Hu C-G, Huang X-H, Hu L. Aust J Chem. 2008;61:310–315.Ingle JR, Busch KW, Kenneth W, Busch MA. Talanta. 2008;75:572–584. doi: 10.1016/j.talanta.2007.11.056.Tanaka H, Matilde S. Chirality. 2008;20:307–312. doi: 10.1002/chir.20439.Yashima E, Katsuhiro M. Macromolecules. 2008;41:3–12.Young BL, Cooks RG. Int J Mass Spectrom. 2007;267:199–204.Li Z-B, Lin J, Sabat M, Hyacinth M, Pu L. J Org Chem. 2007;72:4905–4916. doi: 10.1021/jo0704715.
- 4.(a) Lin J, Zhang HC, Pu L. Org Lett. 2002;4:3297–3300. doi: 10.1021/ol026565c. [DOI] [PubMed] [Google Scholar]; (b) Lee SJ, Lin W. J Am Chem Soc. 2002;124:4554–4555. doi: 10.1021/ja0256257. [DOI] [PubMed] [Google Scholar]; (c) Ahn KH, Ku H-Y, Kim Y, Kim S-G, Kim YK, Son HS, Ku JK. Org Lett. 2003;5:1419–1422. doi: 10.1021/ol034041m. [DOI] [PubMed] [Google Scholar]; (d) Corradini R, Paganuzzi C, Marchelli R, Pagliari S, Sforza S, Dossena A, Galaverna G, Duchateau J Mater Chem. 2005;15:2741–2746. doi: 10.1002/chir.10272. [DOI] [PubMed] [Google Scholar]
- 5.(a) Zhu L, Anslyn EV. J Am Chem Soc. 2004;126:3676–3677. doi: 10.1021/ja031839s. [DOI] [PubMed] [Google Scholar]; (b) Folmer-Andersen JF, Lynch VM, Anslyn EV. J Am Chem Soc. 2005;127:7986–7987. doi: 10.1021/ja052029e. [DOI] [PubMed] [Google Scholar]; (c) Zhu L, Zhong Z, Anslyn EV. J Am Chem Soc. 2005;127:4260–4269. doi: 10.1021/ja0435945. [DOI] [PubMed] [Google Scholar]; (d) Zhu L, Shabbir SH, Anslyn EV. Chem Eur J. 2006;13:99–104. doi: 10.1002/chem.200600402. [DOI] [PubMed] [Google Scholar]; (e) Folmer-Andersen JF, Kitamura M, Anslyn EV. J Am Chem Soc. 2006;128:5652–5653. doi: 10.1021/ja061313i. [DOI] [PubMed] [Google Scholar]
- 6.(a) Chen Y, Shimizu KD. Org Lett. 2002;4:2937–2940. doi: 10.1021/ol026336q. [DOI] [PubMed] [Google Scholar]; (b) Taran F, Gauchet C, Mohar B, Meunier S, Valleix A, Renard PY, Creminon C, Grassi J, Wagner A, Mioskowski C. Angew Chem Int Ed. 2002;41:124–127. doi: 10.1002/1521-3773(20020104)41:1<124::aid-anie124>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]; (c) Li Z, Bütikofer L, Witholt B. Angew Chem Int Ed. 2004;43:1698–1702. doi: 10.1002/anie.200353055. [DOI] [PubMed] [Google Scholar]
- 7.Nieto S, Lynch VM, Anslyn EV, Kim H, Chin J. J Am Chem Soc. 2008;130:9232–9233. doi: 10.1021/ja803443j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voshell SM, Lee SJ, Gagné MR. J Am Chem Soc. 2006;128:12422–12423. doi: 10.1021/ja0647699. [DOI] [PubMed] [Google Scholar]
- 9.(a) Nakanishi K, Berova N, Woody RW. Circular Dichroism Principles and Applications. VCH Publishers, Inc; 1994. [Google Scholar]; (b) Beychok S. Science. 1966;154:1288–1299. doi: 10.1126/science.154.3754.1288. [DOI] [PubMed] [Google Scholar]
- 10.(a) Rodger A, Patel KK, Sanders KJ, Datt M, Sacht C, Hannon MJ. J Chem Soc Dalton Trans. 2002:3656–3663. [Google Scholar]; (b) Uerpman C, Malina J, Pascu M, Clarkson GJ, Moreno V, Rodger A, Grandas A, Hannon MJ. Chem Eur J. 2005;11:1750–1756. doi: 10.1002/chem.200401054. [DOI] [PubMed] [Google Scholar]
- 11.(a) Chin J, Mancin F, ThavaBeriva, Nrajah N, Lee D, Lough A, Chung DS. J Am Chem Soc. 2003;125:15276–15277. doi: 10.1021/ja0387554. [DOI] [PubMed] [Google Scholar]; (b) Kim H-J, Kim H, Jeong EJ, Thavarajah N, Studnicki L, Koprianiuk A, Lough AJ, Suh J, Chin J. J Am Chem Soc. 2005;127:16370–16371. doi: 10.1021/ja055776k. [DOI] [PubMed] [Google Scholar]
- 12.(a) Noyori R. Adv Synth Catal. 2003;345:15–32. [Google Scholar]; (b) Lucet D, Le Gall T, Mioskowski C. Angew Chem Int Ed. 1998;37:2580–2627. doi: 10.1002/(SICI)1521-3773(19981016)37:19<2580::AID-ANIE2580>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]; (c) Bennani YL, Hanessian S. Chem Rev. 1997;97:3161–3195. doi: 10.1021/cr9407577. [DOI] [PubMed] [Google Scholar]
- 13.Yao S, Fang X, Jorgensen KA. Chem Commun. 1998:2547–2548. [Google Scholar]
- 14.Jurs PC, Baken GA, McClelland HE. Chem Rev. 2000;100:2649–2678. doi: 10.1021/cr9800964. [DOI] [PubMed] [Google Scholar]
- 15.Gong G. J Am Stat Assoc. 1986;81:108–113. [Google Scholar]
- 16.Burns JA, Whitesides GM. Chem Rev. 1993;93:2583–2601. [Google Scholar]
- 17.(a) Wiskur SL, Floriano PN, Anslyn EV, McDevitt JT. Angew Chem Int Ed. 2003;42:2070–2072. doi: 10.1002/anie.200351058. [DOI] [PubMed] [Google Scholar]; (b) McCleskey SC, Floriano PN, Wiskur SL, Anslyn EV, McDevitt JT. Tetrahedron. 2003;59:10089–10092. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental procedures, and crystallographic data (CIF). This material is avalaible free of charge via the Internet at http://pubs.acs.org.


