Abstract
Protoporphyrinogen oxidase (PPO, EC 1.3.3.4) catalyzes the six electron oxidation of protoporphyrinogen IX to the fully conjugated protoporphyrin IX. Eukaryotes and Gram-positive bacteria possess an oxygen-dependent, FAD-containing enzyme for this step while the majority of Gram-negative bacteria lack this oxygen-dependent PPO. In E. coli, PPO activity is known to be linked to respiration and the quinone pool. In E. coli SASX38, the knockout of hemG causes loss of measurable PPO activity. HemG is a small soluble protein typical of long chain flavodoxins. Herein purified recombinant HemG was shown to be capable of a menadione-dependent conversion of protoporphyrinogen IX to protoporphyrin IX. Electrochemical analysis of HemG revealed similarities to other flavodoxins. Interestingly, HemG, a member of a class of the long chain flavodoxin family that is unique to the γ-proteobacteria, possesses a 22 residue sequence that, when transferred into E. coli flavodoxin A, produces a chimera that will complement an E. coli hemG mutant, indicating that this region confers PPO activity on the flavodoxin. These findings reveal a previously unidentified class of PPO enzymes that do not utilize oxygen as an electron acceptor thereby allowing γ-proteobacteria to synthesize heme in both aerobic and anaerobic environments.
Keywords: Heme biosynthesis, protoporphyrinogen oxidase, quinol reductase, flavodoxin
Heme is a highly versatile small organic compound that is a key component of most living organisms. As a cofactor it functions in diverse reactions ranging from a gas sensor and transporter of diatomic gasses to serving as an electron carrier in redox-linked reactions such as is found in mixed-function oxidases, catalases or electron transport chains (1-3). In addition, it is now recognized that it is also a regulatory molecule. It has been shown to manage protein expression at the transcriptional (4, 5) and translational levels (6) as well as through protein targeting and degradation (7), it participates in regulating circadian rhythm, lipid metabolism, and gluconeogenesis (8-10) and in prokaryotic organisms heme may even serve to alter pathogenesis by triggering decreased virulence levels and evasion of the host immune response (11, 12).
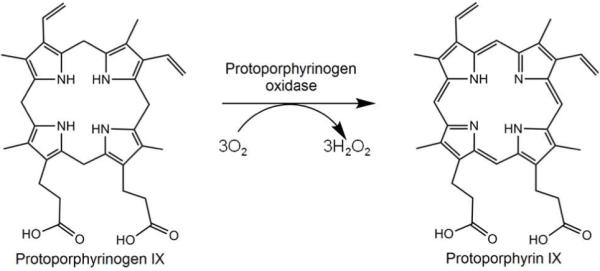
The biosynthesis of heme has been studied in both eukaryotes and prokaryotes. It is a tightly regulated metabolic pathway consisting of seven enzymatic steps from a precursor compound 5-aminolevulinic acid to the end product, protoheme (13). In humans and animals defects in any of these seven enzymes result in clinically distinct diseases, porphyrias, due to an accumulation of pathway intermediates (14). While the system has been extensively studied and is relatively well understood in eukaryotes (15), the pathway is less well-defined in bacteria and archaea (16). Two steps in particular, the penultimate and antepenultimate, have characterized enzymes that require molecular oxygen as an electron acceptor in eukaryotes. In higher organisms and a handful of Gram-negative organisms such as Myxococcus xanthus, the penultimate enzyme, protoporphyrinogen oxidase (PPO), exists as a membrane-associated enzyme that utilizes an FAD cofactor (17, 18) and converts oxygen to hydrogen peroxide during the reaction (scheme 1). This enzyme belongs to an FAD superfamily of proteins that also contains monoamine oxidases and phytoene desaturases (19). A soluble form of the enzyme named HemY is found in Gram-positive bacteria that produce heme and apart from its solubility, it is virtually the same as the eukaryotic enzyme (20). Not surprising, these oxygen-dependent enzymes have not been found in facultative or anaerobic bacteria although it is clear that some mechanism to catalyze this reaction must exist.
Scheme 1. Reaction catalyzed by oxygen-dependent protoporphyrinogen oxidase.
