SUMMARY
Nuclear pore complexes (NPCs) are the sole mediators of transport between the nucleus and the cytoplasm. NPCs have a life cycle: they assemble, disassemble, turn over and age. The molecular mechanisms governing these different vital steps are beginning to emerge, suggesting key roles for the core structural scaffold of the NPC and auxiliary factors in the assembly of this large macromolecular complex, and connections between NPC maintenance, NPC turnover, and ageing of the cell.
INTRODUCTION
The defining characteristic of a eukaryotic cell is the nucleus. The nucleus is delimited by a double membrane bilayer called the nuclear envelope (NE) that is continuous with the endoplasmic reticulum (ER). The NE surrounds and protects the chromatin, forming a barrier that spatially uncouples transcription from translation. The NE also provides a central anchor site for numerous nuclear and cytoplasmic structures, such as the nuclear lamina and the spindle organizer [1].
Trafficking between the nucleoplasm and the cytoplasm occurs through nuclear pore complexes (NPCs), large multi-protein complexes that form selective channels perforating the NE double membrane. NPCs function as gatekeepers of the nucleus [2], allowing the free diffusion of small molecules, while regulating with high specificity the transport of macromolecules. In this review, we focus on the mechanisms and signals that regulate how more than 450 protein molecules assemble to form this efficient molecular machine.
STRUCTURE AND FUNCTION OF THE NPC
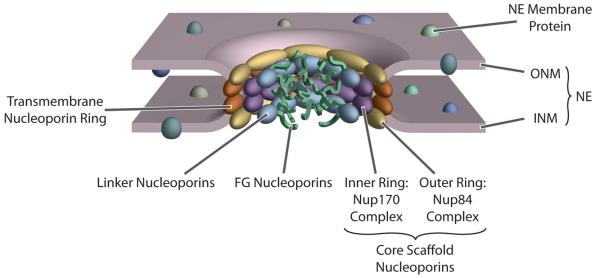
The NE is formed by two concentric membrane layers: the outer nuclear membrane (ONM) faces the cytoplasm and is continuous with the rough ER, while the inner nuclear membrane (INM) faces the nucleoplasm. The only connections between the ONM and INM are at the NPCs (Fig. 1). The NPC itself is a large proteinaceous assembly estimated to range from ~40-70 MDa depending on the organism (reviewed in [2,3]). Approximately 30 distinct proteins, termed nucleoporins, compose the ~450 protein NPC assembly (Table 1). The NPC is a doughnutshaped structure, consisting of eight spokes arranged around a central channel. Peripheral filaments emanate from the core into both the cytoplasm and nucleoplasm.
Figure 1.
Structure of the NPC. Major features of the interphase NPC and its surrounding NE are indicated; see main text for details (map based on [5•]).
Table 1.
Nucleoporins of S. cerevisiae and vertebrate homologs are listed. Nucleoporin composition of the different structural features shown in Figure 1 are also shown (based on [4,5•], see also [12]). Brackets signal paralog proteins in Saccharomyces cerevisiae and point to their vertebrate homolog. A dash indicates that no homolog has yet been described
| S. cerevisiae | Vertebrates | NPC structural feature |
|---|---|---|
| Pom152 | Gp210 | |
| Pom34 | - | Transmembrane ring |
| Ndc1 | Ndcl | |
| - | Pom121 | |
| (Nup170 complex) | ||
| Nup192 | Nup205 | |
| Nup188 | Nup188 | Inner ring |
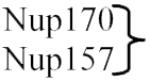 |
Nup155 | |
| (Nup84 complex) | (Nup107-160 complex) | |
| Nup133 | Nup133 | |
| Nup120 | Nup160 | |
| Nup145C | Nup96 | |
| Nup85 | Nup75 | Outer ring |
| Nup84 | Nup107 | |
| Seh1 | Seh1 | |
| Sec13 | Sec13 | |
| - | Nup43 | |
| - | Nup37 | |
| - | ALADIN | |
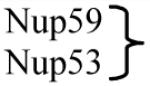 |
Nup35 | Other core nucleoporins |
| Nic96 | Nup93 | Linker nucleoporins |
| Nup82 | Nup88 | |
| Nup159 | Nup214 | |
 |
Nup98 | |
| Nsp1 | Nup62 | FG nucleoporins |
| Nup57 | Nup54 | |
| Nup49 | Nup58/45 | |
| Nup42 | NLP1 | |
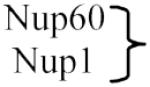 |
Nup153 | |
 |
Tpr | Peripheral nucleoporins |
| Gle2 | Rae1 | |
| - | Nup358 |
A detailed map for the relative position of each nucleoporin in the S. cerevisiae NPC was calculated based on multiple molecular, biochemical and structural data [4,5•]. This map shows that the NPC structure is modular, consisting of a few highly repetitive protein fold types, suggesting that the bulk of the NPC’s structure has evolved through gene duplications from a precursor set encoding only a few proteins. Within its core, the NPC possesses a cage-like scaffold made of proteins with striking similarities to the clathrin-like proteins that coat transport vesicles (e.g. clathrin/adaptins, COPI complex, COPII complex [6,7]), a resemblance that has been borne out in numerous recent crystallographic studies (e.g. [8]). These similarities suggest a common evolutionary origin for NPCs and coated vesicles, suggesting in turn that these proteins functioned as an early membrane-curving mechanism that led to the formation of the internal membrane systems in modern eukaryotes (reviewed in [9]).
