SUMMARY
The Engrailed homeodomain protein is an ‘active’ or dominant transcriptional repressor in cultured cells. In contrast, the Fushi Tarazu homeodomain protein is an activator, both in cultured cells and in Drosophila embryos, where it activates several known target genes, including its own gene. This auto-activation has been shown to depend on targeting to a fushi tarazu enhancer by the Fushi Tarazu homeodomain. We combined Fushi Tarazu targeting and Engrailed active repression in a chimeric regulator, EFE. When EFE is ubiquitously expressed, it overrides endogenous Fushi Tarazu and causes a fushi tarazu mutant phenotype. Normal Fushi Tarazu target genes are affected as they are in fushi tarazu mutants. One such target gene is repressed by EFE even where Fushi Tarazu is not expressed, suggesting that the repression is active. This is confirmed by showing that the in vivo activity of EFE depends on a domain that is required for active repression in culture. A derivative that lacks this domain, while it cannot repress the endogenous fushi tarazu gene, can still reduce the activity of the fushi tarazu autoregulatory enhancer, suggesting that it competes with endogenous Fushi Tarazu for binding sites in vivo. However, this passive repression is much less effective than active repression.
Keywords: engrailed, fushi tarazu, active repression, Drosophila embryogenesis, transcriptional regulation, homeodomain
INTRODUCTION
The creation of dominant negative molecules is a powerful tool for investigating function in vivo, as it can mimic the loss of function of a regulator (Herskowitz, 1987). This is particularly useful under circumstances where specific mutations are unavailable or difficult to obtain. Dominant negative effects can be generated in a number of ways, including the introduction of altered dimerization partners to disrupt function at the protein level (e.g., see Amaya et al., 1991), and the use of antisense nucleic acids (Erickson, 1993; Wagner, 1994) and ribozymes (Uhlenbeck, 1987; Zhao and Pick, 1993) to disrupt expression of specific messenger RNAs. The recent characterization of dominant or ‘active’ transcriptional repressors (see below) provides a potential new approach to generating dominant negative regulators that prevent transcription of specific sets of genes.
A wealth of information about the regulatory interactions that govern early embryogenesis has been obtained in recent years from studies in Drosophila. Not only does this detailed information give us a window into the mechanisms of development, it provides fertile ground for testing the molecular actions of regulatory proteins of various kinds. In particular, the segmentation gene hierarchy, which involves a cascading series of interactions that sequentially subdivide the embryo into patterns of transcription of regulatory genes, provides a useful backdrop for the study of transcriptional mechanisms in vivo. In this work, we use this system to test the action of an engineered active repressor as a dominant negative molecule in vivo.
The product of the Drosophila engrailed (en) locus, En, is a homeodomain-containing protein (HD protein) that acts in much the same manner as the homeotic gene products. It is required for cell fate specification throughout development in the progeny of cells that first express it, and without it, these cell fates are transformed toward those of cells in which it is normally off (Kornberg et al., 1985; Kornberg, 1981; Lawrence and Morata, 1976; Morata et al., 1983; Nüsslein-Volhard and Wieschaus, 1980). From genetic and molecular studies, En is known to activate some genes and to repress others (Eaton and Kornberg, 1990; Harding et al., 1986; Heemskerk et al., 1991; Hooper and Scott, 1989; Tabata et al., 1992), but no direct interactions have yet been established.
En is an active repressor of transcription in cultured cells (Jaynes and O’Farrell, 1991). That is, it is capable of repressing both basal transcription and transcription enhanced by a variety of activators in a manner that requires En-binding sites in the target gene, but does not require competition for activator-binding sites (Jaynes and O’Farrell, 1991; Han and Manley, 1993a). An increasing number of transcription factors are known to possess similar functional properties (Biggin and Tjian, 1989; Forrest et al., 1990; Han and Manley, 1993b; Ip et al., 1991; Johnson and Herskowitz, 1985; Johnson and Krasnow, 1992; Keleher et al., 1992; Licht et al., 1990; Madden et al., 1991; Saha et al., 1993; Sap et al., 1989; Sauer and Jäckle, 1991; Zuo et al., 1991).
The homeodomain (HD) is a sequence-specific DNA-binding domain (Desplan et al., 1988; Ekker et al., 1991; Hoey and Levine, 1988) that is responsible for directing proteins to their appropriate target genes in vivo. This is apparently true even for proteins with closely related HDs that bind to virtually identical sites in vitro. For example, HD swap experiments have shown that the Deformed and Ultrabithorax HDs interact with distinct sets of target genes in vivo (Kuziora and McGinnis, 1989), even though they have identical ‘recognition helices’ that make most of the sequence-specific DNA contacts (Kissinger et al., 1990). In addition, the slight differences that have been found in their sequence preference cannot account for their target discrimination in vivo (Lin and McGinnis, 1992). Thus even closely related HDs can effectively distinguish between target genes, probably due to interactions with other DNA-binding proteins.
Like En, Fushi Tarazu (Ftz) is a HD protein that acts in the segmentation hierarchy (Ingham, 1988). It is expressed in a pair-rule pattern of seven stripes (Carroll et al., 1988) that, in combination with other pair-rule gene products, provides for the proper expression of the segment polarity genes, such as en. (DiNardo and Farrell, 1987, Kassis, 1990). Unlike En, Ftz is a strong activator of transcription in cultured cells (Jaynes and O’Farrell, 1988; Winslow et al., 1989). Consistent with this activity, Ftz activates its own gene in embryos (Hiromi and Gehring, 1987; Pick et al., 1990), apparently through a direct interaction with binding sites in an upstream enhancer (Schier and Gehring, 1992). Ftz is also required to activate the even-numbered en stripes (DiNardo et al., 1988), which lie within its domain of expression (Lawrence et al., 1987), an interaction that is also thought to be direct (Kassis, 1990). In addition, Ftz has a role in limiting wingless (wg) expression. In the absence of Ftz, wg is initiated in stripes that are much broader than normal, extending throughout the normal Ftz domains (Ingham et al., 1988). It is not known whether Ftz interacts directly with the wg gene. These defects in en and wg expression contribute to the pair-rule pattern deletions observed in the cuticles of ftz-mutant embryos (DiNardo and O’Farrell, 1987; Frasch and Levine, 1987; Ingham et al., 1988).
When Ftz is expressed ubiquitously in early embryos from a heat-shock promoter, it causes a phenotype consistent with its normal autoregulatory activity: it expands the domain of endogenous ftz expression, causing expanded en expression in the same cells, and concomitant loss of half of the wg stripes (Ish-Horowicz et al., 1989). The outcome at the end of embryogenesis is a pair-rule cuticular phenotype approximately complementary to that of a ftz mutant (Struhl, 1985). This activity of Ftz can be seen even when its HD has been partially deleted (Fitzpatrick et al., 1992). The effects of ectopic En expression have been previously analyzed only later in embryogenesis, after endogenous en expression has begun. The effects are somewhat similar to those produced at earlier times by ectopic Ftz. En stripes are expanded and wg stripes are repressed, but in this case, all the stripes are affected (Heemskerk et al., 1991).
In this study, we use a HD swap between Ftz and En to uncouple the targeting functions of these proteins from their transcriptional activities of activation and repression, allowing these properties to be independently characterized. When the En HD is replaced by the HD of Ftz, the resulting chimera, EFE, possesses the repression activity of En, as assayed in cultured cells (Jaynes and O’Farrell, 1991). This is consistent with studies that have mapped En repression function to domains outside the HD (Han and Manley, 1993a; Jaynes and O’Farrell, 1991). Although previous work has shown that the Ftz and En HDs bind to similar sequences in vitro (Desplan et al., 1988; Hoey and Levine, 1988), and, indeed, can act through the same consensus binding sites in cultured cells (Jaynes and O’Farrell, 1988), the present study shows that EFE represses normal Ftz target genes in vivo, and that these are distinct from the genes repressed by En. Thus the Ftz HD, in the context of EFE, is apparently sufficient for specifying these targets. The repression activity of EFE in vivo is also shown to depend on an En domain that mediates active repression in cultured cells. Without this active repression domain, the Ftz HD can apparently still compete with endogenous Ftz for binding sites in vivo, but this passive repression is much less effective than active repression. The combined activities of Ftz targeting and En active repression create a potent dominant negative molecule that efficiently mimics a ftz mutant phenotype in vivo.
MATERIALS AND METHODS
Embryo preparation and staining
P-element transformations (Spradling and Rubin, 1982), cuticle preparations (Wieschaus and Nüsslein-Volhard, 1986), and in situ hybridization to fixed embryos (Edgar and O’Farrell, 1990) were performed essentially as described previously. Antibody staining was performed essentially as described (Manoukian and Krause, 1992) using a polyclonal α-En antisera, a kind gift of Charles Girdham and Patrick O’Farrell, that had been prepared against full-length, partially purified GST-tagged En and affinity purified against a His-tagged peptide with the N-terminal 150 amino acids (aa) of En. An alkaline phosphatase coupled secondary antibody (Vector Laboratories) was used both for microscopic examination of fixed embryos, where the BCIP and NBT substrates were used for staining (Boehringer Mannheim), and for quantitation of antibody signals, where p-nitrophenyl phosphate (Sigma) was used as described (Manoukian and Krause, 1992). This substrate was also used to quantify in situ hybridization signals in Fig. 9C. (In situ hybridization utilized an alkaline phosphatase coupled antibody, Boehringer Mannheim, against a DGG-labeled probe.) Incubation times were determined to be in the linear range of the assay by incubating sets of embryos with different signal intensities for various times.
