Abstract
Lentiviral vectors effectively transduce both dividing and non-dividing cells and stably integrate into the genome of the host cell. In this study, we evaluated the usefulness of a lentiviral system for genetic modulation of primary human hepatocyte cultures. Infection with GFP-expressing lentivectors shows that Huh7 and HepG2 cell lines, as well as primary cultures of human hepatocytes, are efficiently transduced by lentiviral vectors. Real-time RT-PCR analyses demonstrate that infection with lentivectors does not alter hepatic hallmarks such as the expression of the nuclear receptors CAR, PXR, RXRα, or HNF4α, or albumin expression. Additionally, infected hepatocytes retain the capacity for CYP3A4 induction in response to treatment with phenobarbital, a uniquely sensitive indicator of hepatic differentiation status. Lentivectors may be used for both over-expression and knockdown analyses in primary hepatocytes, as demonstrated in this study by >200-fold CAR over-expression and knockdown of CAR to less than 40% of endogenous levels, with corresponding effects on CYP2B6 expression. In summary, lentiviral vectors provide a novel methodology by which primary human hepatocytes may be stably genetically manipulated, with minimal effects on the differentiated hepatic phenotype. These approaches offer considerable advantage over current methodologies, providing a valuable alternative for use in pharmacological and toxicological investigations involving primary human hepatocyte models and potentially for cell-based therapeutics to treat hepatic dysfunction in vivo.
Keywords: primary human hepatocytes, lentivirus, differentiation, CAR, siRNA
Introduction
Healthy hepatic function is integral to the disposition and metabolism of a diverse array of endogenous and exogenous substances. However, the liver is the target of many congenital and acquired diseases, with some amenable to gene therapy. Additionally, cultures of primary human hepatocytes serve as vital models for drug metabolism research and as predictors of toxicological outcomes that may result following exposure to xenobiotic agents. The ability to modulate gene expression within the fully differentiated hepatocyte is imperative for the realization of the full potential of hepatic gene therapy, and for directing the hepatocyte model in research investigations.
Development of an ideal methodology for the genetic manipulation of primary human hepatocyte cultures has proven challenging. Primary human hepatocytes are refractory to most common transfection techniques. While adenoviral vectors transduce hepatocytes effectively, the vectors are non-replicating and remain episomal, and thus gene expression is transient [1,2]. Retroviral vectors integrate into the host genome, but most require a round of cell division for the integration event to occur [3–5]. Such viral vectors are therefore ineffectual for stable genetic modulation in quiescent primary hepatocyte cultures [6]. Baculovirus vectors are reasonably efficient for hepatocyte infection, however, previous studies in our laboratory have demonstrated that the baculoviral infection event adversely affects the differentiated hepatic phenotype [7,8], with infection reducing hepatic albumin levels and ablating the phenobarbital induction response; both sensitive indicators of hepatocyte differentiation status [9].
Lentiviral vectors are unique within the retroviral vector family since lentivirus is capable of infecting both dividing and non-dividing cells and stably integrates into the host genome, thereby facilitating long-term transgene expression [10–12]. Several studies have shown that lentiviral vectors effectively transduce the widely used hepatoma cell lines, Huh7 and HepG2 [13–18]. More recently, researchers demonstrated that lentivectors offer high efficiency transduction of primary cultures of human hepatocytes [14,19–21]. While these results are promising, it is essential to evaluate whether the infection event alters the differentiated hepatic phenotype, or otherwise adversely affects hepatocytes in primary culture.
In this investigation, we verify that lentiviral vectors efficiently transduce Huh7 and HepG2 hepatoma-derived cell lines, and demonstrate that transgene expression is preserved through cell division. We also confirm that primary human hepatocyte cultures are effectively transduced by lentivectors and that hepatocytes retain transgene expression for the duration of the culture period. Importantly, the results of our studies demonstrate that, lentiviral infection does not alter mRNA expression levels of albumin or of key nuclear receptors in primary human hepatocytes, including constitutive androstane receptor (CAR), retinoid X receptor alpha (RXRα), pregnane X receptor (PXR), and hepatocyte nuclear factor four alpha (HNF4α). Additionally, we show that lentiviral-infected hepatocytes retain the capacity for cytochrome P450 3A4 (CYP3A4) induction in response to treatment with phenobarbital, responses all consistent with a highly differentiated hepatic phenotype [22]. Further, we demonstrate that a lentiviral infection strategy in hepatocytes is effective for achieving either over-expression or knock down of select RNA expression and resulting target gene responses.