Investigations by the Jacobs group in the 1970’s found that PPO enzymatic activity was found in crude cellular extracts of E. coli and was reported to be membrane-associated (21). The activity was intimately linked to cellular respiration and increased in the presence of terminal electron acceptors such as fumarate or nitrate (22-24). Respiratory enzymes using these substrates all share one thing in common: the usage of quinones, specifically menaquinone-8, as an electron donor (25, 26). These same studies also showed that menaquinone-deficient E. coli were deficient in PPO activity (21). Mutagenesis of E. coli by the Sasarman group resulted in the production of one mutant, SASX38, that was deficient in PPO activity (27). The gene, named hemG, was mapped and later annotated as being responsible for the penultimate step in heme biosynthesis (28). Sequence analysis of the encoded protein reveals it to be a member of the protein family known as long chain flavodoxins. These are small electron transfer proteins containing an FMN cofactor. They are distinct from the small chain flavodoxins in that they possess an additional loop inserted in the fifth beta strand. This insert has previously been implicated in specificity and function between redox partners (29, 30). To date, however, there are no published data for the expression, purification and characterization of HemG. Of particular note, because of HemG’s resemblance to flavodoxins, it has been generally assumed that it may simply function as an electron carrier for a bonafide PPO enzyme, and PPO enzymatic activity has never been reported for HemG (31).
In the current study the HemG protein was expressed, purified and characterized. In addition the nature of the hemG mutation in E. coli SASX38 was identified. EPR-monitored redox titrations of HemG were performed to examine the FMN cofactor’s role in electron transfer and a menadione-dependent PPO activity was identified and characterized. The possible function for the long chain insert loop, which is unique to HemG of the γ-proteobacteria, was also examined. The data presented unequivocally demonstrates that HemG functions as a protoporphyrinogen oxidase and that the PPO activity is conveyed by the long chain insert loop.
Materials and Methods
Bacterial strains and constructed plasmids
The hemG gene was amplified from E. coli genomic DNA and cloned into the NheI and HindIII sites of the 6x-His tag vector pTrchisA (Invitrogen, Carlsbad, CA, USA). The resulting plasmid pTHHemG was then tested for functionality by transforming into the SASX38 cell line that contains a knockout of hemG. Overexpression of recombinant protein was carried out in JM109 cells. Bdellovibrio bacteriovorus Bd2899 was cloned identically to E. coli hemG. The chimeric FldG plasmid was constructed by first cloning the fldA gene of E. coli into pTrchisA and then Quickchange (Stratagene, Jolla, CA, USA) mutagenesis was used to swap the 66 nucleotide insert region to that of hemG.
Protein expression, purification, and characterization
Cells containing pTHHemG were grown for 7 hours at 30 °C in 100 mL Circlegrow (MP Biomedical, Solan, OH, USA) containing 50 μg/mL ampicillin and then transferred to 1 L of the same media and grown for 22 hours at 30° C. Six hours prior to harvesting, the cells were supplemented with riboflavin at a final concentration of 0.75 mg/mL. Cells were then collected by centrifugation and resuspended in a solubilization buffer containing 50 mM Tris-MOPS pH 8.1, 100 mM KCl, 1% sodium cholate, and 10 μg/ml PMSF. The resulting suspension was sonicated three times for 30 s each and centrifuged at 100,000 x g for 30 minutes after which the supernatant was applied to a column containing HisPur Cobalt affinity resin (Thermo Sci., Rockford, IL, USA). The column was washed with solubilization buffer containing 15 mM imidazole and the protein eluted in solubilization buffer containing 300 mM imidazole and 10% glycerol. Concentration determination and spectrophotometric analysis was carried out using a Cary 1G scanning spectrophotometer (Varian, Palo Alto, CA, USA). SDS-page was done using Bio-Rad 4-20% Tris HCl ready made gels (Bio-Rad, Hercules, CA, USA). FPLC was done using an Aktaprime machine equipped with a Hi-prep Sephacryl S-300 column (GE healthcare, Piscataway, NJ, USA) using solubilization buffer.
EPR-monitored Redox Titrations
Redox titrations were carried out at 25° C using a solution containing 100 μM HemG in solubilization buffer (pH 7.0) and the following redox mediator dyes at 50 μM each: Methyl viologen, benzyl viologen, neutral red, safranin O, anthraquinone-2-sulphonate, phenosafranin, anthraquinone-1,5-disulphonate, 2-OH-1,4-naphthoquinone, indigo-disulphonate, and methylene blue. The reductant and oxidant used were sodium dithionite (20 mg/ml) and potassium ferrocyanide (50 mg/ml) respectively. HemG was fully reduced anaerobically and titration points were taken from potentials ranging from ∼-450 to -150 mV by adding small amounts of oxidant. After each addition of oxidant, the sample was stirred and allowed to come to a stable potential, and 250 μl of sample was transferred to an anaerobic EPR tube and frozen in liquid nitrogen. Potentials were obtained using a Ag/AgCl electrode. Reported potentials were then recalculated with respect to NHE values. EPR data was then collected at 9.18 gHz with a microwave power of 20 μW and the intensity of the semiquinone signal was quantified using double integration.