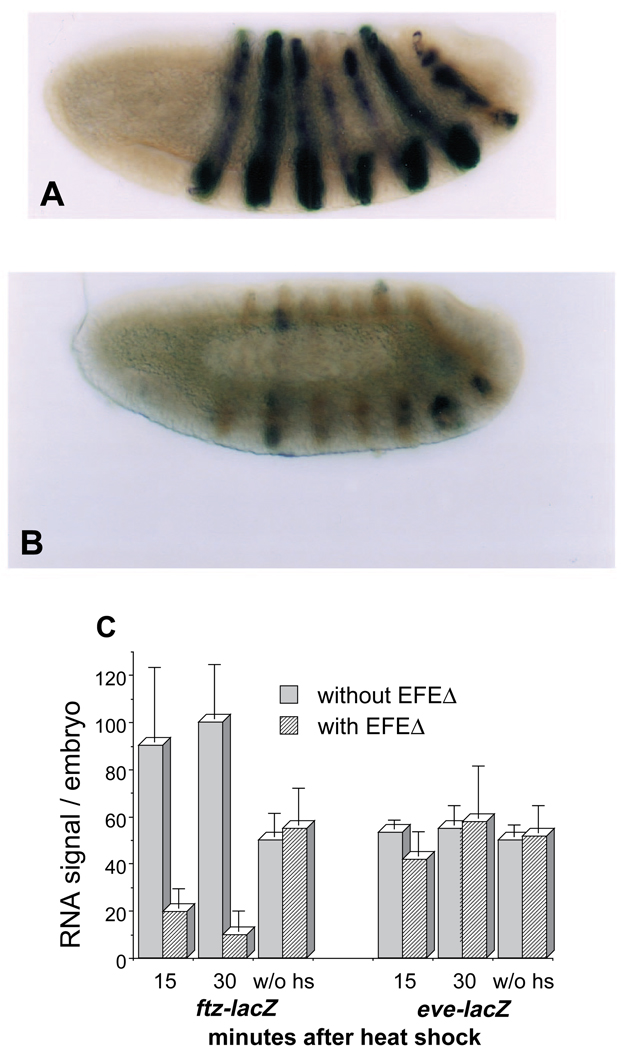
Fig. 9.
(A,B) EFEΔ inhibits the activity of a ftz enhancer in vivo. Embryos carrying a ftz-lacZ transgene, and either (A) lacking or (B) containing the EFEΔ transgene, were double-stained for lacZ RNA (black), as in Fig. 2 except using a lacZ probe, and endogenous Ftz protein (brown), using an α-Ftz monoclonal antibody (kindly provided by Ian Duncan) that does not bind the Ftz HD and therefore does not cross-react with EFEΔ. Embryos were fixed 30 minutes after a 30 minute heat shock beginning between 2.5 and 3 hours of development (30 minute collections). Although the ftz-lacZ transgene gives a range of staining intensities in different embryos, there was a clear shift in the entire range due to repression by EFEΔ, most clearly discernible as a lack of embryos with strong, mostly complete stripes like that shown in A. (C) Quantitation of ftz enhancer activity. Embryos were probed for lacZ RNA expressed from the ftz enhancer, or, as a control, from an eve enhancer (8.0 eve-lacZ of Harding et al., 1989), as in A and B (fixed either 15 or 30 minutes after heat shock, as indicated), and the signal was quantified using a soluble chromogenic substrate, as described in Materials and Methods. A background from non-lacZ RNA-producing embryos processed in parallel was subtracted from each of the values presented. The graph represents the average and range of two separate experiments for the heat-shock-induced values. For the non-heat-shocked values, 2 sets of embryos were collected for each transgenic line at each of the two stages used for the heat-shocked values, and processed in parallel; the values shown are the average and range of these 4 determinations.
Heat shocks were administered to embryos on 35 mm collection plates by floating the plates on 37°C water inside a sealed container, in order to minimize evaporative cooling. Standard heat-shock conditions employed a 15 minute incubation followed by return to a 25°C humidified environment.
Transfections
Cell culture assays for passive and active repression were performed as described (Jaynes and O’Farrell, 1991), using 2 µg per 60 mm culture dish of one of two target genes: T3N6D-33CatA and N6T3D-33CATB (Jaynes and O’Farrell, 1991). Active repression assays with each of these target genes gave qualitatively similar results. The quantitative values quoted in the legend to Fig. 8 were from transfections using the former plasmid, where values were averages from at least two separate transfections, normalized to the activity of a co-transfected reference gene (Jaynes and O’Farrell, 1988; Jaynes et al., 1990). In each case, the range of values was less than 20% of the average. For active repression assays, 0.04 µg of pPAc-GR (Yoshinaga et al., 1993) was used to express the rat glucocorticoid receptor. For passive repression assays, 0.3 µg Ftz expression plasmid pPAc-ftz (Jaynes and O’Farrell, 1988; Winslow et al., 1989) was used. CAT assays, as well as β-galactosidase assays for expression of the co-transfected reference gene pLac82SU (Dorsett et al., 1989), were performed as described (Jaynes and O’Farrell, 1991). Co-transfected plasmids used to express EFE and its derivatives were the same as those used for P-element transformation. Transfections typically used 5 µg of these constructs. Exceptions are noted in the text.
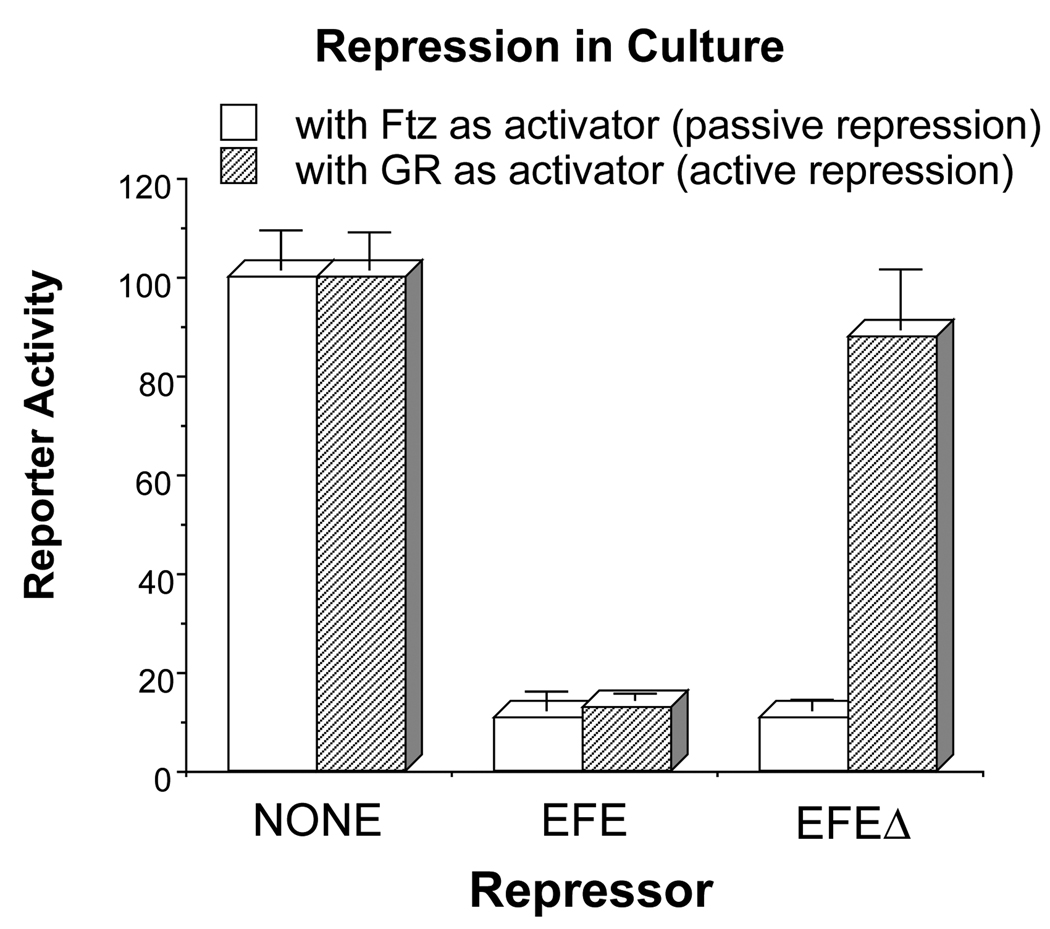
Fig. 8.
EFEΔ can compete for Ftz-binding sites in cultured cells, but cannot actively repress transcription. Drosophila S2 cells were co-transfected with a CAT (chloramphenicol acetyltransferase) reporter plasmid, which contains binding sites for both the glucocorticoid receptor (GR) and the Ftz HD, separated by 40 bp, upstream of a basal promoter, and a plasmid that expresses either Ftz or GR (see Materials and Methods for details). Each of the latter activate reporter expression 50- to 100-fold above the basal level. The ability of either EFE or EFEΔ to repress this activated transcription was determined by co-transfection of an appropriate expression plasmid. The non-repressed level was determined by co-transfection of the same quantity of ‘empty’ parental expression plasmid, which is a P-element transformation vector (see Materials and Methods). CAT activity was determined and normalized to the activity of a co-transfected reference gene (see Materials and Methods for details). The graph represents the average and range of at least 4 independent transfections with 2 different plasmid preparations, in at least 2 separate experiments.
Plasmid constructions and Drosophila strains
To construct the EFE transformation vector, pRK232 (Heemskerk et al., 1991) containing the hsp70 promoter upstream of the wild-type en coding region (Poole et al., 1985) was cut at two unique restriction sites (MluI and BstEII) flanking the En HD. The En HD containing fragment was replaced with the corresponding fragment from pAc-EnFtzHD (Jaynes and O’Farrell, 1991), which contains the 60 aa Ftz HD coding sequence in place of the 60 aa En HD coding region. This plasmid, phs-EFE, was modified using unique restriction sites within the En coding region to yield the deletion derivative phs-EFEΔ, removing codons 169–399. These plasmids were introduced into flies using standard methodologies (Spradling and Rubin, 1982). Homozygous viable insertions on either the second or third chromosome were used in all the analyses. A Drosophila strain carrying a ftz-lacZ transgene on the second chromosome (‘5′B’ of Pick et al., 1990; kindly provided by L. Pick) was crossed with a strain carrying phs-EFEΔ on the third, and appropriate progeny were crossed to give flies that both carried ftz-lacZ and were homozygous for the phs-EFEΔ transgene. A strain carrying an eve-lacZ transgene on the balancer chromosome SM6B (Panzer et al., 1992) was analogously manipulated. These were used in the analysis presented in Fig. 9.