Materials and methods
Materials
Cell lines were purchased from ATCC (Manassas, VA). Dulbecco’s Modified Eagle’s Media (DMEM) + GlutaMAX, Minimum Essential Media (MEM) + Earle’s Salts + L-Glutamine, non-essential amino acids, sodium bicarbonate, sodium pyruvate, HEPES, penicillin, streptomycin, GlutaMAX, and fetal bovine serum (FBS) were purchased from Gibco/Invitrogen (Grand Island, NY). Dexamethasone, insulin, selenium, transferrin, and linoleic acid/albumin were obtained from Sigma (St. Louis, MO). 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) was purchased from BioMol (Plymouth Meeting, PA). Phenobarbital (PB) was purchased from Sigma (St. Louis, MO). All oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). pTracer-CMV2 was purchased from Invitrogen (Carlsbad, CA). pCDH1-MCS1-EF1-copGFP cDNA lentivector, pSIH1-H1-copGFP shRNA lentivector, and pPACKH1 packaging plasmid mix were purchased from System Biosciences (Mountain View, CA). QIAquick Gel Extraction Kit and QIAfilter Plasmid Maxi Kit were obtained from Qiagen (Valencia, CA). Lipofectamine 2000 was purchased from Invitrogen, (Carlsbad, CA). PEG-8000 was purchased from Sigma (St. Louis, MO). Nikon inverted fluorescence microscope was purchased from Nikon USA (Melville, NY), and digital camera was purchased from Diagnostic Instruments (Sterling Heights, MI). TRIzol Reagent was obtained from Invitrogen (Carlsbad, CA). High Capacity cDNA Archive Kit, Assays-on-Demand Gene Expression Products, and ABI 7300 Real-time PCR System were purchased from Applied Biosystems (Foster City, CA).
Cell culture and treatment
293T/17 transformed human embryonic kidney cells were maintained in DMEM + GlutaMAX supplemented with 0.1 mM non-essential amino acids, 0.75 g/L sodium bicarbonate, 1 mM sodium pyruvate, 10 mM HEPES, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. HepG2 and Huh7 human hepatoma-derived cell lines were maintained in MEM + Earle’s Salts + L-Glutamine supplemented with 0.1 mM non-essential amino acids, 0.75 g/L sodium bicarbonate, 1 mM sodium pyruvate, 10 mM HEPES, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. Primary human hepatocyte cultures were obtained through the Liver Tissue Procurement and Distribution System under NIH Contract #N01-DK-9-2310 (refer to Table 1 for hepatocyte donor information). Hepatocytes were isolated by a three-step collagenase perfusion technique and plated on rat-tail collagen as previously described [23]. Hepatocytes were maintained in William’s E Media supplemented with 10 mM HEPES, 2 mM GlutaMAX, 100 units/ml penicillin, 100 μg/ml streptomycin, 25 nM dexamethasone, 10 nM insulin, 5 ng/ml selenium, 5 μg/ml transferrin, and 1% linoleic acid/albumin.
Table 1.
Human hepatocyte donor information.
| Donor | Age | Gender | Ethnicity | Cause of Death |
|---|---|---|---|---|
| HH-A | 54 | M | C | N/A (Resection) |
| HH-B | 45 | M | C | Head Trauma |
| HH-C | 54 | M | C | N/A (Resection) |
| HH-D | 55 | M | C | Cerebrovascular Accident |
| HH-E | 74 | F | C | N/A (Resection) |
| HH-F | 49 | M | C | N/A (Resection) |
| HH-G | 69 | F | C | N/A (Resection) |
| HH-H | 44 | M | C | N/A (Resection) |
| HH-I | 57 | F | C | N/A (Resection) |
Lentiviral cDNA expression vector construction
Human CAR (NM_005122) was initially PCR amplified from human liver cDNA using the primer sequences: F 5′ GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG 3′, R 5′ GATCGATATCTCAGCTGCAGATCTCCTGGAGCCAG 3′ (restriction sites underlined). The amplicon was then digested with Eco RI and Eco RV and cloned by ligation into pTracer-CMV2. The hCAR fragment was subsequently sub-cloned into pCDH1-MCS1-EF1-copGFP cDNA lentivector. Briefly, hCAR was digested from the original vector using Eco RI and Eco RV, electrophoresed through a 0.6% agarose gel, purified using QIAquick Gel Extraction Kit, and ligated into Eco RI and Swa I sites of the pCDH1 vector. Plasmids were purified using QIAfilter Plasmid Maxi Kit.
SiRNA design and lentiviral siRNA expression vector construction
Small interfering RNA target sequences within the hCAR mRNA were identified using Genscript Corporation’s “siRNA Target Finder” software (https://www.genscript.com/ssl-bin/app/rnai) and confirmed by searching a number of other siRNA target sequence identifiers accessible to the public. Template sequences encoding a short hairpin RNA (a stem-loop structure consisting of both the sense and anti-sense strands of the targeted sequence separated by a loop sequence) were generated and cloned into pSIH1-H1-copGFP shRNA lentivector. Briefly, two complementary oligonucleotides (Table 2, restriction sites underlined) were annealled, digested, electrophoresed through a 3% agarose gel, purified using QIAquick Gel Extraction Kit, and ligated into Bam H1 and Eco RI sites of the pSIH1 vector. Plasmids were purified using QIAfilter Plasmid Maxi Kit.
Table 2.