PPO assay
PPO activity was monitored as previously described (32). Reaction mixtures consisted of 50 mM NaH2PO4 (pH 8.0), 0.2% (w/v) Tween 20, 2.5 mM glutathione, 100 nM HemG and varying amounts of protoporphyrinogen IX and menadione. Experiments were also done using mesoporphyrinogen and coproporphyrinogen prepared as for protoporphyrinogen. Activity was monitored at 37° C by following porphyrin fluorescence with a Synergy HTI plate reader (BioTek, Winooski, VT, USA) equipped with 528 nm and 545nm bandpass filters and a 550 nm highpass filter on the excitation light and 635 nm bandpass filter on the emission light. Data (v vs. [S]) was then fitted to equation 1,
where Vapp is the apparent maximum rate and Kapp is the apparent Michaelis-Menton constant.
Results
Protein expression and characterization
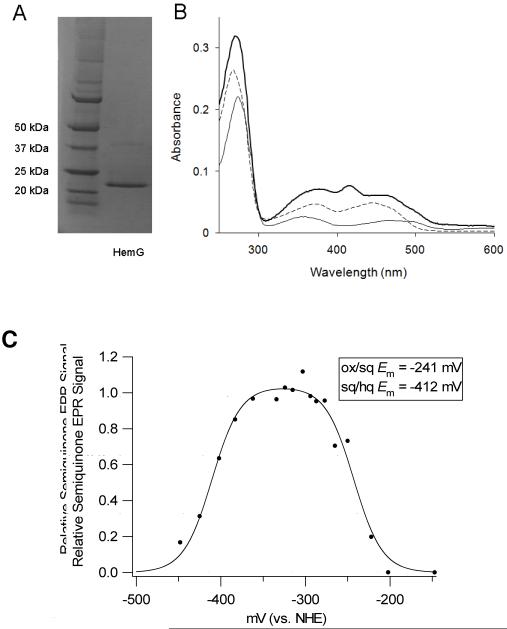
Sequence analysis of HemG from E. coli revealed that it is a member of a protein family found in the γ-proteobacteria that is part of a larger COG known as long chain flavodoxins (figure 1). (33) The hemG gene of E. coli was cloned, expressed and purified as a six histidine-tagged recombinant protein with a yield was approximately 20 mg protein/L culture. A band corresponding to approximately 22 kDa was observed on SDS-PAGE (figure 2A), which is in agreement with the theoretical value of 22.5 kDa calculated from the amino acid sequence. Maldi TOF/TOF mass spectroscopy identified this band as HemG. A faint band located at approximately 40 kDa was identified as HemG and may represent a dimer of the protein. The cofactor was determined to be similar in size to FMN by ESI MS. The recombinant expression plasmid rescued the HemG-deficient strain of E. coli SASX38, which alone cannot grow without the presence of exogenous heme.
Figure 1. Sequence analysis of HemG.
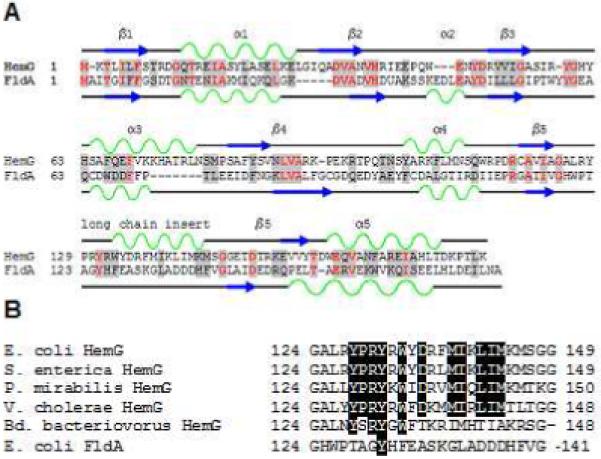
A. Sequence and secondary structure similarity of HemG with Flavodoxin A of E. coli. HemG is predicted by Jpred (33) to possess a typical flavodoxin-fold motif but with one notable difference, the presence of an alpha helical domain within the long chain insert loop. In the diagram similar residues are highlighted in grey, with conserved residues in red. Alpha helices and beta sheets are shown in green and blue respectively for each protein. B. Sequence alignment of the long chain insert loop of E. coli HemG with that of other annotated HemG proteins and FldA. Identical residues within the predicted alpha helix are highlighted. Significant change in this region (i. e. Bd. bacteriovorus) results in loss of function.
Figure 2. Initial characterization of HemG and its cofactor.
A. SDS-page analysis of purified recombinant HemG. Left lane, molecular weight marker; Right lane, purified HemG. Recombinant HemG possesses a major band of approximately 22 kDa with a faint band seen at approximately 40 kDa. Both bands were identified as HemG by MS/MS analysis. B. UV-vis spectrum analysis of purified recombinant HemG. The holoprotein (top line) exhibits characteristic FMN features (dashed line), as well as the presence of a peak at 416 nm which is lacking in E. coli flavodoxin A (bottom line). C. EPR-monitored redox titration of HemG. Data were fitted to the Nernst equation for two consecutive one-electron reduction steps (Eq. 1). A relative semiquinone EPR signal of 1 indicates a sample where all FMN present is in the semiquinone state. Inset: Calculated redox potentials for the oxidized/semiquinone and semiquinone/hydroxyquinone couples of cofactor as determined by Eq. 1.