RESULTS
Replacing the En HD with that of Ftz results in a predictable change in regulatory specificity
A chimeric En protein containing an exact replacement of its 60 aa HD with that of Ftz was previously tested in cell culture assays for transcriptional activity, and was found to be an active repressor (Jaynes and O’Farrell, 1991). The corresponding protein coding region was placed in a P-element transformation vector under heat-shock promoter control, and transgenic lines were established. The effect of ectopic production of the chimeric protein EFE was compared to that of the two parental proteins, En and Ftz. Ftz is a transcriptional activator as assayed in cultured Drosophila cells (Jaynes and O’Farrell, 1988), and has been implicated in a positive feedback loop in early embryos (Hiromi and Gehring, 1987; Pick et al., 1990), apparently acting directly through an enhancer in its own gene (Schier and Gehring, 1992). As described previously (Struhl, 1985), when Ftz is ectopically expressed between 2.5 and 3.5 hours of development, it causes pair-rule deletions in the pattern of cuticular structures formed at the end of embryogenesis that are approximately complementary to those caused by lack of Ftz (Fig. 1B). In contrast, ectopic EFE production between 2.5 and 3 hours mimics the effects of a ftz mutant (Wakimoto et al., 1984), causing pattern deletions predominantly in the ftz-dependent parasegments (Fig. 1C; see below and Fig. 1 legend for details). The effects of ectopic En production have been analyzed previously only in older embryos, after 3 hours of development, when it causes pattern defects in each parasegment (Heemskerk et al., 1991). At earlier times, between 2.5 and 3 hours, it also causes pre dominantly segment polarity-type pattern defects, although occasionally it produces pattern deletions that are close to pair-rule (not shown). These usually take the form of deletions of naked cuticle (Fig. 1D), and are distinct from the defects seen at high frequency in EFE embryos. The mimic of a ftz mutant phenotype by ectopic EFE is consistent with the combination in the EFE molecule of Ftz targeting activity and En dominant repression activity.
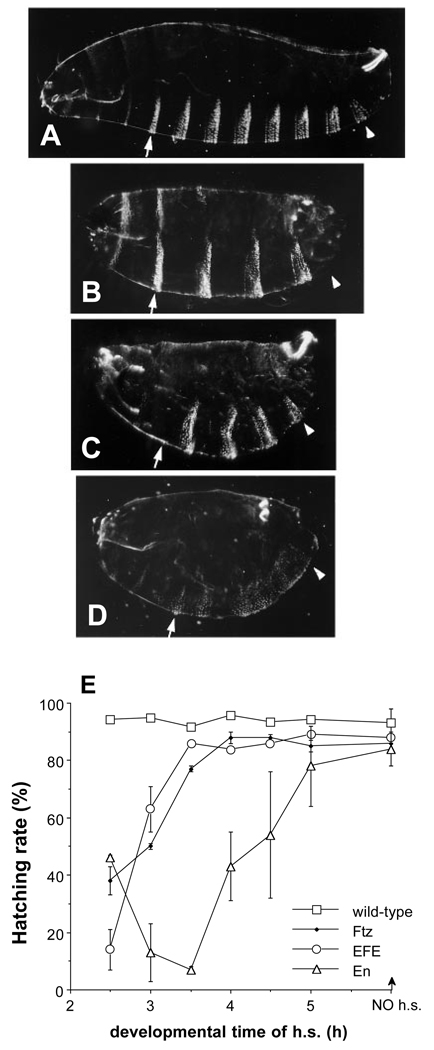
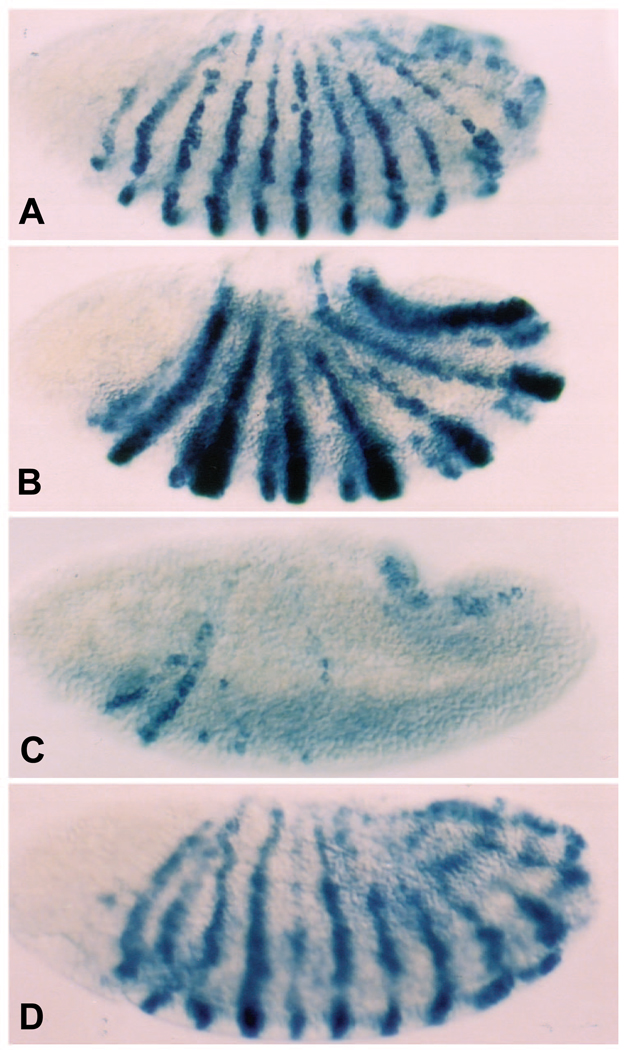
Fig. 1.
(A–D) Cuticular pattern deletions caused by EFE and its parental proteins. Cuticle patterns in (A) wild-type embryos, or 20 hours after transient induction of either (B) Ftz, (C) EFE, or (D) En from heat-inducible transgenes. Embryos were subjected to a 15 minute heat pulse at 37°C (see Materials and Methods) beginning 2.5 hours after the end of a 30 minute egg collection period. Ftz caused deletion of the even-numbered abdominal denticle bands, as well as the first and third thoracic bands, while EFE-induced deletions were essentially complementary. The positions of the A1 (arrow) and A8 (arrowhead) denticle bands are indicated. The EFE pattern is very similar to that of ftz mutants (Wakimoto et al. 1984). EFE pair-rule embryos retain ventral pits posterior to the T3 denticle band but usually lose the Keilin’s organs, which lie just posterior, suggesting that the register of the deletions is very similar, if not identical, to that of ftz mutants (Struhl, 1985). More than 70% of EFE embryos had preferential deletions of ftz-dependent parts of the ventral denticle bands, including the following categories: pair-rule deletions of ftz-dependent bands, 20–30%; deletions of a subset of the ftz-dependent bands, 20–30%; and deletions of all of the ftz-dependent bands plus some additional bands, 15–30%. 10–20% of these cuticles showed normal patterns, most likely due to embryos that were partially developed at the time of egg laying, and therefore escaped the effects of EFE (see part E). In contrast, En caused extensive disorganization of repeating pattern, including polarity reversals. All embryos in all figures are oriented ventral down and anterior to the left. (E) Ectopically expressed EFE causes defects within a time window similar to that of Ftz, but different from that of En. Hatching rates were determined (after 40 hours) following a 15 minute heat pulse beginning at the indicated time after the end of a 30 minute egg collection. The indicated protein was ectopically expressed from a heat-inducible transgene. At least 3 independently heat-shocked plates with at least 100 eggs each were counted (both unhatched eggs and empty egg casings) for each data point. The graph indicates the average and range of the data obtained. Normal embryos hatch after about 24 hours.
As described in Fig. 1, not all of the EFE embryos show simple pair-rule pattern deletions. The most commonly seen less severe phenotype was the fusion of pairs of denticle bands, most often at the edges, with a loss of naked cuticle anterior to the ftz-dependent bands. This is the region most sensitive to loss in response to ubiquitously expressed Ftz protein (Struhl, 1985), as well as in en mutants (Nüsslein-Volhard and Wieschaus, 1980), and probably reflects the loss of wg expression anterior to the even-numbered en stripes. At a lower frequency, we also observed partial loss of ftz-dependent denticle bands without loss of naked cuticle. In an attempt to determine whether a distinct time window exists for these distinct pattern deletions, we performed shorter collections of 15 minutes. In collections corresponding to the earlier half of the previous collections, we observed a predominance of denticle band-only deletions, while in collections corresponding to the later half of previous collections, we observed an increased frequency of denticle band fusions. This suggests that the ftz-dependent denticle bands are most sensitive to a loss of Ftz between 2½ and 2¾ hours of development, while the anteriorly adjacent naked cuticle becomes most sensitive 15 minutes later, which may reflect a temporal order in patterning events within the ftz domain (see Discussion). The earlier collections also contained a higher proportion of more severely affected embryos, suggesting that the odd-numbered parasegments are more sensitive to disruption by EFE at earlier times. Deletions of either non-ftz-dependent denticle bands or additional naked cuticle were observed, usually in different embryos. Although this suggests that there may be a temporal order of sensitivity to the effects of EFE in different non-ftz-dependent portions of severely affected embryos, we have not attempted to further characterize these effects.
The time window during development in which EFE can induce defects was also assessed by determining hatching rates. Both Ftz and EFE, when ectopically expressed, are most effective at producing lethal defects early in development (Fig. 1E). The analysis was not extended to earlier times, since earlier heat shock causes significant lethality in wild-type embryos (not shown). The effectiveness of Ftz and EFE falls off rapidly after 3 hours of development. In contrast, En is more effective later, with a peak around 3.5 hours, and the window extends to much later times. Thus the activity of En appears to be significantly altered when its HD is replaced by that of Ftz. This conclusion is confirmed by results described below.