Sequences of oligonucleotides used to generate pSIH1 lentivectors expressing siRNAs targeted to CAR (restriction sites underlined).
| Template I.D. | Strand 1 | Strand 2 |
|---|---|---|
| CAR 210 | 5′GGTACCGGATCCGCCACAGGCTACCA CTTTAATCTTCCTGTCAGAATTAAAGTG GTAGCCTGTGGCTTTTTGAATT CGAATT CGGTACC 3′ |
5′GGTACCGAATTCGAATTCAAAAAGCCAC AGGCTACCACTTTAATTCTGACAGGAAGA TTAAAGTGGTAGCCTGTGGCGGATCCGG TACC3′ |
| CAR 379 | 5′ACCGGATCCCTGGCATGAGGAAAGAC ATGACTTCCTGTCAGATCATGTCTTTCC TCATGCCAGTTTTTGAATTCGGT3′ |
5′ACCGAATTCAAAAACTGGCATGAGGAA AGACATGATCTGACAGGAAGTCATGTCTT TCCTCATGCCAGGGATCCGGT3′ |
| CAR 572 | 5′ACCGGATCCAGCTCATCTGTTCATCCA TCACTTCCTGTCAGATGATGGATGAACA GATGAGCTTTTTTGAATTCGGT3′ |
5′ACCGAATTCAAAAAAGCTCATCTGTTCA TCCATCATCTGACAGGAAGTGATGGATGA ACAGATGAGCTGGATCCGGT3′ |
| CAR 755 | 5′ACCGGATCCGGAAATCTGTCACATCG TACTCTTCCTGTCAGAAGTACGATGTGA TGACATATTTCCTTTTTGAATTCGGT3′ |
5′ACCGAATTCAAAAAGGAAATCTGTCACA TCGTACTTCTGACAGGAAGAGTACGATGT GACAGATTTCCGGATCCGGT3′ |
Lentiviral production, titering and target cell infection
Lentivirus production and subsequent target cell infections were performed essentially according to manufacturer’s instructions, except that Lipofectamine 2000 was utilized for transfection of the 293T/17 packaging cells with pPACKH1 packaging plasmids and either pCDH1 or pSIH1 expression vectors. Psuedoviral supernatant harvested from packaging cells was either used to infect target cells directly, or was concentrated prior to infection by 10% PEG-8000 precipitation. Viral titer was estimated by infecting 100,000 293T/17 cells with 10-fold dilutions of virus immediately after plating. 48 h post-infection the percentage of GFP+ cells in 3–5 random fields was determined by counting cells at 200× magnification and used to calculate the number of transducing units per milliliter of viral supernatant (TU/ml). GFP expression in infected cells was observed using a Nikon inverted fluorescence microscope, and images were captured using SpotRT software with a digital camera.
Real-time RT-PCR
Primary human hepatocyte RNA was isolated using TRIzol Reagent and converted to cDNA using High Capacity cDNA Archive Kit, both according to manufacturers’ instructions. Real-time RT-PCR was performed using Assays-on-Demand Gene Expression Products according to manufacturer’s protocol. Briefly, 100 ng of cDNA template, 25 μl 2X Taqman Universal Master Mix, and 2.5 μl 20× Target Assay Mix were combined into 50 μl reactions. Each reaction was divided in half and run on an ABI 7300 Real-time PCR System. Real-time RT-PCR data were analyzed using the ΔΔCT Method as previously described [24]. Briefly, CT values for each half-reaction were averaged and this value used in subsequent calculations. Target gene expression levels for each sample were normalized to those of the corresponding 18S endogenous control by calculating the ΔCT [ΔCT = CT target − CT 18S]. Target gene expression levels of samples infected with gene-or siRNA- expressing virus were further normalized to those infected with empty virus by computing the ΔΔCT [ΔΔCT = ΔCT gene/siRNA-exp. virus − ΔCT empty virus]. The average relative fold change of target gene expression levels of samples infected with gene- or siRNA- expressing virus relative to those infected with empty virus was calculated by raising 2 to the −ΔΔCT power [2−ΔΔCT], and is depicted in the bar graphs.
Results
Lentiviral vectors effectively transduce Huh7 and HepG2 cells, as well as primary human hepatocyte cultures
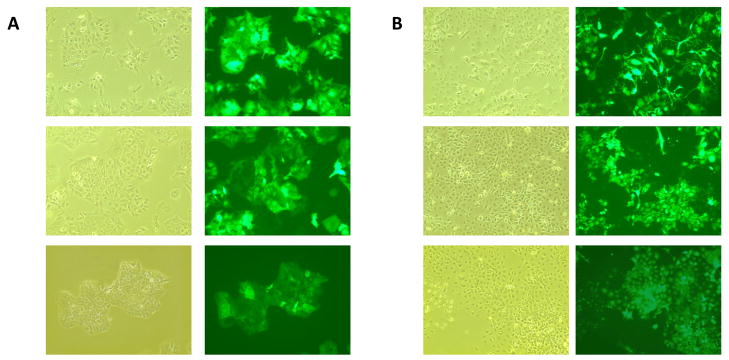
In order to evaluate the usefulness of a lentiviral system for genetic modulation of hepatic models, we first tested the ability of the lentiviral vectors to transduce two commonly used hepatic model cell lines, Huh7 and HepG2. Indeed, when infected with lentiviral vectors engineered to express green fluorescent protein (GFP) at a multiplicity of infection (MOI) of ~5 TU/cell, >95% of Huh7 cells, and >75% of HepG2 cells were estimated to be GFP+ (Figure 1A and 1B, upper panels). These results are consistent with those of previous studies which have reported that Huh7 cells are more efficiently transduced by lentivectors than HepG2 cells [14,16]. To confirm retention of transgene expression throughout cell division, infected Huh7 and HepG2 cells were maintained for over three months, throughout twenty passages, in the absence of any selective agent. Results indicated that GFP expression is retained in close to 100% of the cells in the initial GFP+ Huh7 and HepG2 populations, through cell proliferation and without selective pressures, although there is a decrease in mean fluorescent intensity of the GFP+ cells over time, most markedly in the HepG2 cells (Figure 1A and 1B, middle and lower panels).