There are several classes of nucleoporins (Table 1): transmembrane, core, linker and FG nucleoporins. Transmembrane nucleoporins span the nuclear membrane and presumably help anchor the NPC to the NE. Core nucleoporins make up the Nup84 and Nup170 complexes and form the cage-like scaffold within the NPC. FG nucleoporins are characterized by natively unfolded domains that include multiple phenylalanine-glycine (FG) dipeptide repeats. Linker nucleoporins connect the FG nucleoporins to the core scaffold (Fig. 1).
The transport function of the NPC is mediated mainly by FG nucleoporins. The disordered domains form a cloud filling the NPC thereby occluding most macromolecules from transiting the central tube (Fig. 1). Meanwhile, these same domains provide docking sites for transport factor-cargo complexes and facilitate their passage across the NPC. Because transport is mediated by diffusion and stochastic interactions, without requiring large-scale changes in the NPC’s structure, the NPC thus behaves as a “virtual gate” (reviewed in [10,11]).
Transport across the NPC is fast, energy-dependent, receptor-mediated, and highly regulated (reviewed in [10,11]). The process involves many transport factors, particularly those belonging to a structurally-related family of proteins termed Karyopherins (Kaps). Kaps bind to specific import (NLS) or export (NES) signals in their cargos. The Ran GTPase provides the energy for Kap-mediated transport. In the nucleoplasm, Ran is maintained in its GTP-bound form by chromatin-associated RanGEF, while in the cytoplasm, GTP hydrolysis by Ran is stimulated by RanGAP. Thus, the nucleotide-bound state of Ran also provides a means for the directionality of transport. During import, a Kap binds to its NLS-bearing cargo in the cytoplasm and transits the NPC. Once in the nucleus, RanGTP binds to the Kap, causing cargo release. During export, Kaps bind their NES-bearing cargo in the presence of RanGTP. Once the export complex passes through the NPC into the cytoplasm, conversion of Ran to its GDP-bound state triggers cargo release.
NPC ASSEMBLY INTO AN INTACT NE
NPCs have their own lifespan - they are born, live out lives, age, and can be taken apart and reassembled. The NPC life cycle has been shown to be tightly associated to cell cycle progression and to the life cycle of the NE [12]. During interphase, the NE’s surface area enlarges, concomitant with an increase in the number of NPCs [13-15]. How these NPCs assemble during interphase is still largely an uncharacterized process, but key clues to unraveling this process are now emerging.
Several plausible mechanisms have been discussed for the formation of NPCs during interphase: (i) the newly synthesized components could be assembled and incorporated de novo into a region of the NE devoid of preexisting NPCs.; (ii) newly synthesized components could be assembled into preexisting NPCs, forming enlarged precursors that then split into two daughter NPCs, or (iii) NPCs could be assembled in cytoplasmic membrane systems or vesicular intermediates that can later be fused to the NE. In this last case, the ER is the most probable source of membrane-associated NPC precursors due to its intimate relation with the ONM. For three decades, despite tantalizing clues, no clear evidence has emerged to differentiate between these possibilities. However, a recent breakthrough [16•] showed that in vertebrate cells, NPC assembly occurs in NE regions where no pre-existing NPCs were detected and that the newly formed NPCs did not contain significant amounts of components taken from pre-formed NPCs. Using a cell free NPC insertion assay, D’Angelo et al [16•] were also able to determine that NPCs components were recruited from both sides of the NE. The process also required RanGTP, both a nuclear and cytoplasmic pool of the Kap importin beta, and the Nup107-160 complex (the vertebrate homolog of the yeast core scaffold Nup84 complex) (Table 1). The Nup107-160 complex [17,18], as well as the Nup84 complex [19], is a biochemically distinct entity that can be isolated via extraction of intact NPCs. This raises the possibility that the NPC assembly might function by incorporating some large prefabricated “building blocks” made from nucleoporins connected in biochemically stable subcomplexes. Presumably the nuclear pool of NPC building blocks is maintained by nuclear import, implying that the preexisting NPCs are needed as transporters, but not templates, for assembly of new NPCs into intact NEs.