The UV-visible spectrum of recombinant HemG possesses the expected features associated with protein and the FMN cofactor (figure 2B) along with a feature at 416 nm which probably corresponds to the Soret band of porphyrins, but it is yet to be determined if this is due to bound protoporphyrin IX. This last feature is distinct between the spectra of HemG and that of its closest relative E. coli flavodoxin A, where no band is observed. Gel filtration by FPLC identified a peak corresponding to ∼108 kDa suggesting that HemG exists in solution as a homotetramer.
EPR-monitored redox titrations
The EPR spectra of HemG revealed a signal typical of the FMN semiquinone radical centered at g = 2.00. Using sodium dithionite (potential of approximately -450 mV at pH 7.0) it was possible to fully reduce the FMN. Titrations of HemG included methyl and benzyl viologens, which possess EPR signals that overlap with the FMN semiquinone radical so the data were corrected by subtracting out the signal of the viologens alone. Data obtained at stable and reliable potentials from the double integration of interpretable spectra are shown in figure 2C. The data were fitted to the Nernst equation (equation 2) for two consecutive one electron redox steps:
and normalized to the fitted maximum. E2 and E1 represent the first and second reduction steps of HemG and were determined to be -241 mV and -412 mV respectively. These values (also shown in figure 2C) are comparable to those of standard flavodoxins (34). In typical flavodoxin reactions the semiquinone/hydroxyquinone couple is stabilized so that flavodoxins effectively function as one electron donor/acceptors.
PPO assays
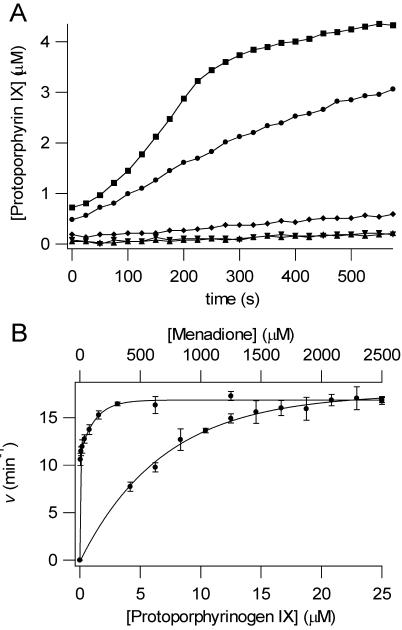
To investigate the role of HemG in the conversion of protoporphyrinogen IX to protoporphyrin IX, HemG was assayed in the presence and absence of the potential electron acceptor, menadione, a soluble analog of menaquinone-8. The results of this assay are shown in figure 3A. Upon addition of excess menadione the activity of HemG increased reaching a rate of roughly half of that found with human PPO (3.71 and 8.54 min-1, respectively). A small amount of light-induced autooxidation occurred as indicated by the assay containing protoporphyrinogen IX only.
Figure 3. Enzymatic conversion of protoporphyrinogen IX to protoporphyrin IX by HemG.
A. PPO assays are shown for human PPO (■), 100 nM HemG + 1 mM menadione (●), 100 nM HemG (◆), 1 mM menadione (▼), and protoporphyrinogen alone (▲). In the absence of menadione HemG is capable of generating only minimal amounts of product when compared to autooxidation. In the presence of saturating menadione concentrations, enzymatic activity is greatly increased to levels that are comparable to human PPO. B. Determination of apparent Michaelis constants for HemG. 100 nM HemG was assayed using varying concentrations of protoporphyrinogen IX (bottom curve) and menadione (top curve). Data were fitted to single rectangular hyperbolae (Eq. 2). Km and Kcat for protoporphyrinogen IX = 7.0 μM and 17.52 min-1; Km and Kcat for menadione = 3.76 μM and 16.87 min-1.
To provide additional evidence that HemG is a bonafide PPO, we carried out the menadione-dependent assay with the closest COG relative of HemG in E. coli, flavodoxin A, which cannot complement SASX38 cells, and FMN alone. Both yielded data equivalent to the rate of autooxidation seen from substrate only (results not shown). Given the close similarity of HemG and FldA, these data indicate that the PPO activity of HemG is not an artifact or side reaction catalyzed by flavodoxins.