Fig 1. GFP expression in Huh7 and HepG2 cells infected with GFP-expressing lentivirus.
Huh7 and HepG2 cells were infected while in suspension with pSIH1 lentiviral vectors at a MOI of ~5 transducing units/cell (TU/cell) and maintained through multiple passages. Cells were imaged at 100× magnification in both phase-contrast and fluorescence microscopy. (A) Upper panel, Huh7 cells at 5 days post infection; middle panel, Huh7 cells after 5 passages; lower panel, Huh7 cells after 20 passages (B) Upper panel, HepG2 cells at 5 days post-infection; middle panel, HepG2 cells after 5 passages; lower panel, HepG2 cells after 20 passages.
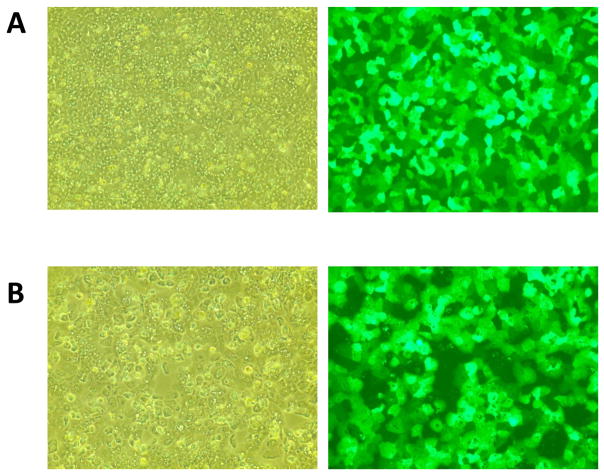
Primary cultures of human hepatocytes are the current gold standard in hepatic models. Thus, subsequent experiments assess the effectiveness of a lentiviral system for genetic manipulation of difficult-to-transfect primary human hepatocyte cultures (refer to Table 1 for hepatocyte donor information). Primary human hepatocytes were infected with GFP-expressing lentivirus at a MOI of ~5 TU/cell. Infected hepatocytes displayed high levels of GFP expression (>75% GFP+ cells) (Figure 2A) and expression of GFP was retained for the duration of the hepatocyte culture time (up to 14 days) (Figure 2B). Microscopic examination revealed that neither the hepatic cell lines nor the primary human hepatocytes exhibited morphological abnormalities upon lentiviral infection.
Fig 2. GFP expression in primary human hepatocyte cultures infected with GFP-expressing lentivirus.
Primary human hepatocyte cultures (HH-C) were infected with pSIH1 lentiviral vectors at a MOI of ~5 TU/cell. Cells were imaged at 100× magnification in both phase-contrast and fluorescence microscopy. (A) Hepatocyte cultures at 2 days post-infection. (B) Hepatocyte cultures at ~2 weeks post-infection.
Lentiviral infection does not affect select markers of the differentiated hepatic phenotype
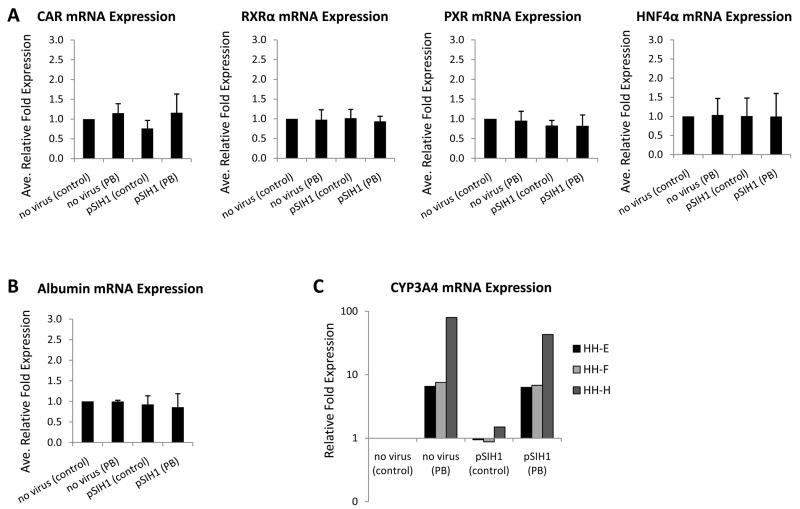
For primary human hepatocyte culture models to most accurately reflect in vivo responses, it is vital that the cultures maintain a highly differentiated status, i.e. that they retain hepatic signature hallmarks such as expression of specific nuclear receptors, hepatic secretory proteins, as well as metabolic enzyme induction capacity. As a genetic manipulation tool, the lentivirus system would be most valuable for use in a primary human hepatocyte model if the infection event does not alter these hepatic characteristics, or otherwise adversely affect the cell. To examine these parameters, we next evaluated whether lentiviral infection would alter an array of hepatic hallmarks, including expression of albumin and the nuclear receptors CAR, RXRα, PXR, and HNF4α, as well as induction of the CYP3A4 gene following phenobarbital treatment. Primary human hepatocyte cultures were infected with lentivectors at a MOI of ~10 TU/cell, a level sufficient to infect >80% of the cells as demonstrated by GFP marker gene expression (data not shown). Under these conditions, we observed no morphological changes in lenti-infected hepatocytes. Additionally, quantitative RT-PCR analyses demonstrated that infection with lentiviral vectors did not alter hepatic mRNA expression of CAR, RXRα, PXR, or HNF4α (Figure 3A). Albumin mRNA expression remained similarly unchanged by viral infection (Figure 3B). Although variable between donors as documented previously [25], induction of CYP3A4 upon phenobarbital treatment, was preserved in lentivirally-infected hepatocytes of individual donors (Figure 3C).