Genetic dissection of NPC assembly in the yeast Saccharomyces cerevisiae has also provided clues towards the mechanism of NPC assembly. In a screen for defective NPC assembly, npa (nuclear pore assembly) mutants were isolated that corresponded to Ran, RanGEF, RanGAP, Ran transport cofactor NTF2, and Kap95 (the homolog of vertebrate importin beta) [20-22]. The loss of proper localization of nucleoporin components in these mutants [20,22] led the authors to propose that Kap95 and Ran mutants are blocking the same stage of NPC assembly. However, this blocking effect seemed to happen in different cellular compartments. Ran cycle mutants showed a characteristic accumulation of nucleoporin-containing small membrane vesicles in the cytoplasm [20], whereas Kap95 loss of function mutants presented accumulation of extended membrane sheets [22]. Regardless, both point to an antagonistic effect on membrane dynamics. Thus, it has been proposed that cytoplasmic Kap95 might function to inhibit the fusion of the nucleoporin-containing vesicles until they reach the NE where RanGTP binds Kap95 [20,22]. This would disrupt Kap95’s inhibition and allow fusion of the vesicular NPC precursor to the NE. Thus, a balance of Kap95/importin beta and RanGTP at the NE seem to be required for NPC assembly into an intact NE in yeast, as happen in vertebrate cells (see above and [16•]).
In addition to Kap95, other transport factors have been implicated in NPC assembly. Kap121 has been proposed to help target Nup53 to the NPC, where it localizes to the core scaffold component Nup170 [23]. In yeast, both Nup170 and Nup53 have paralogs: Nup157 and Nup59 respectively. Over-expression of the c-terminal domain of Nup170 [24•] led to detection of putative cytoplasmic and NE associated intermediates of NPC assembly. These putative intermediates were also detected upon reversible depletion of NUP170 (the Nup157 paralog), as well as upon reversible depletion of the transmembrane nucleoporins Pom34 or Pom152 in a double deletion of the paralogous genes NUP59 and NUP53 [25••,26••].
Makio et al and Onischenko et al [25••,26••] both used an elegant in vivo approach to compare the fate of preexisting and newly synthesized nucleoporins during these genetic blocks of new NPC assembly. This was done by tagging nucleoporins with Dendra, a fluorescent protein that undergoes an irreversible photoconversion from green to red fluorescent states in response to blue or UV light [27]. For the first time in living cells, the pool of nucleoporins present at the point of photoconversion (red) could be distinguished from those newly synthesized after exposure (green). These experiments confirmed that there are at least two different pools of nucleoporins forming functional NPC intermediates. The first pool accumulated in cytoplasmic foci and was enriched in nucleoporins that are found mainly on the cytoplasmic side of NPCs. Also found in this first pool were core scaffold and transmembrane localized nucleoporins. The second pool localized to structures at the nuclear side of the INM; the second pool also included core and transmembrane nucleoporins, but instead of cytoplasmic nucleoporins it was enriched in nucleoporins found on the nuclear face of the NPC. In either case, these accumulating precursors resemble those seen in the mutants impaired in Kap95/importin beta or Ran function, implying that the precursors are formed through blockage of the NPC assembly pathway [20,22]. Together, these intermediates help assemble the NPC de novo from both sides of the yeast NE.