As an additional test, we cloned, expressed and purified the long chain flavodoxin BD2899 from Bdellovibrio bacteriovorus that is currently annotated as a PPO. This protein has high homology to HemG including the long chain insert region (fig. 2B). However, this organism possesses the typical HemY PPO in an operon with other heme synthesizing enzyme genes and there exist no experimental data to support a role for this HemG-related flavodoxin in heme synthesis. Interestingly, while the insert region of the Bd. bacteriovorus protein is more similar to that found in HemG than in FldA, it lacks an acidic residue at position 136 and a methionine at position 144 that are conserved in all other HemGs. When expressed in the E. coli SASX38, this protein did not complement the hemG deletion.
HemG PPO activity was found to be specific for protoporphyrinogen. Neither mesoporphyrinogen nor coproporphyrinogen were oxidized by HemG. Kinetic parameters for HemG were determined for both protoporphyrinogen IX and menadione. These data are shown in figure 3B, and the curves were fitted to the Michaelis equation (eq. 2). The Km and Kcat for protoporphyrinogen IX were found to be 7.0 μM and 17.52 min-1 respectively, and the Km and Kcat for menadione were 3.76 μM and 16.87 min-1. The Kcat/Km for HemG was 2.5. The kinetic parameters, along with those of other well-classified PPOs are shown in table 1 (35-38). The Kcat of HemG is slightly higher than most PPO enzymes, with the exception of Desulfovibrio gigas PPO, and the Km for substrate falls within the range of other PPOs. HemG also has a high affinity for its electron acceptor, menadione. By comparison, the reported Km of mouse PPO for oxygen is 125 μM (37).
Table I.
Comparison of kinetic parameters for HemG and various PPOs
Role of the long chain insert loop
The E. coli strain SASX38 lacks functional PPO activity and was reported to be deficient in HemG, although the nature of the mutation was never identified. In the current work sequence analysis of the SASX38 hemG gene revealed a deletion from nucleotides 152 to 470. This results in a predicted HemG protein that remains in frame but is lacking 106 internal amino acids, including the long chain insert loop. This long chain loop has been previously reported as being responsible for activity, specificity, and/or recognition of redox partners (29, 30). Unique to HemG, this region is predicted to contain an alpha helix (figure 1). Since long chain insert loops have previously been implicated in specificity and function of flavodoxins, this region of HemG was examined by mutating the long chain insert loop of E. coli flavodoxin A to that of HemG. The resulting chimeric clone, FldG, was expressed in SASX38 cells deficient in HemG activity and complementation (see Supplementary Material) was obtained indicating that in vivo the chimera can contains PPO activity. Expression and purification of the FldG protein was performed as described for HemG. Spectral analysis revealed no feature at 416 nm and PPO assays of purified FldG showed no activity in the presence or absence of menadione.
Discussion
The lack of an identifiable protoporphyrinogen oxidase in most Gram-negative bacteria represents a significant gap in our current understanding of prokaryotic heme synthesis. These organisms synthesize heme and contain all other enzymes within the pathway so clearly an enzyme with PPO activity must exist. Dependence upon non-enzymatic conversion of protoporphyrinogen IX in vivo would be untenable since it would be unregulated and the accumulated highly reactive product would be toxic to the cell. Data presented herein clearly demonstrates that HemG is a previously uncharacterized form of PPO that is distinct from currently identified oxygen-dependent bacterial HemY PPOs. Unlike other known PPOs, HemG is a flavodoxin-based enzyme that employs menaquinone, rather than oxygen, as an electron acceptor. This is appropriate since E. coli is a facultative anaerobe and would be unable to synthesize heme in the absence of oxygen if it possessed a HemY type of PPO.
Due to the hydrophobicity of menaquinone and the nature of these assays, the soluble form menadione was used, which lacks the long aliphatic side chain not involved in redox properties. Our finding that HemG utilizes menadione in vitro is consistent with previous research reported by others that was done with cell extracts of E. coli (21-24). These authors found that PPO activity was respiratory chain associated (with fumarate, nitrate or oxygen as terminal electron acceptors) and the activity was localized to the membrane fraction. This location would be anticipated since the respiratory chain components, including the menaquinone-8 pool, are membrane localized. Thus, the coupled reaction will be one where the soluble HemG interacts with menaquinone at the membrane.
The proposition that a flavodoxin has evolved to gain function is not without precedent. WrbA, another protein similar to E. coli flavodoxin, possesses a quinone reductase activity much like HemG (39, 40). It appears that this protein represents an evolutionary link between flavodoxins and eukaryotic NAD(P)H/quinone oxidoreductases (41). Like HemG, it also exists as a homotetramer and the regions adjacent to the four FMN binding sites form an active site complex in the center of the quaternary structure (42, 43). Another interesting property of WrbA and other quinone reductases in general is their ability to transfer two electrons in a ping-pong mechanism in contrast to flavodoxins which do one electron reactions. This avoids the generation of semiquinone radicals that can lead to harmful reactive oxygen species (44). It will be of interest to determine if HemG also has the ability to function as a two electron transfer protein. Such an ability would decrease by half the number of turnovers required to oxidize the porphyrinogen.