Fig 3. mRNA expression levels of hepatic hallmarks in cultures of primary human hepatocytes infected with lentivirus.
Cultures of primary human hepatocytes from three individual donors (HH-E, HH-F, and HH-H) were infected with pSIH1 lentiviral vectors at a MOI of ~10 TU/cell. Hepatocytes were treated with 500 μM PB for 24 h. RNA was extracted 3–4 days post-infection, converted to cDNA, subjected to real-time RT-PCR, and the data analyzed using the ΔΔCT method to determine relative mRNA expression levels. (A) mRNA expression levels of the hepatic nuclear receptors CAR, RXRα, PXR and HNF4α; values are mean +/− S.D. of the three hepatocyte donors. (B) Albumin mRNA expression levels; values are mean +/− S.D. of the three hepatocyte donors. (C) mRNA expression levels of CYP3A4 hepatic enzyme; values are individual expression levels for each of the three donors.
Effectiveness of the lentiviral system for both over-expression and knockdown of CAR
In our studies, lentiviral vectors were demonstrated to efficiently transduce primary human hepatoctye cultures with little effect on the assessed hepatic-specific responses and expression of hepatic hallmarks. We next evaluated the usefulness of the lentiviral genetic modulation system for over-expression and knockdown analyses in primary human hepatocyte cultures.
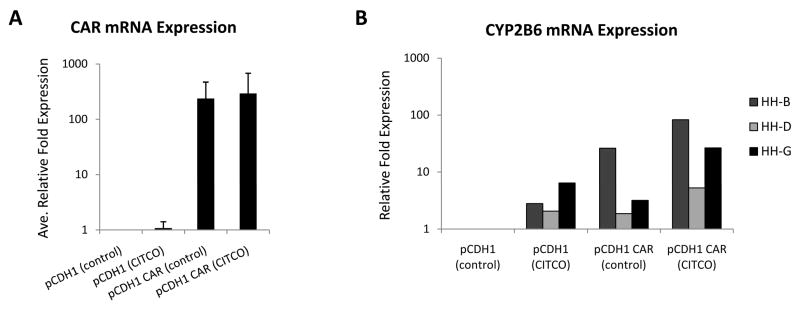
For CAR over-expression assessment, hepatocytes were infected with lentivirus expressing CAR. Quantitative RT-PCR results showed that CAR mRNA expression levels were increased >200-fold in cells infected with CAR-expressing lentivirus (Figure 4A). Whether such over-expression was functionally relevant was assessed by measuring mRNA expression levels of the CAR target gene, CYP2B6 (Figure 4B). Consistent with previous findings, human hepatocytes exhibited inter-individual variability in the level of CYP2B6 induction in response to treatment with the human CAR agonist, CITCO (2-fold to 6-fold) [25,26]. Hepatocytes infected with CAR-expressing lentivirus exhibited a highly variable increase in basal CYP2B6 expression levels (2-fold to 26-fold). CYP2B6 induction in hepatocytes infected with CAR-expressing lentivirus was only slightly enhanced upon CITCO treatment compared to that of hepatocytes infected with empty lentivirus (empty virus induction: HH-B 3-fold, HH-D 2-fold, HH-G 6-fold, and CAR-expressing virus induction: HH-B 3-fold, HH-D 2.5-fold, and HH-G 9-fold). These results indicate that some portion of the over-expressed CAR pool is engaged in cycling between the cytosolic and nuclear compartments, and due to its constitutive activity, that fraction that equilibrates to the nucleus participates in transactivation of endogenous gene targets similarly as would the resident activated receptor.
Fig 4. CAR and CYP2B6 mRNA expression in primary human hepatocyte cultures infected with CAR-expressing lentivirus.
Primary human hepatocyte cultures from three individual donors (HH-B, HH-D, and HH-G) were infected with lentviral vectors expressing CAR or pCDH1 empty lentivector. Hepatocytes were treated with 100 nM CITCO for 24–48 h. RNA was extracted 2–5 days post-infection, converted to cDNA, subjected to real-time RT-PCR, and the data analyzed using the ΔΔCT method to determine relative mRNA expression levels. (A) CAR mRNA expression levels; values are mean +/−S.D. of the three hepatocyte donors. (B) CYP2B6 mRNA expression levels; values are individual expression levels for each of the three donors.