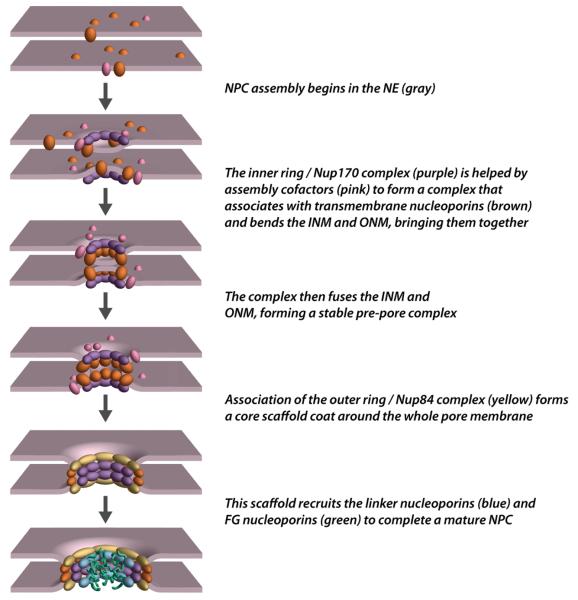
Taking together all that is now known, we can propose a mechanism for NPC assembly into intact NEs (Fig. 3). First, transmembrane nucleoporins and components which form the inner ring in mature NPCs congregate on both sides of the NE, probably starting the process of bending the ONM and INM. Perhaps to accomplish this, Nup170 homologs carry a predicted membrane binding amphipathic alpha-helix, called an ALPS motif [28]. Thus Nup170 homologs, inner ring components which are directly associated with the central NPC transmembrane ring, could also interact directly with the NE membranes through the ALPS motif, bending the ONM and INM, as well as stabilizing the resulting NE pore. Additionally, the membrane nucleoporins may require association of the Nup170 complex proteins in order to initiate the formation of the NE pore. A similar pre-pore arrangement has been described in the fungus Aspergillus nidulans, where transmembrane nucleoporins and Nup170 form the minimal scaffold required for the rest of the NPC to reform at the end of mitosis ([29•] and see below). Members of the Nup84 complex and paralogous nucleoporins Nup53 and Nup59 (all proteins comprising or closely associated with the core scaffold) have also been shown to contain ALPS motifs and thereby are also potentially able to directly interact with membranes (although none of them have actually been shown to do so in vivo) [28,30]. The association of Nup170 and Nup157 with these NPC components [5•] would allow the formation of a membrane coating structure, namely the core scaffold, around the NE pore that would enable the final stabilization of the newly formed pre-pore. To complete the NPC assembly, the remaining nucleoporins would incorporate into the pre-pore scaffold with the FG nucleoporins likely being the last components assembled. However, this scenario is unlikely to be comprehensive and more work is required to understand the exact timing and molecular mechanisms governing NPC insertion into the intact NE.
Figure 3.
Proposed Path of Interphase NPC Assembly; see main text for details.
NON-NUCLEOPORIN FACTORS IMPLICATED IN NPC ASSEMBLY
One exciting possibility that has been raised is the role of non-nucleoporin factors in the NPC’s formation. We have already mentioned how Ran, Ran cofactors and the Kaps appear to aid NPC assembly [20,22,23]. Other recently-described candidates for assembly factors include the ER protein Apq12 [31], and members of the reticulons (RTNs) and Yop1/DP1 protein families [32•]. RTNs and Yop1/DP1 proteins are of particular interest, as they can bend membranes and have functions in both tubular ER maintenance [33-35] and postmitotic NE shaping [36]. Dawson et al [32•] showed that yeast RTNs are required for proper NPC formation, and vertebrate RTNs are required for in vitro de novo pore formation. As RTNs also genetically interact with transmembrane nucleporins [32•], perhaps RTNs can aid the transmembrane nucleoporins during the initial membrane fusion of NPC assembly or facilitate the stabilization of the nascent pore (Fig. 3). No direct interaction has been reported between RTNs and nucleoporins, supporting a transient role for RTNs in NPC assembly. Perhaps other factors are also involved in the NPC formation, but as in the case of RTNs, their role would be transient, as they do not seem to be present in the mature NPC.
NPC DISASSEMBLY AND ASSEMBLY DURING MITOSIS
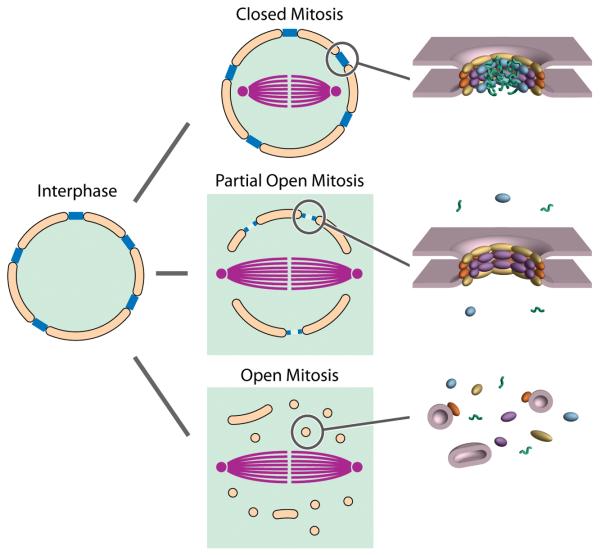
In eukaryotes undergoing closed mitosis (e.g. the budding yeast Saccharomyces cerevisiae), the NE and NPCs remain intact throughout the entire cell cycle. However in eukaryotes undergoing open mitosis, the NE and NPCs disassemble completely at the onset of mitosis (prophase) and reassemble around the segregated chromosomes at the end of mitosis (telophase) (Fig. 2). While a closed mitosis is often cited as characteristic of lower eukaryotes [37], the degree to which a mitosis is closed or open varies markedly among plants and metazoans [38]; for example in the filamentous ascomycete Aspergillus nidulans the NE remains mostly intact throughout the cell cycle but there is a partial disassembly of the NPC during mitosis. These “remnant” NPCs are freely permeable to the assembling spindle components and also provide the framework for the dispersed nucleoporins to reassemble at the end of mitosis. In some metazoans such as Drosophila the mitotic spindles form through open fenestrae in a degenerate NE while the NPCs disassemble to various degrees [39]. We would therefore argue that many organisms utilize mechanisms somewhere along a continuum between “open” and “closed” mitosis. These intermediate states of assembly may serve as important models for understanding the tenets of NPC assembly, particularly as the mechanisms guiding interphase NPC assembly and telophase NPC reassembly after open mitosis share common elements.
