Abstract
We used mechanical trapping of molecular interactions to demonstrate a highly parallel approach to measure the kinetics of biomolecular interactions. This approach consumes 25 fmol of material per measurement and permits 320 measurements in a single experiment. We measured association and dissociation curves for the interactions of 6-His and T7 epitope tags with their antibodies, from which we determined the off rates, on rates, and dissociation constants.
Mapping physical interactions among biological macromolecules is a powerful way to understand their function, and there is wide interest in high-throughput methods of interaction detection.1, 2 However, quantitative characterization of interactions is limited by throughput in conventional methods.3 We recently described a microfabricated device that enables highly parallel measurements of binding constants, thus enabling quantitative characterization of equilibrium parameters.4 There is also significant interest in determining interaction kinetic parameters. The most common method for measuring on and off rates uses surface plasmon resonance (SPR) to detect changes in refractive index as molecules bind to a surface.5, 6 However, this approach suffers from a lack of robustness to changes in buffer refractive index and surface coating efficiency and requires specialized detection equipment as well as highly purified proteins due to sensitivity to nonspecific binding.7
To implement mechanically induced trapping of molecular interactions (MITOMI), our chip uses “buttons,” microfluidic valves that cover part of the bottom surface of a channel but do not completely block flow. Buttons are used for blocking during surface chemistry steps, which deposit an antibody to one of the interaction partners (B) on the surface, confined to the area under the buttons. After B and its interaction partner (P) are introduced, closing the button traps the chemical equilibrium state during subsequent washing steps, until the interaction can be detected fluorescently. As we now show, modification of the buttons’ function to make nonequilibrium measurements allows for calculating the chemical kinetics of the interactions studied.
Assuming that the area under the button is initially covered with B, and that the experiment begins with the introduction of P, the rate of change of the concentration of the dimer BP is modeled by the binary interaction equation
| (1) |
If P is fluorescently labeled such that F, the fluorescence measured under the button, is proportional to [BP], under the assumption that both [P] and [B]0=[B]+[BP] remain constant,
| (2) |
Solving for F as a function of time,
| (3) |
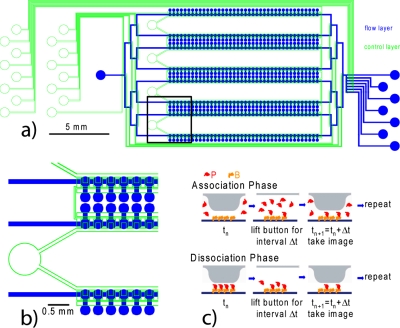
These conditions are realized in the “Kinetichip,” a modified version of the MITOMI chips (Fig. 1). The key feature is that the button control lines are wide enough over their entire length to allow for a fast response to actuation. The button response speeds were characterized by monitoring fluorescence changes under buttons of a chip whose flow lines were loaded with fluorescein, which was displaced by button closure. The sum of the rise time and fall time for a single cycle of button opening and closing averaged 0.14 s. Therefore, the total opening and closing time is negligible if the button is opened for an interval of more than a few seconds.
Figure 1.
(a) Schematic of Kinetichip. (b) Blowup of boxed region in (a). Each unit cell has a button for controlling surface chemistry and trapping interacting proteins. Each pair of rows of buttons is controlled from a single external port. The chip is designed such that the widened button control lines never cross a flow line, where they would create unwanted valves. (c) Cartoon of button functionality. During the association phase of the experiment, the button is raised for a specific amount of time, during which proteins, P, in solution can associate with a bound protein, B, on the surface. Then an image is taken, and the process is repeated, creating time points for an association curve. For the dissociation phase, the process is reversed.