EFE represses Ftz-activated genes in the embryo
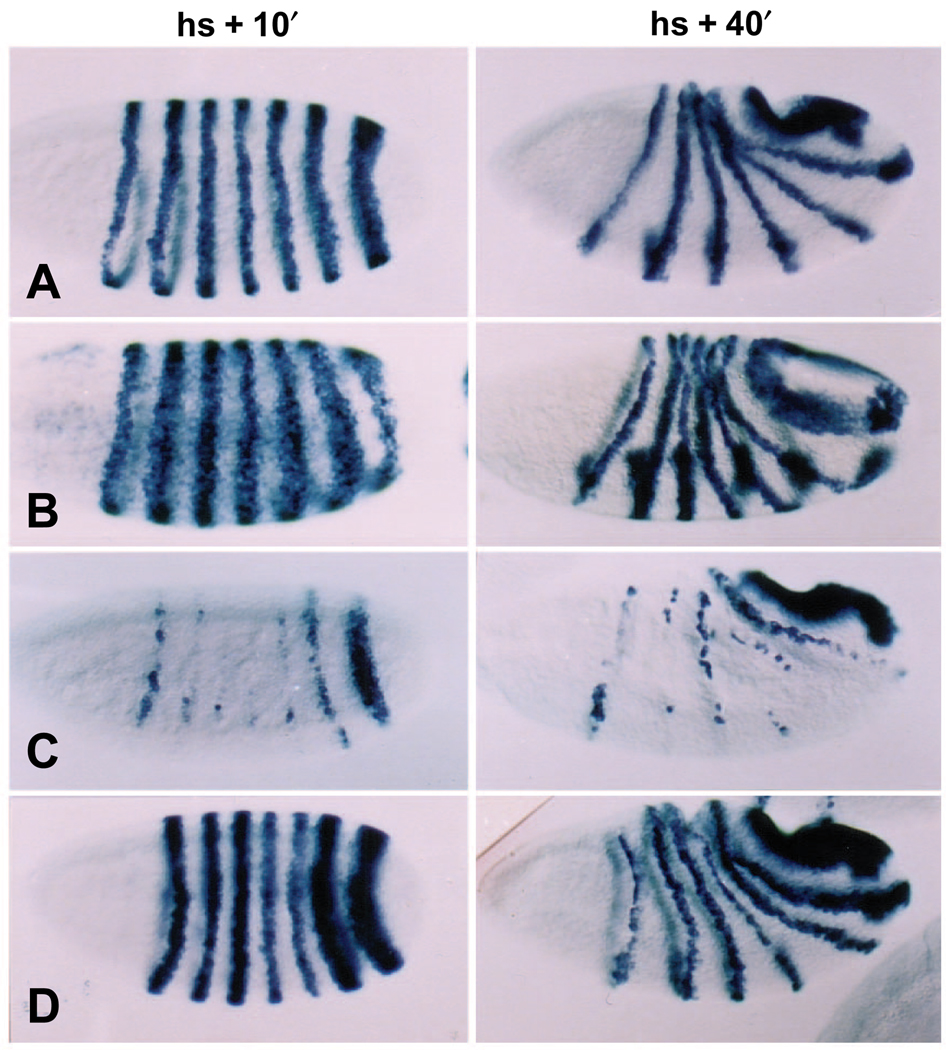
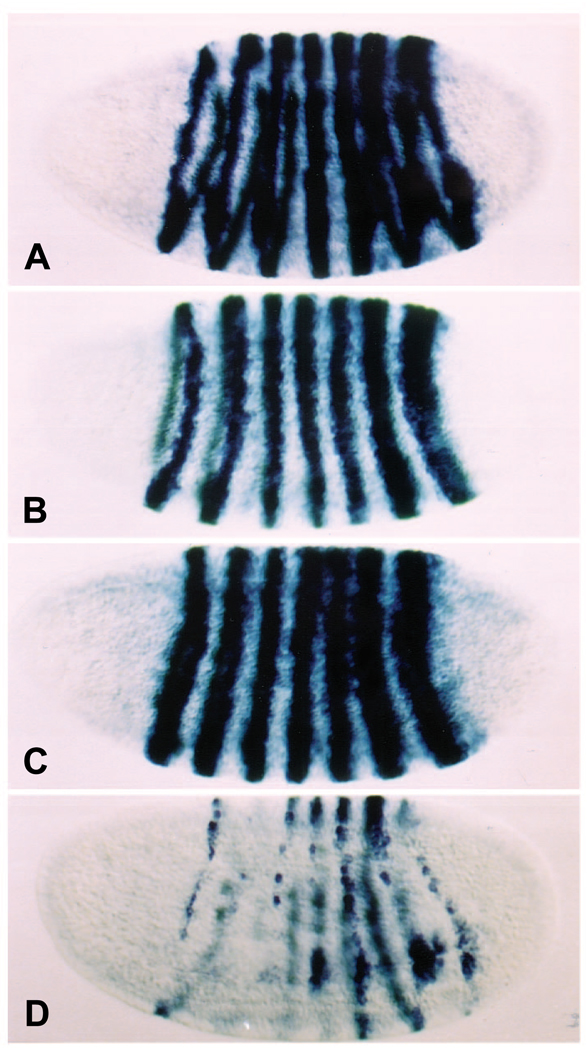
Ectopic EFE production causes a rapid loss of endogenous ftz gene expression. Following a 15 minute heat pulse between 2.5 and 3 hours of development (the ‘standard heat shock’), at a time when ftz expression has been established in 7 broad, sharply defined stripes, hs-EFE embryos fixed within 10 minutes of the end of the heat shock show a dramatic reduction in ftz RNA levels (Fig. 2C). In contrast, wild-type embryos heat shocked in parallel show normal ftz expression, consistent with the fact that such embryos develop normally. This rapid loss of ftz expression suggests that the endogenous ftz gene is repressed directly by EFE. This activity depends on the Ftz HD, since hs-En embryos show only slight repression of ftz stripes 4 and/or 5, and only in some embryos (Fig. 2D). This activity is also distinct from that of ectopic Ftz, which, as has been shown previously (Ish-Horowicz et al., 1989), expands late ftz stripes (Fig. 2B). Following global repression by EFE, ftz expression partially recovers in a characteristic pattern (Fig. 2C, ‘hs + 40min.’), with stripe 7 recovering completely, stripe 6 recovering in a narrow stripe, with some loss ventrally, stripe 3 recovering only dorsally, stripe 1 recovering ventrally (and variably), and the other stripes recovering only patchy expression, mostly dorsally.
Fig. 2.
ftz gene expression is repressed strongly by EFE, but not by either parental protein. The pattern of ftz RNA expression was monitored in embryos at two times after induction (as in Fig. 1) of (A) no transgene, (B) hs-Ftz, (C) hs-EFE or (D) hs-En. Embryos were fixed either 10 minutes (column 1) or 40 minutes (column 2) after induction, and stained by in situ hybridization (as described in Materials and Methods) using a probe to ftz RNA sequences outside the HD. The apparent background staining in B, column 1, is due at least in part to cross-reaction of the probe with ftz RNA sequences expressed from the Ftz transgene.
The pattern of ftz repression is distinct from that observed previously in response to ablation of ftz RNA using an antisense ribozyme construct (Zhao et al., 1993), in which stripes 3 and 7 were relatively sensitive to repression. This distinction between the patterns may indicate that the mechanism of repression by EFE is not solely a passive competition for endogenous Ftz sites, which might be expected to result in a pattern of repression more similar to that caused by ablation of ftz RNA.
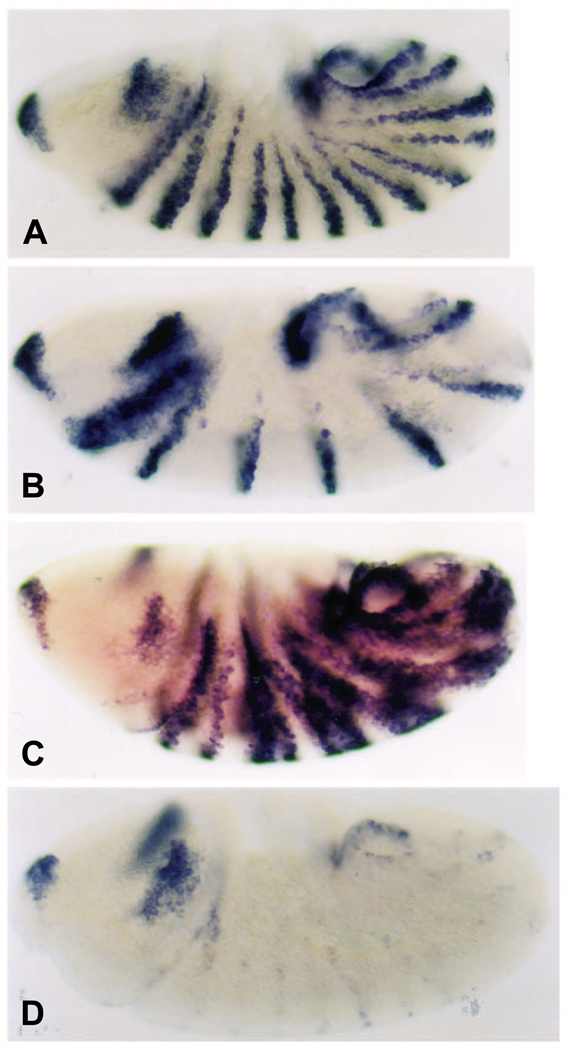
In addition to repressing ftz, EFE prevents activation of the engrailed gene (en), a Ftz target gene that is immediately downstream in the segmentation gene hierarchy. Not only does EFE repress the en stripes that are ftz dependent, the even-numbered stripes, but it also prevents activation of the odd-numbered en stripes, which lie outside the ftz domain of expression (Fig. 3C). Although this effect could be indirect, it suggests that EFE may repress en not only by repressing endogenous ftz, but also directly, in regions where en is activated by factors other than Ftz. This is a second indication that EFE may be acting as an active repressor in vivo, consistent with its transcriptional activity in cultured cells. In contrast, Ftz expands the even-numbered en stripes (Fig. 3B), as previously described (Ish-Horowicz et al., 1989), while En has little effect (Fig. 3D).
Fig. 3.
EFE represses both even- and odd-numbered en stripes. The patterns of en RNA were detected (as in Fig. 2, except using an en probe) in embryos 40 minutes following induction (as in Fig. 1) of either (A) no transgene, (B) hs-Ftz, (C) hs-EFE, or (D) hs-En. Ubiquitous expression from both the En- and EFE-producing transgenes was observed at earlier times after induction, with RNA from the En transgene persisting somewhat longer (not shown). Note that both the even- and odd-numbered en stripes are repressed by EFE.