Figure 2.
Schematic Illustrating the Different Types of Mitosis. During mitosis, the mitotic spindle (purple) forms while the NE (orange) and NPCs (blue) are retained to differing degrees, resulting in the nucleoplasm (green) being retained only in the closed form of mitosis. To the right are shown which major features of the NPC are still intact, and which are dispersed.
Returning to mitosis in Aspergillus nidulans, the NPC is partially disassembled at the onset of mitosis [40], leading to the dispersion of 14 nucleoporins while 12 other nucleoporins [41], remain attached to the NE as a remnant pore structure. Interestingly, the Aspergillus Nup84 subcomplex, though normally part of this remnant framework, was shown to be dispensable, leaving a minimal framework comprising the three transmembrane nucleoporins (AnPom152, AnPom34 and AnNdc1), AnNup170 and AnGle1 [29•]. Remarkably, a triple deletion of all the known transmembrane nucleoporins of the fungus is fully viable [29•]. How can the outer and inner nuclear membranes be fused and an NPC assembled without the presence of any transmembrane nucleoporins, particularly in light of the yeast results discussed above? Although the presence of an as-yet unidentified Aspergillus transmembrane nucleoporin cannot be ruled out, the evidence so far points to an overlapping role of core and transmembrane nucleoporins in NPC pore structure maintenance. It is also possible that other accessory factors that are not dedicated nucleoporins assist the NPC (as discussed above).
The process of NE disassembly and reassembly during open mitosis has been comprehensively discussed in a recent Current Opinions in Cell Biology review [42]. Disassembly of the NPC in open mitosis is a fast process, such that it has been difficult to obtain the precise timing of its ordered steps. However, using in vivo time-lapse microscopy analysis of 11 GFP-tagged nucleoporins corresponding to members of 8 different NPC subcomplexes [43•], it was possible to determine that disassembly is highly synchronous, and moreover it is not simply the reverse sequence of NPC assembly (see below). Phosphorylation of certain key nucleoporins at the onset of mitosis seems to be the signal that triggers the destabilization of protein interactions within the NPC, leading to its deconstruction [44-46]. There is also strong evidence of known mitotis-specific kinases being involved in nucleoporin phosphorylation. Examples are the cyclin-dependent kinase Cdk1 in yeast [47,48] and the Aspergillus nidulans kinase NIMA, which is required for the partial disassembly of the NPC (see above and [40]). Rather that passively waiting for telophase, the resulting disassembled nucleoporins play specific roles in ensuring the proper progress of mitosis. For example, in certain metazoans, the Nup107-160 complex (Table 1) associates with kinetochores and appears to be required for correct assembly of the mitotic spindle [49,50]. NPC components may also help to recruit NE disassembly factors, one of which - the COPI complex - is a vesicle coating complex structurally related to clathrin (and NPCs). It has been suggested that the COPI complex plays a key role in vesiculating the NE membranes [51,52].