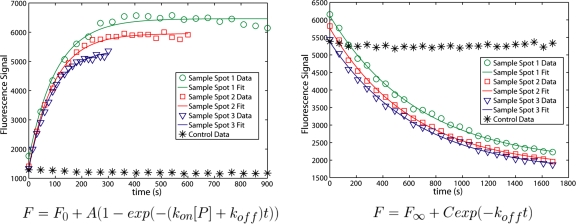
As a proof of principle, we studied the interactions of penta-His epitope antibody (Qiagen 34440) with a 6-His epitope tag, and T7 epitope antibody (Novagen 69968) with its epitope tag (MASMTGGQQMG). We used eGFP made by an in vitro transcription∕translation (ITT) kit (Roche 3335461001) with the appropriate tag at the C terminus, and concentration determined by fluorescence. A Kinetichip was prepared with the appropriate antibody playing the role of B immobilized under the button with a biotin-neutravidin interaction. With buttons closed, unpurified eGFP ITT reaction diluted with phosphate buffered saline (PBS) was introduced, and the buttons were raised for a predetermined time interval. The chip was then imaged using an Arrayworx scanner (Applied Precision) with 480 nm excitation filter, 530 nm emission filter, or a TECAN LS Reloaded scanner with 488 nm excitation laser, 535 nm emission filter. During the scan, fresh eGFP was introduced to maintain a constant [P]. The process was repeated to generate time points for an association curve [Figs. 1c, 2]. Then, with the spot under each button loaded with eGFP, the eGFP solution was washed out with PBS. To generate time points for a dissociation curve, the buttons were raised, an image was taken with PBS flowing during the scan to wash out dissociated eGFP and maintain [P]=0, and the process was repeated.
Figure 2.
Sample data from T7 epitope tag experiments. These data are from four different spots during a single experiment. With multiple independently controlled buttons, the time resolution can be tuned to best capture the exponential decay, as in the association phase of the experiment (left panel). The horizontal axis represents the cumulative time the button is raised, except for the control data points, which are arbitrarily plotted on the same time axis as for Sample 1, to demonstrate the lack of change in signal. The dissociation curves in the right panel are from the same spots as the association curves.
The images were analyzed with GenePixPro 3.0, finding the median fluorescence signal in a selected area in each spot for each time point. The association time points were fit to Eq. 3 using SigmaPlot 10.0 or OriginPro 7.5. For the dissociation phase, with [P]=0, Eq. 2 reduces to a simple exponential decay. With koff determined by fitting to the dissociation curves, and [P] known, kon and Kd=koff∕kon are calculated from the association fits.
In all experiments, during both the association and dissociation phases, one of the button valves remained closed. This provided a control to demonstrate that P is excluded from binding while the button remains closed (during association), and that decay in signal from photobleaching and escape of protein from under the closed button is negligible (during dissociation). Signal from these control spots was flat during both phases (Fig. 2).
To validate results, the kinetic constants were also measured by SPR (Biacore) with HBS-EP (0.01M HEPES, pH 7.4, 0.15M NaCl, 3 mM EDTA) as running buffer, and the ITT-expressed eGFP dialyzed into the running buffer (Table 1). The dialysis was necessary to eliminate nonspecific interactions, which caused a bulk resonance shift that dominated the specific interaction signals in an experiment with unpurified ITT reaction. We suspected that differences between the Biacore and Kinetichip results could be due to differences in buffer conditions, which can significantly affect the on and off rates.8 To test this, a Kinetichip experiment was run on the T7 interaction, with the eGFP diluted in HBS-EP buffer, and with the same buffer used as the wash buffer when continuing from the association to the dissociation phase. Although the on-rate was not substantially affected, this change moved the off rate much closer to agreement, suggesting that the buffer conditions were an important, although not the sole, factor.
Table 1.
Summary of results and comparison to Biacore results. Kinetichip on rates and dissociation constants were calculated spot by spot and then averaged. Biacore kinetic constants were calculated by a global fit to a concentration series of association and dissociation curves, with dissociation constants determined simply by dividing off rate by on rate. This method did not generate standard deviations, but analysis of the curve-by-curve variation when fitting to the curves individually suggests standard deviations on the order of 10−4 s−1, 104 M−1 s−1, and 5 nM for the His results, and 2×10−3 s−1, 5×104 M−1 s−1, and 30 nM for the T7 results.