The data obtained from the chimeric FldG in which the long chain insert of HemG is substituted for the analogous position of flavodoxin A presents possibly the most intriguing aspect of HemG. By replacing the long chain insert loop of E. coli flavodoxin A with the corresponding segment of HemG we were able to complement a PPO-lacking cell line. This clearly indicates a transfer of function from HemG to a flavodoxin normally only capable of transferring electrons between redox partners. No in vitro activity with the chimera could be seen in the presence of menadione, but in vivo complementation can only occur if function is present. Lack of in vitro activity probably results from the absence of a normal flavodoxin A redox partner to accept electrons during the reaction. The most compelling reason for this is that the loop region is responsible for binding and oxidation of the tetrapyrrole but does not confer reoxidation of the cofactor by quinones. From the complementation results, it is clear that FldG is still able to interact with a physiological redox partner of FldA in vivo so that it can turn over.
Computational modeling of HemG results in a predicted structure that is similar to the typical flavodoxin motif. The predicted structure generated using 3D-JIGSAW is shown in figure 4 (45). The FMN binding pocket is formed by the 50’s and 90’s loops, and the long chain insert loop extends behind this pocket presenting the possible interaction site for FMN and substrate, further supporting the role of the long chain insert loop.
Figure 4. Predicted Structure of HemG.
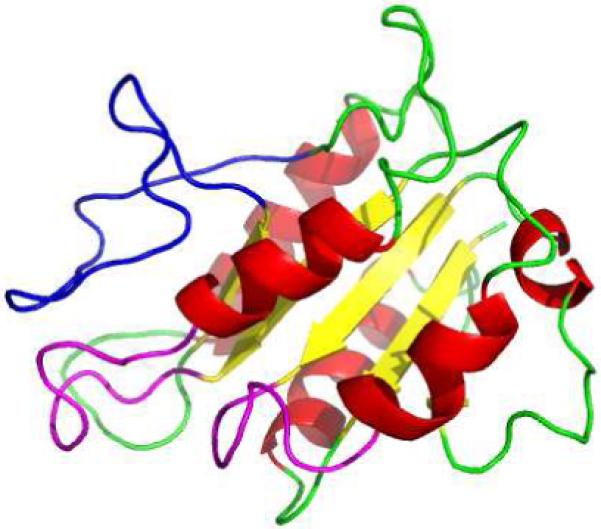
Structure was generated using 3D-JIGSAW (45). FMN-binding loops and the long chain insert loop are colored purple and blue respectively. The long chain insert loop sits above the binding pocket formed by the two loops allowing possible interaction with FMN.
Interestingly, the conversion of protoporphyrinogen IX to protoporphyrin IX appears to be an activity where Gram-negative bacteria have scavenged and adapted other proteins to do the job. Myxococcus species contain an FAD-containing, oxygen dependent enzyme essentially identical to eukaryotic PPOs (17) whereas Desulfovibrio gigas is reported to carry out catalysis using a heterotrimer complex (38) that remains to be characterized. HemG itself is only present in γ-proteobacteria and so the identity of PPO in the remaining Gram negative organisms remains unknown. Thus, HemG’s role as a protoporphyrinogen oxidase fills only a portion of the gap in our understanding of this point of the pathway.
There are many unanswered questions concerning HemG, its function, and the role of the long chain insert. Since it is a relatively small protein, it will be of interest to determine how it binds the substrates protoporphyrinogen and menaquinone and catalyzes the reaction. Ongoing studies aimed at determining the X-ray crystal structure of HemG will help to answer these questions.
Supplementary Material
Abbreviations
- PPO
protoporphyrinogen oxidase
Footnotes
This work was supported by the National Institutes of Health [DK32303 to H.A.D.].