The HD swap causes a re-targeting of repression activity
In addition to conferring on EFE the ability to repress the ftz gene, the HD swap directs En repression activity away from the eve gene. When En is ectopically expressed using standard heat-shock conditions, endogenous eve stripe expression is rapidly lost. In contrast, neither wild-type, hs-ftz, nor hs-EFE embryos show this loss (Fig. 4). These results are consistent with eve being a direct target of En, a possibility consistent also with the previous observation that eve expression persists longer than normal in en mutant embryos (Harding et al., 1986), in those cells that usually express En. Repression of eve at this time in development, somewhat surprisingly, does not cause a loss of odd-numbered en stripes, which are normally sensitive to loss of eve function (DiNardo et al., 1987), although we do see a reduction in intensity in these stripes in many hs-En embryos (e.g., stripe 5 in Fig. 3D). This result is consistent with the recent finding that only early eve expression is absolutely required for activation of these en stripes, although later eve expression enhances their activation (M. Fujioka, J. Jaynes, and T. Goto, submitted). This, and the fact that EFE is ineffective at repressing eve, implies that the loss of odd-numbered en stripes in hs-EFE embryos cannot be explained by a loss of eve expression, adding weight to the possibility that this loss is due to direct, active repression of en by EFE.
Fig. 4.
eve expression is repressed by ectopic En, but not by EFE. The patterns of eve RNA were detected (as in Fig. 2, except using an eve probe) in embryos 20 minutes after heat induction (as in Fig. 1) of either (A) no transgene, (B) hs-Ftz, (C) hs-EFE, or (D) hs-En. Ftz and EFE had little or no effect. En repressed all eve stripes, with stripes 5, 6, and 1 being least affected. Thus En repression activity is re-targeted away from the eve gene by the HD swap.
The distinct regulatory specificities of EFE and En are further seen in their divergent effects on wingless (wg) expression. When ectopically induced, Ftz represses wg in odd-numbered parasegments (Fig. 5B), while En represses all of the wg stripes (Fig. 5D). In contrast, EFE has an effect similar to that of a ftz mutant: wg is derepressed, often throughout the ftz domain (Fig. 5C). This occurs quite rapidly, with ectopic expression appearing within 20 minutes of the end of a standard (15 minute) heat shock. This is roughly coincident with the normal time of induction of wg expression in its 14-stripe pattern within the segmented region of the trunk. However, it is delayed enough following EFE production that it could be an indirect effect due to repression of a wg repressor. This possibility is consistent with indirect regulation of wg by Ftz, via activation of the same (hypothetical) wg repressor (see Discussion).
Fig. 5.
The effect of ectopic EFE on wg expression is distinct from that of En and Ftz. The patterns of wg RNA were detected (as in Fig. 2, except using a wg probe) in embryos 40 minutes after induction (as in Fig. 1) of either (A) no transgene, (B) hs-Ftz, (C) hs-EFE, or (D) hs-En. wg is repressed by Ftz (in odd-numbered parasegments) and by En, while it is strongly de-repressed by EFE in the Ftz-dependent (even-numbered) parasegments, as it is in ftz-mutant embryos.
An active repression domain is required for repression by EFE in vivo
In order to determine whether repression by EFE in vivo depends on the active repression function of En, or whether it might be due solely to competition for Ftz-binding sites in target genes, we examined the effects of a derivative of EFE that could no longer actively repress. We constructed a deletion derivative (EFEΔ) by removing an En-derived region containing both an active repression domain previously identified using cell culture assays (Han and Manley, 1993a; Jaynes and O’Farrell, 1991), as well as a conserved motif from the N-terminal half of En. Deletion of both of these regions was necessary to completely abolish active repression activity in culture (S. T. S. and J. B. J., unpublished observations; see below). Since the Ftz HD appears to be responsible for targeting EFE to the ftz gene, this deletion would not be expected to affect the ability of EFE to compete for Ftz target sites.
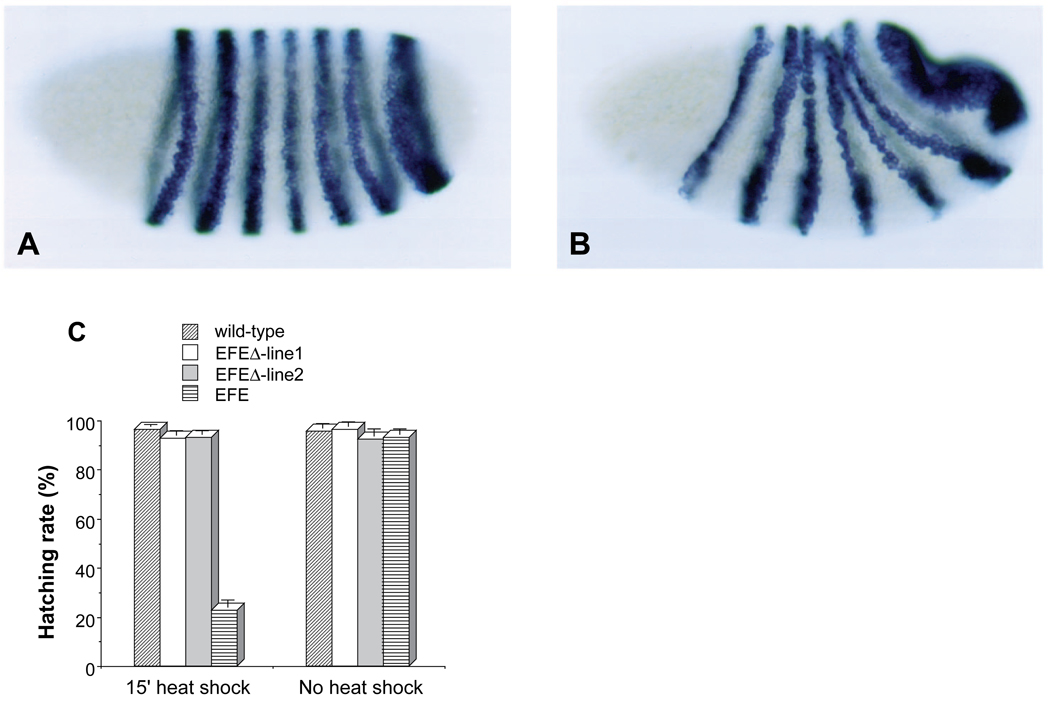
First, we examined the ability of EFEΔ to repress endogenous ftz gene expression. Following a standard (15 minute) heat shock, transgenic embryos showed no reduction of ftz RNA levels relative to wild-type controls (not shown), in strong contrast to the effect of EFE (Fig. 2C). Following a 30 minute induction of EFEΔ, there was a slight reduction in ftz expression, followed by a recovery to normal levels (Fig. 6A,B). Next, we examined whether EFEΔ retained the ability to disrupt embryogenesis. In two independent lines, EFEΔ was unable to significantly affect the hatching rate of embryos following a standard heat shock (Fig. 6C). In contrast, EFE reduced the hatching rate below 30%. Even with a 30 minute heat shock, EFEΔ was unable to significantly reduce hatching rates below those of wild-type embryos (not shown), suggesting that the observed transient reduction of ftz expression has no lasting phenotypic consequence.
Fig. 6.
Repression of ftz requires an active repression domain from En. In situ hybridization to ftz RNA in EFEΔ embryos was carried out as in Fig. 2 following a 30 minute heat shock beginning between 2.5 and 3 hours of development (30 minute collections). Embryos were fixed either (A) 10 minutes or (B) 40 minutes after the end of heat shock. EFEΔ embryos showed a slight repression initially (compare to Fig. 2A), followed by full recovery. (C) EFEΔ is innocuous in vivo. Hatching rates were determined for the wild-type recipient strain, and for transgenic derivatives carrying either an EFE- or an EFEΔ -producing transgene either without heat shock, or following a 15 minute heat shock beginning 2.5 hours after the end of a 30 minute egg collection.
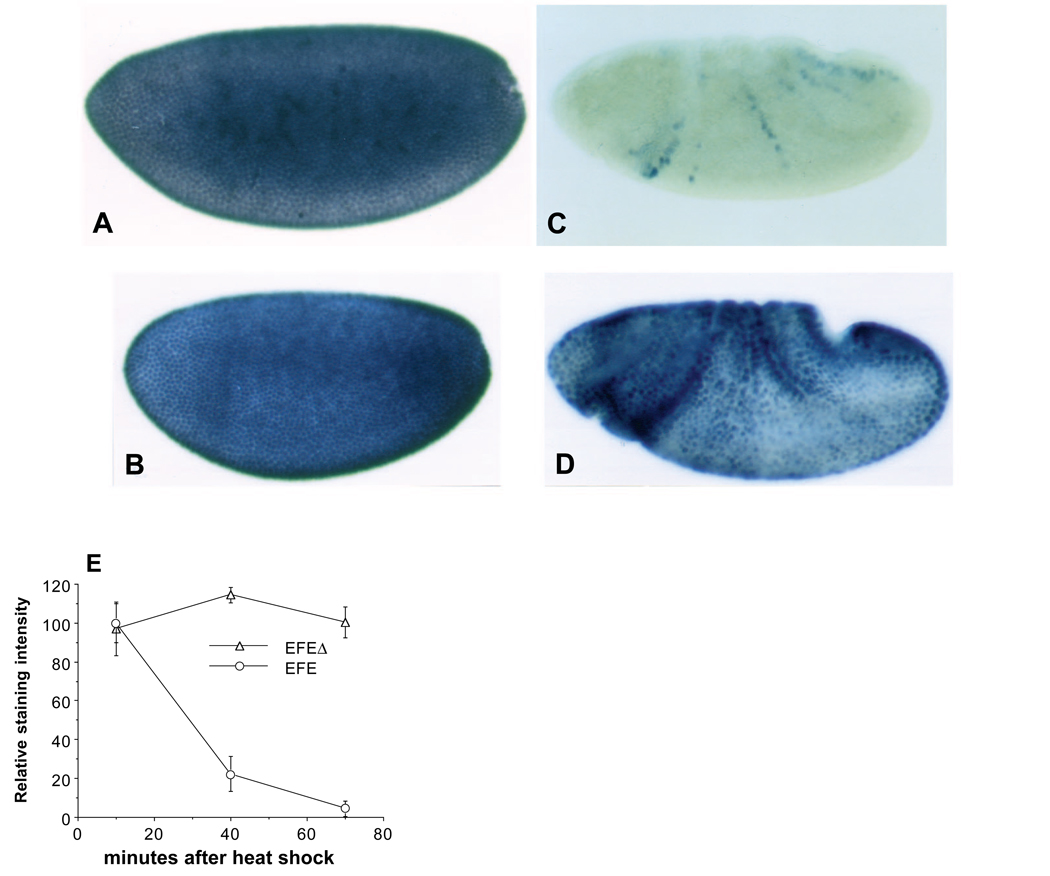
In situ hybridization revealed that EFEΔ mRNA was expressed following heat induction at a level similar to that of EFE mRNA, and that the two decayed with similar kinetics, becoming undetectable after about 30 minutes (not shown). To determine whether EFEΔ was expressed effectively, we employed an antiserum directed against the N-terminal portion of En, which is present in both EFE and EFEΔ. Staining with this antibody preparation showed a similar strong, nuclear signal in both EFE and EFEΔ embryos 10 minutes after induction. While EFE decayed rapidly, EFEΔ protein was more stable, persisting in nuclei for at least 1 hour (Fig. 7A–D). Quantitation of the signal confirmed that EFEΔ is expressed at the same level immediately following induction, and persists considerably longer than EFE (Fig. 7E). These data show that the inability of EFEΔ to repress ftz and to disrupt embryogenesis is not due to a lack of its accumulation or persistence in nuclei.