At the end of mitosis the NE is rebuilt around the segregated chromosomes, and NPCs are re-assembled in a stepwise process [43•]. Recently, an important player in NPC post-mitotic reassembly has been discovered - the protein ELYS/Mel28, which is associated in interphase with the Nup107-160 complex [53-55]. During mitosis, importin beta (the vertebrate Kap95 homologue) has been shown to bind the Nup107-160 complex, regulating its interactions with chromatin and other nucleoporins [56,57]. However the high concentration of RanGTP found around chromatin at the end of mitosis [58] cancels the inhibitory effect of importin beta on nucleoporins [56,59]. ELYS/Mel28 associates with chromatin via its DNA-binding AT hook motifs [60] and recruits the Nup107-160 complex [53,55,61], transmembrane nucleoporins and associated NE membrane [62,63] to the reassembly sites for the NPCs. At this point, in a strikingly similar mechanism to that recently proposed for interphase assembly (see above and [24•-26••]),nucleoporins Nup155 and Nup53/35 are required for NE membrane fusion and NPC assembly progression [64,65]. Nup155 and Nup53/35 are ALPS motif containing homologs to the yeast Nup170 and Nup53 respectively (Table 1). It is tempting to speculate that these nucleoporins are mediating the bending and stabilization of NE membranes around the nascent NPC scaffold along with the Nup107-160 complex [57]. Finally, complexes containing linker and FG nucleoporins are incorporated into the now NE-embedded NPC, thereby restoring the selectivity of the NE even before the final incorporation of the full set of FG nucleoporins [43•] (Fig. 3). It remains an open question if during interphase NPC assembly chromatin plays a role in defining the points in the NE where new NPCs will be inserted, as has been shown to happen during mitotic NPC reassembly.
NPC TURNOVER
Many cells do not divide for prolonged periods of time, such that no new NPCs are added by cell division or recycled during mitosis. For example, in human neurons NPCs must retain functionality in a cell that persists for decades without division. However, defective NPCs could lead to catastrophic mixing of nuclear and cytoplasmic contents. What mechanisms ensure the functional integrity of the NPC? How are components of the NPC repaired or renewed? Once assembled and fully functional, does the NPC adjust to changes in cellular function?
Recent evidence suggests that the cell doesn’t wait for NPC damage - rather, the NPC is a dynamic structure that is constantly exchanging its components. The rates of exchange vary greatly ranging from seconds for some peripheral components to days for core scaffold nucleoporins such as the Nup107-160 complex in dividing mammalian cells [66]. As a result, no one component, even if defective, would remain in the NPC long enough to significantly affect its function (bearing also in mind the tremendous redundancy between nucleoporins).
This observed turnover suggests that even when the NPC is not being regularly disassembled at mitosis, the expression and exchange of components would rejuvenate NPCs in older cells. However, things might not be quite so simple. During mitosis in the yeast Saccharomyces, the original “mother” cell buds off a new “daughter” cell. Surprisingly, septin proteins and Bud6 form a barrier between newly-dividing cells, facilitating asymmetric segregation of “old” NPCs and associated ageing factors to the nuclei of mother cells, while “rejuvenating” daughter cells by targeting newly formed NPCs to the daughter NE [67••]. In a similar vein, NPC component expression is not always maintained over the lifespan of a non dividing cell. In both post-mitotic cells found in C. elegans and in differentiated mammalian cells [68••], expression and turnover for scaffold nucleoporins is greatly diminished, or even perhaps absent. As a result, age-related deterioration of linker nucleoporins (Table 1) compromise the gating process for these NPCs over time. It is possible that some stable non dividing cells stop turning over their NPC components years before cell death. If so, the last line of defense for NPC components would be protection from oxidative damage. As the ageing process causes standard molecular maintenance pathways to fail, these components will begin to show significant damage later in life. These findings open up a whole new area of investigation into links between aging and maintenance of the NPC’s gating function.
PERSPECTIVES
Recent publications have begun to elucidate the mechanisms of NPC biogenesis. Because the core scaffold of the NPC is structurally related to vesicle coating complexes, it is likely that NPC assembly bears mechanistic similarities to coated vesicle formation (especially in regards to curving membranes). It is now understood that key transmembrane and core scaffold nucleoporins are required to form a pre-pore structure to which the rest of the NPC components are recruited. Moreover it has finally been established that NPCs can assemble de novo without requiring pre-existing templates. However, almost nothing is known about the pathways that regulate assembly and disassembly of NPCs. Clearly, regulation do exist for NPC formation, as demonstrated by the rate increase of NPC assembly in response to both cell division and metabolic changes in the cell [67••,69]. Other important questions remain to be answered. What marks the site for insertion of a new NPC into the interphase NE? What role does peripheral chromatin play in NPC biogenesis? Does membrane lipid composition influence NPC assembly? Is there any point during interphase NPC assembly when a pore exists without the FG nup barrier? If so, how does this affect the membrane selective permeability?
Once formed, the NPC is a fluid structure with components turning over to varying degrees. The formation and maintenance of the NPC is tightly linked to the growth of its cell; when a cell stops dividing, NPC biogenesis and turnover seems to slow or even stop. As the cell begins to age, so too does the NPC. Excitingly, as we continue to understand more about the birth, life and death of NPCs at a molecular level, so too do we understand how these events affect the life cycle of the cell as a whole.