Classification: Biochemistry, Enzymology
References
- 1.Lukin JA, Ho C. The structure--function relationship of hemoglobin in solution at atomic resolution. Chem Rev. 2004;104:1219–1230. doi: 10.1021/cr940325w. [DOI] [PubMed] [Google Scholar]
- 2.Michel H, Behr J, Harrenga A, Kannt A. Cytochrome c oxidase: structure and spectroscopy. Annu Rev Biophys Biomol Struct. 1998;27:329–356. doi: 10.1146/annurev.biophys.27.1.329. [DOI] [PubMed] [Google Scholar]
- 3.Munro AW, Girvan HM, McLean KJ, Cheesman MR, Leys D. Heme and Hemoproteins. In: Warren MJ, Smith AG, editors. Tetrapyrroles: Birth, Life and Death. Landes Bioscience and Springer Science + Business Media; 2009. pp. 160–183. [Google Scholar]
- 4.Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, Taketo MM, Yamamoto M, Igarashi K. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahara T, Sun J, Igarashi K, Taketani S. Heme-dependent up-regulation of the alpha-globin gene expression by transcriptional repressor Bach1 in erythroid cells. Biochem Biophys Res Commun. 2004;324:77–85. doi: 10.1016/j.bbrc.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 6.Kramer G, Cimadevilla JM, Hardesty B. Specificity of the protein kinase activity associated with the hemin-controlled repressor of rabbit reticulocyte. Proc Natl Acad Sci U S A. 1976;73:3078–3082. doi: 10.1073/pnas.73.9.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwai K, Drake SK, Wehr NB, Weissman AM, LaVaute T, Minato N, Klausner RD, Levine RL, Rouault TA. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc Natl Acad Sci U S A. 1998;95:4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raspe E, Duez H, Mansen A, Fontaine C, Fievet C, Fruchart JC, Vennstrom B, Staels B. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43:2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 10.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 11.Cho HY, Cho HC, Kim YM, Oh JI, Kang BS. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J Biol Chem. 2009 doi: 10.1074/jbc.M808905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medlock AE, Dailey HA. Regulation of mammalian heme biosynthesis. In: Warren MJ, Smith AG, editors. Tetrapyrroles: Birth, Life and Death. Landes Bioscience and Springer Science + Business Media; 2009. pp. 116–127. [Google Scholar]
- 14.Hift RJ, Meissner PN, Corrigall AV, Ziman MR, Petersen LA, Meissner DM, Davidson BP, Sutherland J, Dailey HA, Kirsch RE. Variegate porphyria in South Africa, 1688-1996--new developments in an old disease. S Afr Med J. 1997;87:722–731. [PubMed] [Google Scholar]
- 15.Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006;1763:723–736. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Panek H, O’Brian MR. A whole genome view of prokaryotic haem biosynthesis. Microbiology. 2002;148:2273–2282. doi: 10.1099/00221287-148-8-2273. [DOI] [PubMed] [Google Scholar]
- 17.Corradi HR, Corrigall AV, Boix E, Mohan CG, Sturrock ED, Meissner PN, Acharya KR. Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen. J Biol Chem. 2006;281:38625–38633. doi: 10.1074/jbc.M606640200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch M, Breithaupt C, Kiefersauer R, Freigang J, Huber R, Messerschmidt A. Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis. EMBO J. 2004;23:1720–1728. doi: 10.1038/sj.emboj.7600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dailey TA, Dailey HA. Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases, and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus. J Biol Chem. 1998;273:13658–13662. doi: 10.1074/jbc.273.22.13658. [DOI] [PubMed] [Google Scholar]
- 20.Dailey TA, Meissner P, Dailey HA. Expression of a cloned protoporphyrinogen oxidase. J Biol Chem. 1994;269:813–815. [PubMed] [Google Scholar]
- 21.Jacobs NJ, Jacobs JM. Quinones as hydrogen carriers for a late step in anaerobic heme biosynthesis in Escherichia coli. Biochim Biophys Acta. 1978;544:540–546. doi: 10.1016/0304-4165(78)90328-8. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs JM, Jacobs NJ. The late steps of anaerobic heme biosynthesis in E. coli: role for quinones in protoporphyrinogen oxidation. Biochem Biophys Res Commun. 1977;78:429–433. doi: 10.1016/0006-291x(77)91272-4. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs NJ, Jacobs JM. Fumarate as alternate electron acceptor for the late steps of anaerobic heme synthesis in Escherichia coli. Biochem Biophys Res Commun. 1975;65:435–441. doi: 10.1016/s0006-291x(75)80112-4. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs NJ, Jacobs JM. Nitrate, fumarate, and oxygen as electron acceptors for a late step in microbial heme synthesis. Biochim Biophys Acta. 1976;449:1–9. doi: 10.1016/0005-2728(76)90002-5. [DOI] [PubMed] [Google Scholar]
- 25.Hastings SF, Kaysser TM, Jiang F, Salerno JC, Gennis RB, Ingledew WJ. Identification of a stable semiquinone intermediate in the purified and membrane bound ubiquinol oxidase-cytochrome bd from Escherichia coli. Eur J Biochem. 1998;255:317–323. doi: 10.1046/j.1432-1327.1998.2550317.x. [DOI] [PubMed] [Google Scholar]