Fig. 7.
(A–D) Both EFE and EFEΔ are localized to nuclei, and EFEΔ is more stable. Embryos were heat shocked for 15 minutes to induce transgene expression, then fixed and stained with α-En (affinity purified using the N-terminal 150 aa of En, present in both EFE and EFEΔ ) and with alkaline phosphatase (AP) coupled 2° antibodies. AP activity was first quantified using the soluble-product-producing substrate PNP (Manoukian and Krause, 1992) (results shown in E), then the embryos were stained as in Fig. 2 (final step). (A) EFE expression 10 minutes after induction, (B) EFEΔ expression at the same time, (C) staining of EFE transformants 55 minutes after induction, leaving only a trace of endogenous en stripes (consistent with the RNA analysis of Fig. 3C), (D) EFEΔ staining 55 minutes after induction. In spite of its persistence, EFEΔ is unable to repress ftz expression or disrupt normal embryogenesis. (E) EFEΔ is expressed at the same level as EFE, and is more stable. α -En antibody staining was quantified as described in A-D. Both proteins were initially expressed at equal levels, but EFEΔ was more stable. A background value (amounting to 7% at the first time point and 10% at the second and third) was subtracted from the numbers plotted based on parallel staining of control embryos.
Deletion of the active repression domain does not affect competition for DNA-binding sites in cultured cells
Based on previous results, we hypothesized that EFEΔ would have reduced or abolished active repression activity in culture, and, since the HD has been shown to be sufficient for sequence-specific DNA binding (Desplan et al., 1988), that it would retain the ability to bind to target sites. To test this, we used an established co-transfection assay in Drosophila S2 cells (Jaynes and O’Farrell, 1988; Jaynes and O’Farrell, 1991). Previous analyses had shown (Jaynes and O’Farrell, 1991) that En and EFE could repress activated transcription of a reporter gene without competing for binding sites with a sequence-specific activator such as the glucocorticoid receptor (GR). This activity was termed active repression. In contrast, both these active repressors and HD-containing proteins with no active repression function could repress by competing for activator-binding sites (Jaynes and O’Farrell, 1988; Jaynes et al., 1990; Jaynes and O’Farrell, 1991). This competition can thus serve as an in vivo measure of DNA-binding activity. In this assay, we used Ftz as the activator, since its activation function on a HD-binding site-containing target gene has been shown to be repressible by simple competition (Jaynes and O’Farrell, 1988), and since EFE and EFEΔ contain the Ftz HD. The results (Fig. 8) show that EFEΔ has lost most, or all, of its active repression function, since it does not significantly repress GR-activated expression, while EFE represses this expression very effectively. In contrast, both EFE and EFEΔ are equally effective in repressing Ftz-activated expression. As previously shown, EFE can repress basal level expression of a reporter gene 2- to 3-fold, but only when the reporter contains HD-binding sites, another characteristic of active repression (Jaynes and O’Farrell, 1991). EFEΔ, however, does not repress basal level expression of the reporter, whether or not it contains HD-binding sites (not shown). These data demonstrate that EFEΔ has selectively lost the ability to actively repress transcription, while retaining the ability to compete for binding sites on the DNA.
EFEΔ is targeted to the ftz autoregulatory enhancer in vivo
Since EFEΔ retains DNA-binding activity in culture and can accumulate in embryonic nuclei, it is somewhat surprising that it is virtually inert in vivo. Based on previous analyses of Ftz autoregulation (Han et al., 1993; Schier and Gehring, 1992), and on our indications of the targeting ability of EFE, we would predict that EFEΔ would compete for Ftz-binding sites within the ftz gene. We therefore tested whether partial competition might be occurring, to an extent that allows EFE to occupy enough sites to actively repress ftz, but not enough to allow EFEΔ to completely prevent auto-activation by Ftz. For this we used a reporter transgene driven by a 1.75 kb ftz autoregulatory enhancer (Pick et al., 1990), testing whether the level of RNA from the lacZ reporter was reduced in doubly transgenic embryos by induction of EFEΔ. The reporter was clearly repressed relative to endogenous Ftz protein (Fig. 9A,B) following a 30 minute heat shock. While this repression by EFEΔ was not complete, quantitation of the in situ hybridization signal indicated that the degree of repression was at least 5-fold (Fig. 9C). Without heat shock, there was no significant difference in reporter expression levels between EFEΔ and control flies. As suggested by the quantitation in Fig. 9C, heat shock itself caused a 2-fold increase in RNA levels from the transgene, which followed a slight reduction immediately after heat shock (not shown). In fact, we have observed a similar effect on endogenous ftz RNA levels (not shown). Thus it appears that ftz expression, once established, is subject to positive feedback, and that this positive feedback may be partially Ftz independent, since increased expression can follow a brief decrease. The effect of EFEΔ is both to prevent this increase, and to repress ftz enhancer expression below the level seen without heat shock. We have also examined the effect of EFEΔ on a 300 bp ‘minimal’ ftz upstream enhancer (Han et al., 1993), with similar results (data not shown). Control experiments with an eve enhancer-lacZ transgene showed no repression by EFEΔ (Fig. 9C). These data suggest that EFEΔ can compete for Ftz-binding sites in vivo, but that this passive repression is much less effective than active repression by EFE at inactivating the endogenous ftz gene (see Discussion).
DISCUSSION
An engineered active repressor generates a predicted mutant phenotype
In this study, we have examined the feasibility of creating a dominant negative version of a transcriptional activator by converting it to an active repressor. We demonstrated that such a regulator can generate a close approximation of the expected mutant phenotype, that of loss-of-function of the activator, by the two criteria of target gene activities and external morphology. The extensive base of knowledge available from molecular studies of early Drosophila embryogenesis allowed us to monitor effects on the known regulatory circuitry so as to test the specificity and mode of action of the engineered regulator. We found that, in vivo, the chimeric regulator is accurately targeted by the activator’s DNA-binding domain and that it acts as an active repressor.
Specifically, we found that the Ftz HD is capable of targeting the chimeric active repressor EFE to Ftz target genes in vivo. This combination of Ftz targeting and active repression efficiently generates a ftz mutant-like phenotype. A considerable degree of specificity is demonstrated by the similarity of this phenotype to that of a ftz mutant, both in the morphology of cuticular structures and in the effects on known ftz target genes. Specificity is further indicated by the fact that neither parental protein, Ftz or En, is able to approximate this phenotype (Fig. 1).
The re-targeting was accomplished by replacement of the 60 aa En HD with that of Ftz (Jaynes and O’Farrell, 1991). These HDs differ at 31 aa, of which one is in the recognition helix (helix 3) at position 2, and they bind to similar sequences in vitro (Desplan et al., 1988; Hoey and Levine, 1988). In addition, they are both able to regulate transcription through the same consensus binding sites in cell culture transfection experiments. A distinct change in regulatory specificity in vivo is seen in a comparison of the target genes affected following ectopic expression of either the chimeric protein or the parental En protein in early Drosophila embryos. Ftz has previously been implicated in a direct positive feedback on its own expression, through an upstream enhancer element (Pick et al., 1990; Schier and Gehring, 1992). The chimeric protein, EFE, strongly represses this Ftz target gene, i.e. the ftz gene itself, while En, with its own HD, is unable to do so. The repression of ftz by EFE occurs very rapidly (we see clear repression 5 minutes after a 7 minute heat shock), suggesting that it occurs via a direct interaction.
That the HD swap has effected a true re-targeting, rather than simply an increased activity, is shown by the fact that ectopically expressed En strongly repress eve, while EFE does not (Fig. 4). The eve gene has previously been implicated as a target of En, since eve expression persists abnormally in en mutants, in cells that normally turn on en as eve expression fades (Harding et al., 1986). Thus, the HD swap re-targets the protein away from one gene (eve) and toward another (ftz), in a manner consistent with the normal targeting activities of the parental proteins in the embryo. Since this difference in targeting activity in vivo is not accompanied by a clear difference in DNA-binding specificity in vitro (Desplan et al., 1988), the results suggest that targeting in vivo is accomplished by recognizing both specific DNA sequences and other DNA-bound proteins. Both types of recognition interaction can apparently be mediated by the Ftz HD itself. Similar inferences have been drawn previously from comparisons of the in vivo targeting activities of other HD proteins (Kuziora and McGinnis, 1989; Lai et al., 1992; Lin and McGinnis, 1992; Mann and Hogness, 1990), although in some cases conserved regions immediately flanking the HD have also been implicated (Gibson et al., 1990; Lin and McGinnis, 1992; Mann and Hogness, 1990).
An additional indication that the HD swap results in a global change in regulatory specificity comes from a determination of when during embryogenesis defects can be induced. The time window of effectiveness of EFE is similar to that of ectopically expressed Ftz, and unlike that of En (Fig. 1E). Both EFE and Ftz are ineffective after 3.5 hours of development, while En is most effective at 3.5 hours. Thus, in vivo efficacy appears to depend on the HD. This might be due to a temporal restriction in the expression or activation of a partner protein for targeting by the Ftz HD. A possible candidate for such a partner is FTZ-F1, identified as a DNA-binding activity that decreases after roughly 3–4 hours of development (Ueda et al., 1990). Cloned factors belonging to the nuclear receptor superfamily, which are thought to correspond to this binding activity (Lavorgna et al., 1991; Ohno and Petkovich, 1992), bind in vitro to sites within both the ftz zebra-stripe element and the upstream enhancer (Han et al., 1993).