Acknowledgments
We thank Kelli Mullin for her enormous help in editing and polishing the manuscript. We also are deeply grateful to Patrick Lusk and Megan King for critically reading the manuscript. J F-M is supported by a Postdoctoral Fellowship from the Ministerio de Educacion of Spain. MR is supported by NIH grants RR022220, GM062427, and GM071329.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the period of review, have been highlighted as:
•of special interest
••of outstanding interest
- 1.D’Angelo MA, Hetzer MW. The role of the nuclear envelope in cellular organization. Cell Mol Life Sci. 2006;63:316–332. doi: 10.1007/s00018-005-5361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 3.Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- 4.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- •5.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405.Using diverse sources of proteomic and structural experimental data, for the first time the authors were able to calculate a structure for the whole yeast NPC where the position of each nucleoporin within the overall architecture of the NPC is resolved. Major structural features of the NPC and their nucleoporin composition are described, revealing a close evolutionary relationship between the structures of membrane coating complexes and the NPC.
- 6.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci U S A. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field MC, Dacks JB. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr Opin Cell Biol. 2009;21:4–13. doi: 10.1016/j.ceb.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 11.Peters R. Translocation through the nuclear pore: Kaps pave the way. Bioessays. 2009 doi: 10.1002/bies.200800159. [DOI] [PubMed] [Google Scholar]
- 12.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T, Imamoto F, Imamoto N. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci. 2006;119:4442–4451. doi: 10.1242/jcs.03207. [DOI] [PubMed] [Google Scholar]
- 14.Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winey M, Yarar D, Giddings TH, Jr., Mastronarde DN. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by threedimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •16.D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196.In this study the authors showed that NPC assembly into an intact NE can occur de novo, independently of preexisting NPCs. They also showed that assembly is achieved from both sides of the NE and requires (at least) RanGTP, Importin beta and the Nup107-160 complex.
- 17.Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan KJ, McCaffery JM, Wente SR. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J Cell Biol. 2003;160:1041–1053. doi: 10.1083/jcb.200209116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan KJ, Wente SR. Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 2002;3:17. doi: 10.1186/1471-2156-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan KJ, Zhou Y, Wente SR. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol Biol Cell. 2007;18:886–898. doi: 10.1091/mbc.E06-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lusk CP, Makhnevych T, Marelli M, Aitchison JD, Wozniak RW. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J Cell Biol. 2002;159:267–278. doi: 10.1083/jcb.200203079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Flemming D, Sarges P, Stelter P, Hellwig A, Bottcher B, Hurt E. Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. J Cell Biol. 2009 doi: 10.1083/jcb.200810016. In press.Distinct roles in NPC assembly are unravelled for the N-terminal beta-propeller and C-terminal alpha-solenoid domains of Nup170. Negative stain EM structures for both domains are also defined.
- ••25.Onischenko EA, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the NDC1-interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol. 2009 doi: 10.1083/jcb.200810030. In press.This paper defines the direct interactions established between the transmembrane nucleoporin Ndc1 and other transmembrane and core nucleoporins. Together with reference [26••], describes the accumulation of distinct reversible cytoplasmic and nucleoplasmic nucleoporin precursors for NPC assembly and shows the key role of transmembrane and core nucleoporins
- ••26.Makio T, Stanton LH, Lin CC, Goldfarb DS, Weis K, Wozniak RW. The yeast nucleoporins Nup170 and Nup157 are essential for nuclear pore complex assembly. J Cell Biol. 2009 doi: 10.1083/jcb.200810029. In press.Using a combination of genetic and cell biological analyses, this study shows the requirement of the paralogous proteins Nup170 and Nup157 for the first steps of NPC assembly into an intact NE. It identifies, together with reference [25••], different subsets of cytoplasmic and nucleoplasmic nucleoporin precursors for NPC formation.
- 27.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 28.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- •29.Liu HL, De Souza CP, Osmani AH, Osmani SA. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell. 2009;20:616–630. doi: 10.1091/mbc.E08-06-0628.This paper reports the surprising finding that deletion of the three transmembrane nucleoporins of Aspergillus nidulans is not deleterious, and defines the minimal framework of nucleoporins necessary to NPC reassembly after mitosis in this fungus.
- 30.Patel SS, Rexach MF. Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol Cell Proteomics. 2008;7:121–131. doi: 10.1074/mcp.M700407-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Scarcelli JJ, Hodge CA, Cole CN. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol. 2007;178:799–812. doi: 10.1083/jcb.200702120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •32.Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174.Genetic and in vitro experiments identify a role of Reticulon and Yop1/DP1 proteins in the initial steps of NPC assembly during interphase in yeast and vertebrate cells.