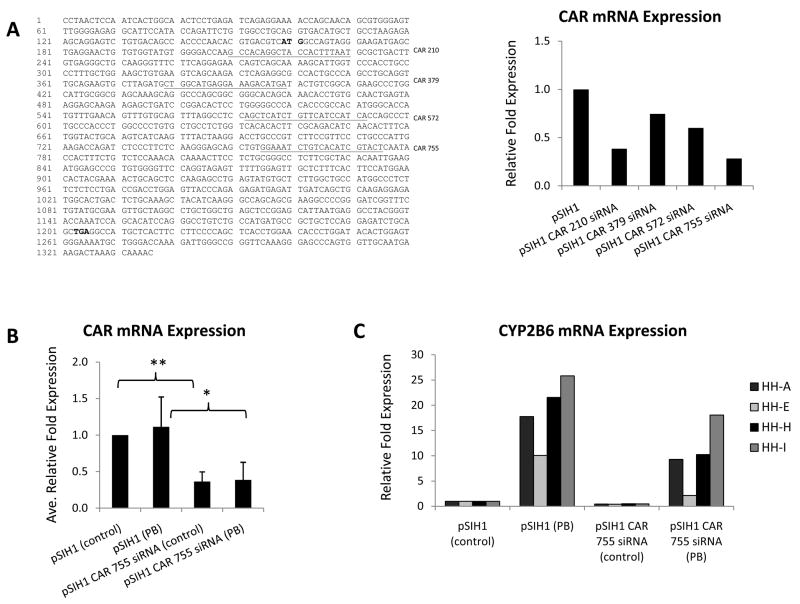
For CAR knockdown analysis, primary human hepatocytes were infected with lentivirus expressing four unique siRNAs targeted to CAR at specific locations along the mRNA. Quantitative RT-PCR analyses indicated that the siRNA targeted to CAR at position 755 exerted the most effective knockdown (Figure 5A). CAR mRNA expression was reduced to ~40% of endogenous cellular levels in hepatocytes infected with lentivirus expressing the siRNA against CAR at position 755 at a MOI of ~3 TU/cell (virus sufficient to infect >60% of the cells) (Figure 5B). Functional significance of the CAR knockdown was confirmed by assessing mRNA expression levels of CYP2B6 in the presence and absence of phenobarbital. Results demonstrated that CAR knockdown led to a corresponding ~50% decrease in both basal and phenobarbital-induced mRNA expression levels of CYP2B6 (Figure 5C).
Fig 5. CAR and CYP2B6 mRNA expression in cultures of primary human hepatocytes infected with lentivirus expressing siRNA against CAR.
Cultures of primary human hepatocytes from four individual donors (HH-A, HH-E, HH-H, and HH-I) were infected with pSIH1 lentivectors expressing siRNA against CAR at various positions. RNA was extracted 4–7 days post-infection, converted to cDNA, subjected to real-time RT-PCR, and the data analyzed using the ΔΔCT method to determine relative mRNA expression levels. (A) Left panel, schematic showing targeting locations of the CAR siRNAs assessed for CAR knockdown capability. Underlined sequence indicates siRNA target location; bolded sequence denotes translation initiation and termination codons. Right panel, graph illustrating the effect of the siRNA lentivectors on CAR transcript expression (pSIH1 = empty lentivector control). (B) CAR mRNA expression levels from hepatocytes infected with pSIH1 empty lentivector or pSIH1 CAR 755 siRNA (MOI of ~3 TU/cell) and treated with 500 μM PB for 24–48 h; values are mean +/− S.D. of the four hepatocyte donors (* denotes p <.05, ** denotes p <.0,1 by Student’s t-test, one-tailed, two sample unequal variance). (C) CYP2B6 mRNA expression levels from hepatocytes infected with pSIH1 empty lentivector or pSIH1 CAR 755 siRNA (MOI of ~3 TU/cell) and treated with 500 μM PB for 24–48 h; values are individual expression levels from each of the four donors.
Discussion
Previous investigations demonstrated that lentiviral vectors efficiently transduce the hepatic model cell lines Huh7 and HepG2 [13–18], as well as cultures of primary human hepatocytes [14,19–21], with Nguyen and colleagues reporting an MOI of 5 TU/cell as sufficient for transduction of ~75% of the primary hepatocyte culture [20]; all findings with which our results are consistent. Although these investigators demonstrated that lentivectors transduce primary human hepatocyte cultures with high efficiency, they did not evaluate whether the infection event alters hepatic signature hallmarks, such as expression of nuclear receptors or secreted proteins, or metabolic enzyme induction.