Repression in vivo by EFE is active
En acts as a dominant or active transcriptional repressor in cultured cells, and represses several target genes in embryos (Eaton and Kornberg, 1990; Harding et al., 1986; Hooper et al., 1989), although direct interaction with these target genes in vivo has not been demonstrated. Cell culture experiments had shown that the active repression function of En could be conferred by regions separate from the HD (Han and Manley, 1993a; Jaynes and O’Farrell, 1991), and that the EFE chimera was also an active repressor (Jaynes and O’Farrell, 1991). When EFE is expressed in 2.5-3 hour old embryos, it represses the normal Ftz target genes ftz and en (Fig 2 and Fig 3). Repression of en occurs even where Ftz is not expressed, i.e., where en is activated by other factors (DiNardo et al., 1987). These activities require a domain of En that is also required for active repression in cell culture assays (Fig 6, Fig 7, Fig 8). Together, these results suggest that in vivo, EFE possesses the targeting activity of Ftz and the active repression activity of En.
The pattern of ftz repression is distinct from that seen following an overall reduction in ftz RNA levels caused by expression of an antisense ribozyme (Zhao et al., 1993). In that case, the differential sensitivity of stripes was attributed to a differential requirement for autoactivation, possibly correlating with the order of establishment of ftz stripes. The EFE effect is quite different, with some stripes that were very sensitive to loss of Ftz being less sensitive to EFE repression. The pattern of EFE repression may relate to its ability to actively repress transcription, an activity that is likely to be affected by the ‘context’ of binding sites, i.e. the nearby binding of other regulators, in a different way than is activation by Ftz, leading to different relative sensitivities of different stripes to active repression by EFE vs. a simple loss of auto-activation. In any case, these results are consistent with previous studies which suggested that there are significant differences in how different ftz stripes are regulated (Binari and Perrimon, 1994; Dearolf et al., 1989; Han et al., 1993; Ueda et al., 1990).
The pattern deletions caused by EFE, although clearly exhibiting a strong bias toward ftz-dependent regions (see Fig. 1 legend), are sometimes less severe and sometimes more severe than the pair-rule deletions caused by ftz mutants. The details of these phenotypes can be instructive with regard to the regulatory interactions and functions of ftz and its target genes. In less severely affected embryos, there is a preferential loss of naked cuticle anterior to even-numbered en stripes (relative to that adjacent to odd-numbered en stripes) that parallels a similar effect in en mutants. Mutations which are thought to eliminate en function, leaving that of the en-like gene invected (inv) intact, cause fusion of pairs of denticle bands due to loss of naked cuticle anterior to even-numbered en stripes, but only partial loss of the naked cuticle that forms anterior to (and within) odd-numbered en stripes (Nüsslein-Volhard and Wieschaus, 1980). Even-numbered en stripes form before the odd-numbered stripes (Weir et al., 1988), and inv is activated later than en (in an en-like pattern; Coleman et al., 1987). It seems that en/inv function is required earlier in the even-numbered stripes, which form first, than in the odd-numbered stripes. Interestingly, en is required to maintain wg expression in the adjacent cell row (Martinez Arias et al., 1988), and wg is activated first (anteriorly) adjacent to the incipient odd-numbered en stripes, and only later next to the even-numbeed en stripes (Baker, 1988). In EFE embryos, odd-numbered en stripes are transiently lost, re-appearing between 3.5 and 4 hours of development. These embryos apparently go on to exhibit a ftz-mutant like pair-rule phenotype as shown in Fig. 1. Thus the wg stripes that are activated first appear to be less dependent on early en expression, where transient repression of en by EFE has little phenotypic consequence.
The detailed time course of sensitivity of different pattern elements to the effects of EFE, as outlined in Results, suggests that different Ftz target genes in different portions of each Ftz stripe may require Ftz at different times, with the presumptive denticle band region apparently becoming independent of Ftz earlier than the anteriorly adjacent naked cuticle, which depends on the even-numbered en stripes.
The deletion derivative of EFE, EFEΔ, has several intriguing properties. Consistent with previous suggestions that the HD is sufficient for effective competition for binding sites (Jaynes and O’Farrell, 1988; Jaynes et al., 1990; Jaynes and O’Farrell, 1991), EFEΔ retains the ability to passively repress transcription in cultured cells, apparently by competing with the Ftz activator for binding sites. However, it is unable to repress transcription activated by the glucocorticoid receptor, which binds to distinct sites within the reporter gene. Two features are deleted in EFEΔ that may contribute to the loss of repression activity in vivo. The first is an active repression domain as defined by transient expression studies in cultured cells (Han and Manley, 1993a; Jaynes and O’Farrell, 1991). The second is a conserved motif found in all known En-class HD proteins (Logan et al., 1992), which we have recently found is shared with other classes of HD proteins (J. B. J. and A. Mazo, unpublished observations).
Active repression is much more effective in vivo than passive repression
Not only is EFEΔ able to interact with HD-binding sites in culture, as indicated by its ability to passively repress transcription (Fig. 8), but it is more stable in embryonic nuclei than EFE (Fig. 7). Therefore, it was somewhat surprising that it is unable to cause more than a weak, transient reduction in ftz gene expression. We tested whether EFEΔ is still targeted to the ftz gene in vivo by monitoring the activity of an enhancer specifically implicated in Ftz autoregulation (Han et al., 1993; Pick et al., 1990; Schier and Gehring, 1992). We found that a 1.7 kb enhancer region (Fig. 9), as well as a 300 bp subregion, is repressed significantly by EFEΔ, presumably by competing for the multiple Ftz-binding sites that they contain (Han et al., 1993; Schier and Gehring, 1992). These results suggest that while EFEΔ is able to significantly reduce Ftz autoregulation, this does not severely affect endogenous gene expression. This may be due to contributions from several effects, including the following: i) the competition for Ftz HD-binding sites by EFEΔ is probably not complete, ii) Ftz has the ability to activate its own gene through a HD-independent mechanism (Fitzpatrick et al., 1992), and iii) direct Ftz auto-activation may not be crucial to the maintenance of endogenous ftz gene expression. Whatever the reasons, competition for Ftz-binding sites by EFEΔ appears not to be an effective means of repression of the endogenous ftz gene. In contrast, repression by EFE is very effective, apparently due to its ability to actively repress transcription. The mechanisms of active repression are not known. However, it may be mediated by alterations in chromatin structure (Roth et al., 1992), or by direct interaction with the basal transcriptional machinery (Herschbach et al., 1994), or both.
Re-targeting the En active repressor by fusing it with a het-erologous targeting activity may be a generally useful method of inactivating specific sets of genes in vivo. In fact, a recent study suggests that the N-terminal half of En, which can confer active repression function to the GR DNA-binding domain (Jaynes and O’Farrell, 1991), is active in the hematopoietic lineage of mice when fused with the myb DNA-binding domain (Badiani et al., 1994). Our results indicate that such retargeting can be very effective, since EFE is capable of efficiently generating a ftz mutant-like phenotype. However, when expressed ubiquitously, EFE represses at least one Ftz target gene, en, outside the ftz domain. Although cross-regulatory interactions among segment polarity genes are apparently able to later restore en expression in these regions, so that few phenotypic changes occur outside the ftz domain, this result suggests that an engineered active repressor can be effective in vivo even without directly competing for activator-binding sites.
The effects produced by a specifically targeted active repressor can illuminate complex regulatory relationships in vivo. For example, our results suggest that Ftz may act as an activator in early embryos on all its direct target genes. This follows from the fact that re-targeting an active repressor to these genes mimics a loss of Ftz function. The normal repression of wg expression by Ftz is likely to be an indirect effect, through activation of a repressor of wg. Thus, this kind of regulatory manipulation has useful applications in a variety of contexts, both for understanding normal gene function and, potentially, for intervening in cases of aberrant function.
Acknowledgments
We thank Patrick H. O’Farrell, in whose laboratory this work was initiated, for advice and critical comments on the manuscript. We thank Tadaatsu Goto, Alexander Mazo and Jay Rothstein for critical comments on the manuscript, and Tadaatsu Goto, Miki Fujioka, Jill Heemskerk and Steve DiNardo for support and technical advice.