- 33.De Craene JO, Coleman J, Estrada de Martin P, Pypaert M, Anderson S, Yates JR, 3rd, Ferro-Novick S, Novick P. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 35.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 36.Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol. 2008;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. 3rd Edition Garland Publishing Inc. edn; New York and London: 1989. [Google Scholar]
- 38.Heath IB. Variant mitoses in lower eukaryotes: indicators of the evolution of mitosis? Int. Rev. Cytol. 1980;64:1–80. doi: 10.1016/s0074-7696(08)60235-1. [DOI] [PubMed] [Google Scholar]
- 39.Stafstrom JP, Staehelin LA. Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo. Eur J Cell Biol. 1984;34:179–189. [PubMed] [Google Scholar]
- 40.De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 41.Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kutay U, Hetzer MW. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol. 2008;20:669–677. doi: 10.1016/j.ceb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43.Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026.This work demonstrates that NPC disassembly and reassembly during mitosis are not the reversal of each other in mammalian cells. The kinetic behavior of eight different nucleoporins is shown during mitosis using time-lapse microscopy analysis.
- 44.Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, Mattaj I. A role for gp210 in mitotic nuclear-envelope breakdown. J Cell Sci. 2008;121:317–328. doi: 10.1242/jcs.022525. [DOI] [PubMed] [Google Scholar]
- 45.Glavy JS, Krutchinsky AN, Cristea IM, Berke IC, Boehmer T, Blobel G, Chait BT. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107-160 subcomplex. Proc Natl Acad Sci U S A. 2007;104:3811–3816. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- 47.Lusk CP, Waller DD, Makhnevych T, Dienemann A, Whiteway M, Thomas DY, Wozniak RW. Nup53p is a target of two mitotic kinases, Cdk1p and Hrr25p. Traffic. 2007;8:647–660. doi: 10.1111/j.1600-0854.2007.00559.x. [DOI] [PubMed] [Google Scholar]
- 48.Onischenko EA, Gubanova NV, Kiseleva EV, Hallberg E. Cdk1 and okadaic acid-sensitive phosphatases control assembly of nuclear pore complexes in Drosophila embryos. Mol Biol Cell. 2005;16:5152–5162. doi: 10.1091/mbc.E05-07-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Prunuske AJ, Fager AM, Ullman KS. The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev Cell. 2003;5:487–498. doi: 10.1016/s1534-5807(03)00262-4. [DOI] [PubMed] [Google Scholar]
- 52.Prunuske AJ, Liu J, Elgort S, Joseph J, Dasso M, Ullman KS. Nuclear envelope breakdown is coordinated by both Nup358/RanBP2 and Nup153, two nucleoporins with zinc finger modules. Mol Biol Cell. 2006;17:760–769. doi: 10.1091/mbc.E05-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galy V, Askjaer P, Franz C, Lopez-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol. 2006;16:1748–1756. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 55.Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 58.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 59.Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 60.Kimura N, Takizawa M, Okita K, Natori O, Igarashi K, Ueno M, Nakashima K, Nobuhisa I, Taga T. Identification of a novel transcription factor, ELYS, expressed predominantly in mouse foetal haematopoietic tissues. Genes Cells. 2002;7:435–446. doi: 10.1046/j.1365-2443.2002.00529.x. [DOI] [PubMed] [Google Scholar]
- 61.Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 63.Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol Biol Cell. 2008;19:1753–1762. doi: 10.1091/mbc.E07-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodenas E, Klerkx EP, Ayuso C, Audhya A, Askjaer P. Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev Biol. 2009;327:399–409. doi: 10.1016/j.ydbio.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 66.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- ••67.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212.This paper describes the existence of a septin-dependent diffusion barrier that compartmentalizes the nuclear envelope during budding in S. cerevisiae. The barrier limits the translocation of pre-existing nuclear pores into the bud and ensures the asymmetric segregation of nuclear age-determinants between mother and daughter cells.
- ••68.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037.Here the authors provide evidence that in postmitotic metazoan cells, nucleoporins forming the core scaffold of the NPC are neither expressed nor turned over. As a consequence, age related oxidative damage is accumulated in scaffold nucleoporins, correlated with leaky NPCs and the loss of the nuclear permeability barrier.
- 69.Maul GG, Deaven L. Quantitative determination of nuclear pore complexes in cycling cells with differing DNA content. J Cell Biol. 1977;73:748–760. doi: 10.1083/jcb.73.3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]