- 26.Wissenbach U, Kroger A, Unden G. The specific functions of menaquinone and demethylmenaquinone in anaerobic respiration with fumarate, dimethylsulfoxide, trimethylamine N-oxide and nitrate by Escherichia coli. Arch Microbiol. 1990;154:60–66. doi: 10.1007/BF00249179. [DOI] [PubMed] [Google Scholar]
- 27.Sasarman A, Chartrand P, Lavoie M, Tardif D, Proschek R, Lapointe C. Mapping of a new hem gene in Escherichia coli K12. J Gen Microbiol. 1979;113:297–303. doi: 10.1099/00221287-113-2-297. [DOI] [PubMed] [Google Scholar]
- 28.Sasarman A, Letowski J, Czaika G, Ramirez V, Nead MA, Jacobs JM, Morais R. Nucleotide sequence of the hemG gene involved in the protoporphyrinogen oxidase activity of Escherichia coli K12. Can J Microbiol. 1993;39:1155–1161. doi: 10.1139/m93-174. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Llano J, Maldonado S, Bueno M, Lostao A, Angeles-Jimenez M, Lillo MP, Sancho J. The long and short flavodoxins: I. The role of the differentiating loop in apoflavodoxin structure and FMN binding. J Biol Chem. 2004;279:47177–47183. doi: 10.1074/jbc.M405792200. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Llano J, Maldonado S, Jain S, Lostao A, Godoy-Ruiz R, Sanchez-Ruiz JM, Cortijo M, Fernandez-Recio J, Sancho J. The long and short flavodoxins: II. The role of the differentiating loop in apoflavodoxin stability and folding mechanism. J Biol Chem. 2004;279:47184–47191. doi: 10.1074/jbc.M405791200. [DOI] [PubMed] [Google Scholar]
- 31.Dailey HA. Terminal steps of haem biosynthesis. Biochem Soc Trans. 2002;30:590–595. doi: 10.1042/bst0300590. [DOI] [PubMed] [Google Scholar]
- 32.Shepherd M, Dailey HA. A continuous fluorimetric assay for protoporphyrinogen oxidase by monitoring porphyrin accumulation. Anal Biochem. 2005;344:115–121. doi: 10.1016/j.ab.2005.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steensma E, Heering HA, Hagen WR, Van Mierlo CP. Redox properties of wild-type, Cys69Ala, and Cys69Ser Azotobacter vinelandii flavodoxin II as measured by cyclic voltammetry and EPR spectroscopy. Eur J Biochem. 1996;235:167–172. doi: 10.1111/j.1432-1033.1996.00167.x. [DOI] [PubMed] [Google Scholar]
- 35.Corrigall AV, Siziba KB, Maneli MH, Shephard EG, Ziman M, Dailey TA, Dailey HA, Kirsch RE, Meissner PN. Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus subtilis. Arch Biochem Biophys. 1998;358:251–256. doi: 10.1006/abbi.1998.0834. [DOI] [PubMed] [Google Scholar]
- 36.Dailey TA, Dailey HA. Human protoporphyrinogen oxidase: expression, purification, and characterization of the cloned enzyme. Protein Sci. 1996;5:98–105. doi: 10.1002/pro.5560050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira GC, Dailey HA. Mouse protoporphyrinogen oxidase. Kinetic parameters and demonstration of inhibition by bilirubin. Biochem J. 1988;250:597–603. doi: 10.1042/bj2500597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klemm DJ, Barton LL. Purification and properties of protoporphyrinogen oxidase from an anaerobic bacterium, Desulfovibrio gigas. J Bacteriol. 1987;169:5209–5215. doi: 10.1128/jb.169.11.5209-5215.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grandori R, Khalifah P, Boice JA, Fairman R, Giovanielli K, Carey J. Biochemical characterization of WrbA, founding member of a new family of multimeric flavodoxin-like proteins. J Biol Chem. 1998;273:20960–20966. doi: 10.1074/jbc.273.33.20960. [DOI] [PubMed] [Google Scholar]
- 40.Patridge EV, Ferry JG. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J Bacteriol. 2006;188:3498–3506. doi: 10.1128/JB.188.10.3498-3506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carey J, Brynda J, Wolfova J, Grandori R, Gustavsson T, Ettrich R, Smatanova IK. WrbA bridges bacterial flavodoxins and eukaryotic NAD(P)H:quinone oxidoreductases. Protein Sci. 2007;16:2301–2305. doi: 10.1110/ps.073018907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrade SL, Patridge EV, Ferry JG, Einsle O. Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli. J Bacteriol. 2007;189:9101–9107. doi: 10.1128/JB.01336-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorman J, Shapiro L. Crystal structures of the tryptophan repressor binding protein WrbA and complexes with flavin mononucleotide. Protein Sci. 2005;14:3004–3012. doi: 10.1110/ps.051680805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deller S, Macheroux P, Sollner S. Flavin-dependent quinone reductases. Cell Mol Life Sci. 2008;65:141–160. doi: 10.1007/s00018-007-7300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Contreras-Moreira B, Bates PA. Domain fishing: a first step in protein comparative modelling. Bioinformatics. 2002;18:1141–1142. doi: 10.1093/bioinformatics/18.8.1141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.