REFERENCES
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- Baker N. Localization of transcripts from the wingless gene in whole Drosophila embryos. Development. 1988;103:289–298. doi: 10.1242/dev.103.2.289. [DOI] [PubMed] [Google Scholar]
- Biggin MD, Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989;58:433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- Carroll SB, DiNardo S, O’Farrell PH, White RA, Scott MP. Temporal and spatial relationships between segmentation and homeotic gene expression in Drosophila embryos, distributions of the fushi tarazu, engrailed, Sex combs reduced, Antennapedia, and Ultrabithorax proteins. Genes Dev. 1988;2:350–360. doi: 10.1101/gad.2.3.350. [DOI] [PubMed] [Google Scholar]
- Coleman KG, Poole SJ, Weir MP, Soeller WC, Kornberg T. The invected gene of Drosophila: sequence analysis and expression studies reveal a close kinship to the engrailed gene. Genes Dev. 1987;1:19–28. doi: 10.1101/gad.1.1.19. [DOI] [PubMed] [Google Scholar]
- Dearolf CR, Topol J, Parker CS. The caudal gene product is a direct activator of fushi tarazu transcription during Drosophila embryogenesis. Nature. 1989;341:340–343. doi: 10.1038/341340a0. [DOI] [PubMed] [Google Scholar]
- Desplan C, Theis J, O’Farrell PH. The sequence specificity of homeodomain-DNA interaction. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, O’Farrell PH. Establishment and refinement of segmental pattern in the Drosophila embryo, spatial control of engrailed expression by pair-rule genes. Genes Dev. 1987;1:1212–1225. doi: 10.1101/gad.1.10.1212. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Sher E, Heemskerk JJ, Kassis JA, O’Farrell PH. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature. 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Viglianti GA, Rutledge BJ, Meselson M. Alteration of hsp82 gene expression by the gypsy transposon and suppressor genes in Drosophila melanogaster. Genes Dev. 1989;3:454–468. doi: 10.1101/gad.3.4.454. [DOI] [PubMed] [Google Scholar]
- Eaton S, Kornberg TB. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 1990;4:1068–1077. doi: 10.1101/gad.4.6.1068. [DOI] [PubMed] [Google Scholar]
- Edgar B, O’Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–480. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker SC, Young KE, von Kessler DP, Beachy PA. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 1991;10:1179–1186. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RP. The use of antisense approaches to study development. Devel. Genetics. 1993;14:251–257. doi: 10.1002/dvg.1020140402. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick VD, Percival-Smith A, Ingles CJ, Krause HM. Homeodomain-independent activity of the fushi tarazu polypeptide in Drosophila embryos. Nature. 1992;356:610–612. doi: 10.1038/356610a0. [DOI] [PubMed] [Google Scholar]
- Forrest D, Munoz A, Raynoschek C, Vennstrom B, Beug H. Requirement for the C-terminal domain of the v-erbA oncogene protein for biological function and transcriptional repression. Oncogene. 1990;5:309–316. [PubMed] [Google Scholar]
- Frasch M, Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987;1:981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Gibson G, Schier A, LeMotte P, Gehring WJ. The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell. 1990;62:1087–1103. doi: 10.1016/0092-8674(90)90386-s. [DOI] [PubMed] [Google Scholar]
- Han K, Manley JL. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993a;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Manley JL. Transcriptional repression by the Drosophila Even-skipped protein, definition of a minimal repression domain. Genes Dev. 1993b;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Han W, Yu Y, Altan N, Pick L. Multiple proteins interact with the fushi tarazu proximal enhancer. Mol. Cell. Biol. 1993;13:5549–5559. doi: 10.1128/mcb.13.9.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K, Rushlow C, Doyle HJ, Hoey T, Levine M. Cross-regulatory interactions among pair-rule genes in Drosophila. Science. 1986;233:953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- Harding K, Hoey T, Warrior R, Levine M. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 1989;8:1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S, Kostriken R, O’Farrell PH. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature. 1991;352:404–410. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschbach BM, Arnaud MB, Johnson AD. Transcriptional repression directed by the yeast α2 protein in vitro. Nature. 1994;370:309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Hiromi Y, Gehring WJ. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987;50:963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- Hoey T, Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988;332:858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Baker NE, Martinez AA. Regulation of segment polarity genes in the Drosophila blastoderm by fushi tarazu and even-skipped. Nature. 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [published erratum appears in Nature 1988; 335, 744]. [DOI] [PubMed] [Google Scholar]
- Ip YT, Kraut R, Levine M, Rushlow CA. The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell. 1991;64:439–446. doi: 10.1016/0092-8674(91)90651-e. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D, Pinchin SM, Ingham PW, Gyurkovics HG. Autocatalytic ftz activation and metameric instability induced by ectopic ftz expression. Cell. 1989;57:223–232. doi: 10.1016/0092-8674(89)90960-4. [DOI] [PubMed] [Google Scholar]
- Jaynes JB, O’Farrell PH. Activation and repression of transcription by homoeodomain-containing proteins that bind a common site. Nature. 1988;336:744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes JB, Vincent J, O’Farrell PH. Genetics of Pattern Formation and Growth Control. New York: Wiley-Liss Inc; 1990. Drosophila Homeodomain-containing Proteins Regulate Transcription; pp. 47–64. [Google Scholar]
- Jaynes JB, O’Farrell PH. Active repression of transcription by the Engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985;42:237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Johnson BF, Krasnow MA. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeo domain proteins. Genes & Dev. 1992;6:2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- Kassis JA. Spatial and temporal control elements of the Drosophila engrailed gene. Genes Dev. 1990;4:433–443. doi: 10.1101/gad.4.3.433. [DOI] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution, a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Sidén I, O’Farrell PH, Simon M. The engrailed locus of Drosophila, in situ localization of transcripts reveals compartment-specific expression. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- Kornberg T. Compartments in the abdomen of Drosophila and the role of the engrailed locus. Dev. Biol. 1981;86:363–372. doi: 10.1016/0012-1606(81)90194-9. [DOI] [PubMed] [Google Scholar]
- Kuziora MA, McGinnis W. A homeodomain substitution changes the regulatory specificity of the Deformed protein in Drosophila embryos. Cell. 1989;59:563–571. doi: 10.1016/0092-8674(89)90039-1. [DOI] [PubMed] [Google Scholar]
- Lai J-S, Cleary MA, Herr W. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeo domain. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- Lavorgna G, Ueda H, Clos J, Wu C. FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of fushi tarazu. Science. 1991;252:848–851. doi: 10.1126/science.1709303. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Morata G. Compartments in the wing of Drosophila, a study of the engrailed gene. Dev. Biol. 1976;50:321–337. doi: 10.1016/0012-1606(76)90155-x. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, Macdonald P, Struhl G. Borders of parasegments in Drosophila embryos are delimited by the fushi tarazu and even-skipped genes. Nature. 1987;328:440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Licht JD, Grossel MJ, Figge J, Hansen UM. Drosophila Krüppel protein is a transcriptional repressor. Nature. 1990;346:76–79. doi: 10.1038/346076a0. [DOI] [PubMed] [Google Scholar]
- Lin L, McGinnis W. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev. 1992;6:1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- Logan C, Hanks MC, Noble-Topham S, Nallainathan D, Provart NJ, Joyner AL. Cloning and sequence comparison of the mouse, human, and chicken engrailed genes reveal potential functional domains and regulatory regions. Devel. Genetics. 1992;13:345–358. doi: 10.1002/dvg.1020130505. [DOI] [PubMed] [Google Scholar]
- Madden SL, Cook DM, Morris JF, Gashler A, Sukhatme VP, Rauscher FI. Transcriptional repression mediated by the WT1 Wilms tumor gene product. Science. 1991;253:1550–1553. doi: 10.1126/science.1654597. [DOI] [PubMed] [Google Scholar]
- Mann RS, Hogness DS. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- Manoukian AS, Krause HM. Concentration-dependent activities of the even-skipped protein in Drosophila embryos. Genes Dev. 1992;6:1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- Martinez Arias A, Baker NE, Ingham PW. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development. 1988;103:157–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- Morata G, Kornberg T, Lawrence PA. The phenotype of engrailed mutations in the antenna of Drosophila. Dev. Biol. 1983;99:27–33. doi: 10.1016/0012-1606(83)90250-6. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Ohno CK, Petkovich M. FTZ-F1β, a novel member of the Drosophila nuclear receptor family. Mech. Development. 1992;40:13–24. doi: 10.1016/0925-4773(93)90084-b. [DOI] [PubMed] [Google Scholar]
- Panzer S, Fong A, Beckendorf SK. Genetics notes: new lacZ marked balancer. DIN. 1992;6:1. [Google Scholar]
- Pick L, Schier A, Affolter M, Schmidt GT, Gehring WJ. Analysis of the ftz upstream element, germ layer-specific enhancers are independently autoregulated. Genes Dev. 1990;4:1224–1239. doi: 10.1101/gad.4.7.1224. [DOI] [PubMed] [Google Scholar]
- Poole SJ, Kauvar LM, Drees B, Kornberg T. The engrailed locus of Drosophila, structural analysis of an embryonic transcript. Cell. 1985;40:37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Roth SY, Shimizu M, Johnson L, Grunstein M, Simpson RT. Stable nucleosome positioning and complete repression by the yeast alpha-2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- Saha S, Brickman JM, Lehming N, Ptashne M. New eukaryotic transcriptional repressors. Nature. 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- Sap J, Munoz A, Schmitt J, Stunnenberg H, Vennstrom B. Repression of transcription mediated at a thyroid hormone response element by the v-erb-A oncogene product. Nature. 1989;340:242–244. doi: 10.1038/340242a0. [DOI] [PubMed] [Google Scholar]
- Sauer F, Jäckle H. Concentration-dependent transcriptional activation or repression by Krüppel from a single binding site. Nature. 1991;353:563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- Schier AF, Gehring WJ. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature. 1992;356:804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:341–347. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Struhl G. Near-reciprocal phenotypes caused by inactivation or indiscriminate expression of the Drosophila segmentation gene ftz. Nature. 1985;318:677–680. doi: 10.1038/318677a0. [DOI] [PubMed] [Google Scholar]
- Tabata T, Eaton S, Kornberg TB. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- Ueda H, Sonoda S, Brown JL, Scott MP, Wu C. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev. 1990;4:624–635. doi: 10.1101/gad.4.4.624. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck OC. A small catalytic oligoribonucleotide. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Wagner RW. Gene inhibition using antisense oligodeoxynucleotides. Nature. 1994;372:333–335. doi: 10.1038/372333a0. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT, Turner FR, Kaufman TC. Defects in embryogenesis in mutants associated with the Antennapedia gene complex of Drosophila melanogaster. Dev. Biol. 1984;102:147–172. doi: 10.1016/0012-1606(84)90182-9. [DOI] [PubMed] [Google Scholar]
- Weir MP, Edgar BA, Kornberg T, Schubiger G. Spatial regulation of engrailed expression in the Drosophila embryo. Genes Dev. 1988;2:1194–1203. doi: 10.1101/gad.2.9.1194. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila, A Practical Approach. Oxford, England: IRL Press; 1986. pp. 199–228. [Google Scholar]
- Winslow GM, Hayashi S, Krasnow M, Hogness DS, Scott MP. Transcriptional activation by the Antennapedia and fushi tarazu proteins in cultured Drosophila cells. Cell. 1989;57:1017–1030. doi: 10.1016/0092-8674(89)90340-1. [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1993;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Pick L. Generating loss-of-function phenotypes of the fushi tarazu gene with a targeted ribozyme in Drosophila. Nature. 1993;365:448–450. doi: 10.1038/365448a0. [DOI] [PubMed] [Google Scholar]
- Zuo P, Stanojevic D, Colgan J, Han K, Levine M, Manley JL. Activation and repression of transcription by the gap proteins hunchback and Krüppel in cultured Drosophila cells. Genes Dev. 1991;5:254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]