| koff±SD (s−1) | kon±SD (M−1 s−1) | Kd±SD (nM) | n | |
|---|---|---|---|---|
| His Kinetichip | 3.3×10−4±2.8×10−5 | 4.4×104±6.5×103 | 7.5±0.73 | 240 |
| His Biacore | 3.0×10−4 | 9.8×104 | 3.1 | |
| T7 Kinetichip | 1.9×10−3±6.6×10−4 | 4.4×104±1.0×104 | 45±14 | 492 |
| T7 Biacore | 8.1×10−3 | 1.1×105 | 72 | |
| T7 Kinetichip (HBS-EP buffer) | 5.7×10−3±8.5×10−4 | 4.3×104±8.6×103 | 130±33 | 120 |
Although not explored in as much depth as with the 6-His and T7 epitope interactions, the interaction between the c-Myc epitope tag (EQKLISEEDL) and its antibody (Sigma B7554) was also characterized by Kinetichip, yielding koff=2.7×10−3±2.5×10−4 s−1, kon=1.4×104±2.9×103 M−1 s−1, and Kd=195±49 nM.
The Kinetichip has several advantages over SPR methods, including the ability to use unpurified protein. The Biacore experiment consumed about 200 μl of 4 μM protein for a single measurement, while a typical Kinetichip experiment used 20 μl of 0.4 μM protein to perform 320 measurements, representing four orders of magnitude reduction in the amount of protein per experiment. Another advantage is that the images were taken with microarray scanners instead of a device dedicated specifically to SPR detection, though with recent development of microfluidic SPR devices,9 a hybrid approach incorporating buttons may also be useful. Additionally, the Kinetichip is compatible with MITOMI protein-protein interaction platforms in which the proteins are expressed in situ during the experiment.10 Each unit cell of the chip has a DNA chamber connected by a neck to the interaction chamber [Fig. 1b]. DNA templates can be arrayed on a microarray slide, and the chip aligned over them and annealed so that each DNA chamber contains a unique template for an on-chip ITT reaction. In this way, an array of proteins bound under the buttons can be created for kinetic interrogation by a protein introduced from off chip. A Kinetichip of the size used here can measure the interaction kinetics of one protein against as many as 320 partners. The combination of high throughput, low amount of material used, and ability to accommodate unpurified protein makes this method an attractive tool for proteomics.
Acknowledgments
The authors would like to thank Michael Eckart for assistance with Biacore experiments, the Stanford Microfluidics Foundry for chip fabrication, and Sebastian Maerkl, Doron Gerber and Thomas Snyder for valuable discussions. This work was supported by the NSF NanoCEMMS center (2007-06015-03) and the NIH Director’s Pioneer award (0D000251).
References
- Arifuzzaman M., Maeda M., Itoh A., Nishikata K., Takita C., Saito R., Ara T., Nakahigashi K., Huang H. C., Hirai A., Tsuzuki K., Nakamura S., Altaf-Ul-Amin M., Oshima T., Baba T., Yamamoto N., Kawamura T., Ioka-Nakamichi T., Kitagawa M., Tomita M., Kanaya S., Wada C., and Mori H., Genome Res. 16, 686 (2006). 10.1101/gr.4527806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish J. R., Gulyas K. D., and R. L.Finley, Jr., Curr. Opin. Biotechnol. 17, 387 (2006). 10.1016/j.copbio.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Jones R. B., Gordus A., Krall J. A., and MacBeath G., Nature (London) 439, 168 (2006). 10.1038/nature04177 [DOI] [PubMed] [Google Scholar]
- Maerkl S. J. and Quake S. R., Science 315, 233 (2007). 10.1126/science.1131007 [DOI] [PubMed] [Google Scholar]
- Morton T. A. and Myszka D. G., Methods Enzymol. 295, 268 (1998). 10.1016/S0076-6879(98)95044-3 [DOI] [PubMed] [Google Scholar]
- Campbell C. T. and Kim G., Biomaterials 28, 2380 (2007). 10.1016/j.biomaterials.2007.01.047 [DOI] [PubMed] [Google Scholar]
- Karlsson R., J. Mol. Recognit. 17, 151 (2004). 10.1002/jmr.660 [DOI] [PubMed] [Google Scholar]
- Andersson K., Areskoug D., and Hardenborg E., J. Mol. Recognit. 12, 310 (1999). [DOI] [PubMed] [Google Scholar]
- Luo Y., Yu F., and Zare R. N., Lab Chip 8, 694 (2008). 10.1039/b800606g [DOI] [PubMed] [Google Scholar]
- Gerber D., Maerkl S. J., and Quake S. R., Nat. Methods 6, 71 (2009). 10.1038/nmeth.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]