To determine whether lentiviral infection may adversely affect the differentiated hepatic character, we measured expression levels of a number of hepatic-enriched nuclear receptors subsequent to lentiviral infection. The nuclear receptors CAR, PXR, RXRα, and HNF4α were selected for evaluation as they regulate expression of a myriad of hepatic genes encoding, generally, drug metabolizing enzymes (such as cytochromes P450, sulfotransferases, UDP glucuronosyltransferases, and glutathione S-transferases), drug transporters (such as multidrug-resistance proteins, multidrug resistance-associated proteins, and organic anion-transporting proteins), as well as other nuclear receptors [27,28]. Our data demonstrate that the levels of these hepatic nuclear receptors remain constant despite lentiviral transduction at a MOI of ~10 TU/cell (virus sufficient to infect >80% of hepatocytes), therefore indicating that hepatic gene regulation by these transcription factors is not impaired by the infection event. This conclusion is further confirmed by the retention of albumin expression levels in lentivector-infected hepatocyte cultures. In contrast, albumin expression was adversely affected by baculoviral infection of primary hepatocytes [9]. Importantly, phenobarbital-mediated induction of CYP3A4, a sensitive indicator of hepatocyte differentiation status [22], was fully preserved in lentivirally-transduced cells.
Results from this study show that lentivectors may be employed for both over-expression and knockdown investigations in primary human hepatocyte cultures (as demonstrated here by CAR modulation), with the functional significance of these manipulations apparent by measure of effects on downstream target gene expression. Since the lentivectors integrate into the genome of the host cell, duration of transgene expression is limited in theory only by the length of time that hepatocytes can be maintained in culture. Our results demonstrate that lentivirus-infected Huh7 and HepG2 cell lines maintain robust GFP transgene expression for over three months in culture, spanning twenty passages (duration of experiment). Likewise, transgene expression is retained for at least 14 days post-infection in primary cultures of human hepatocytes. Further, we identified a novel small interfering RNA (siRNA) sequence capable of knocking down CAR to less than ~40% of endogenous levels, with specific knock-down of CAR mRNA expression remaining evident at 7 days post-infection. More lengthy time-course studies to confirm preservation of transgene and siRNA expression are underway.
Our results demonstrate that CAR can be knocked down to less than ~40% of endogenous cellular levels using a siRNA targeted to CAR at position 755 expressed from a lentivector. The data in Figure 5 represent the CAR mRNA expression levels from the entire hepatocyte culture, of which ~60% of the cells were infected. It is possible that the ~40% of uninfected hepatocytes may account for the remaining CAR expression, suggesting that this siRNA may approach close to 100% knockdown efficiency in infected cells. Experiments are currently underway in our laboratory to enrich for the infected population by sorting the cells by FACS. Recently, Chen and colleagues identified a siRNA capable of knocking down CAR to ~30% [29]. However, the knockdown experiment was performed in Caco-2 cells transiently transfected with both a CAR expression vector and the siRNA against CAR. Thus, to our knowledge, our report is the first to demonstrate direct knockdown of endogenous CAR.
This study demonstrates that lentiviral vectors may be utilized successfully to infect Huh7 cells, HepG2 cells, and, importantly, cultures of primary human hepatocytes. Data presented herein show that lentiviral infection does not alter hepatic markers including expression of albumin and an array of hepatic nuclear receptors. Further, infected hepatocytes retain the capacity for CYP3A4 induction by phenobarbital treatment, a response that is maintained only in highly differentiated hepatocytes. Both over-expression and knockdown analyses may be successfully undertaken using a lentiviral system in primary human hepatocyte cultures. Therefore, lentiviral vectors offer a powerful methodology for achieving stable genetic modulation of primary human hepatocytes, in the absence of perceived adverse effect on expression of the differentiated hepatic phenotype. These strategies offer a promising approach for both pharmacological and toxicological investigations within the primary hepatocyte model, as well for ultimate use in ex vivo gene therapy applications intended for intervention of hepatic disease.
Acknowledgments
The authors would like to thank Dr. Matthew Stoner for critical discussions and Ms. Denise Weyant for expert technical assistance. This research was supported by a USPHS grant from the National Institute of General Medicine (GM066411).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ilan Y, Saito H, Thummala NR, Chowdhury NR. Adenovirus-mediated gene therapy of liver diseases. Semin Liver Dis. 1999;19:49–59. doi: 10.1055/s-2007-1007097. [DOI] [PubMed] [Google Scholar]
- 2.Li Q, Kay MA, Finegold M, Stratford-Perricaudet LD, Woo SL. Assessment of recombinant adenoviral vectors for hepatic gene therapy. Hum Gene Ther. 1993;4:403–409. doi: 10.1089/hum.1993.4.4-403. [DOI] [PubMed] [Google Scholar]
- 3.Ferry N, Duplessis O, Houssin D, Danos O, Heard JM. Retroviral-mediated gene transfer into hepatocytes in vivo. Proc Natl Acad Sci U S A. 1991;88:8377–8381. doi: 10.1073/pnas.88.19.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller DG, Adam MA, Miller AD. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamashita M, Emerman M. Retroviral infection of non-dividing cells: old and new perspectives. Virology. 2006;344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Wolff JA, Yee JK, Skelly HF, Moores JC, Respess JG, Friedmann T, Leffert H. Expression of retrovirally transduced genes in primary cultures of adult rat hepatocytes. Proc Natl Acad Sci U S A. 1987;84:3344–3348. doi: 10.1073/pnas.84.10.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci U S A. 1995;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandig V, Hofmann C, Steinert S, Jennings G, Schlag P, Strauss M. Gene transfer into hepatocytes and human liver tissue by baculovirus vectors. Hum Gene Ther. 1996;7:1937–1945. doi: 10.1089/hum.1996.7.16-1937. [DOI] [PubMed] [Google Scholar]
- 9.Beck NB, Sidhu JS, Omiecinski CJ. Baculovirus vectors repress phenobarbital-mediated gene induction and stimulate cytokine expression in primary cultures of rat hepatocytes. Gene Ther. 2000;7:1274–1283. doi: 10.1038/sj.gt.3301246. [DOI] [PubMed] [Google Scholar]
- 10.Bukrinsky MI, Haggerty S, Dempsey MP, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 13.Cao YZ, Friedman-Kien AE, Huang YX, Li XL, Mirabile M, Moudgil T, Zucker-Franklin D, Ho DD. CD4-independent, productive human immunodeficiency virus type 1 infection of hepatoma cell lines in vitro. J Virol. 1990;64:2553–2559. doi: 10.1128/jvi.64.6.2553-2559.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowolik CM, Yee JK. Preferential transduction of human hepatocytes with lentiviral vectors pseudotyped by Sendai virus F protein. Mol Ther. 2002;5:762–769. doi: 10.1006/mthe.2002.0603. [DOI] [PubMed] [Google Scholar]
- 15.Liao W, Ning G. Knockdown of apolipoprotein B, an atherogenic apolipoprotein, in HepG2 cells by lentivirus-mediated siRNA. Biochem Biophys Res Commun. 2006;344:478–483. doi: 10.1016/j.bbrc.2006.03.164. [DOI] [PubMed] [Google Scholar]
- 16.Nash KL, Jamil B, Maguire AJ, Alexander GJ, Lever AM. Hepatocyte-specific gene expression from integrated lentiviral vectors. J Gene Med. 2004;6:974–983. doi: 10.1002/jgm.591. [DOI] [PubMed] [Google Scholar]
- 17.Seppen J, Rijnberg M, Cooreman MP, Oude Elferink RP. Lentiviral vectors for efficient transduction of isolated primary quiescent hepatocytes. J Hepatol. 2002;36:459–465. doi: 10.1016/s0168-8278(01)00308-7. [DOI] [PubMed] [Google Scholar]
- 18.Sirin O, Park F. Regulating gene expression using self-inactivating lentiviral vectors containing the mifepristone-inducible system. Gene. 2003;323:67–77. doi: 10.1016/j.gene.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Giannini C, Morosan S, Tralhao JG, Guidotti JE, Battaglia S, Mollier K, Hannoun L, Kremsdorf D, Gilgenkrantz H, Charneau P. A highly efficient, stable, and rapid approach for ex vivo human liver gene therapy via a FLAP lentiviral vector. Hepatology. 2003;38:114–122. doi: 10.1053/jhep.2003.50265. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen TH, Oberholzer J, Birraux J, Majno P, Morel P, Trono D. Highly efficient lentiviral vector-mediated transduction of nondividing, fully reimplantable primary hepatocytes. Mol Ther. 2002;6:199–209. doi: 10.1006/mthe.2002.0653. [DOI] [PubMed] [Google Scholar]
- 21.Selden C, Mellor N, Rees M, Laurson J, Kirwan M, Escors D, Collins M, Hodgson H. Growth factors improve gene expression after lentiviral transduction in human adult and fetal hepatocytes. J Gene Med. 2007;9:67–76. doi: 10.1002/jgm.1000. [DOI] [PubMed] [Google Scholar]
- 22.Sidhu JS, Liu F, Omiecinski CJ. Phenobarbital responsiveness as a uniquely sensitive indicator of hepatocyte differentiation status: requirement of dexamethasone and extracellular matrix in establishing the functional integrity of cultured primary rat hepatocytes. Exp Cell Res. 2004;292:252–264. doi: 10.1016/j.yexcr.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. Use of human hepatocytes to study P450 gene induction. Methods Enzymol. 1996;272:388–401. doi: 10.1016/s0076-6879(96)72044-x. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Moreau A, Maurel P, Vilarem MJ, Pascussi JM. Constitutive androstane receptor-vitamin D receptor crosstalk: consequence on CYP24 gene expression. Biochem Biophys Res Commun. 2007;360:76–82. doi: 10.1016/j.bbrc.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 27.Eloranta JJ, Meier PJ, Kullak-Ublick GA. Coordinate transcriptional regulation of transport and metabolism. Methods Enzymol. 2005;400:511–530. doi: 10.1016/S0076-6879(05)00028-5. [DOI] [PubMed] [Google Scholar]
- 28.Tirona RG, Kim RB. Nuclear receptors and drug disposition gene regulation. J Pharm Sci. 2005;94:1169–1186. doi: 10.1002/jps.20324. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Zhang J, Baker SM, Chen G. Human constitutive androstane receptor mediated methotrexate induction of human dehydroepiandrosterone sulfotransferase (hSULT2A1) Toxicology. 2007;231:224–233. doi: 10.1016/j.tox.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]