Summary
Agrobacterium tumefaciens uses a type IV secretion (T4S) system composed of VirB proteins and VirD4 to deliver oncogenic DNA (T-DNA) and protein substrates to susceptible plant cells during the course of infection. Here, by use of the Transfer DNA Immuno-Precipitation (TrIP) assay, we present evidence that the mobilizable plasmid RSF1010 (IncQ) follows the same translocation pathway through the VirB/D4 secretion channel as described previously for the T-DNA. The RSF1010 transfer intermediate and the Osa protein of plasmid pSa (IncW), related in sequence to the FiwA fertility inhibition factor of plasmid RP1 (IncPα), render A. tumefaciens host cells nearly avirulent. By use of a semi-quantitative TrIP assay, we show that both of these ‘oncogenic suppressor factors’ inhibit binding of T-DNA to the VirD4 substrate receptor. Both factors also inhibit binding of the VirE2 protein substrate to VirD4, as shown by coimmunoprecipitation and bimolecular fluorescence complementation assays. Osa fused to the green fluorescent protein (GFP) also blocks T-DNA and VirE2 binding to VirD4, and Osa-GFP colocalizes with VirD4 at A. tumefaciens cell poles. RSF1010 and Osa interfere specifically with VirD4 receptor function and not with VirB channel activity, as shown by (i) TrIP and (ii) a genetic screen for effects of the oncogenic suppressors on pCloDF13 translocation through a chimeric secretion channel composed of the pCloDF13-encoded MobB receptor and VirB channel subunits. Our findings establish that a competing plasmid substrate and a plasmid fertility inhibition factor act on a common target, the T4S receptor, to inhibit docking of DNA and protein substrates to the translocation apparatus.
Introduction
Agrobacterium tumefaciens uses the VirB/D4 type IV secretion (T4S) system to translocate DNA and protein effector molecules to plant cells during infection (Zhu et al., 2000; Cascales and Christie, 2003). The VirB/D4 T4S system is assembled from 11 VirB proteins, termed the mating pair formation (Mpf) proteins (Lessl et al., 1993), and the VirD4 coupling protein (T4CP) (Ding et al., 2003; Gomis-Ruth et al., 2004), also termed the substrate receptor (Atmakuri et al., 2003; Cascales and Christie, 2004a). This system is a bona fide conjugation apparatus, as shown by its capacity to deliver oncogenic transfer DNA (T-DNA) as well as the mobilizable plasmid RSF1010 (IncQ) to target cells through a cell-contact-dependent mechanism (Buchanan-Wollaston et al., 1987; Fullner, 1998). Studies of the VirB/D4 T4S system and of related conjugation systems, e.g. F (IncF), RP4 (IncPα), R388 (IncW) and pKM101 (IncN), have identified several early steps of conjugation (Lawley et al., 2004; Christie and Cascales, 2005; Shroder and Lanka, 2005). DNA transfer initiates when the relaxase and auxiliary factors bind at the origin of transfer (oriT) sequence of a DNA substrate to form the relaxosome (Pansegrau and Lanka, 1996). The relaxosome catalyses the single-stranded (ss) cleavage of the DNA strand destined for transfer, whereupon the relaxase remains covalently bound to the 5′ end of ssDNA substrate (Lessl and Lanka, 1994). The DNA substrate then docks at the T4CP via translocation signals carried by the relaxase and, for some systems, other processing components of the relaxosome (Simone et al., 2001; Schroder et al., 2002; Llosa et al., 2003; Beranek et al., 2004; Vergunst et al., 2005). Finally, the T4CP acts together with the Mpf proteins, e.g. the VirB subunits, to mediate transfer of the nucleoprotein particle across the cell envelope (Cascales and Christie, 2004a). Conjugation machines also translocate protein substrates independently of DNA. For example, the A. tumefaciens VirB/D4 T4S system delivers several effector proteins, e.g. VirE2, VirE3 and VirF, to susceptible plant cells during infection (Vergunst et al., 2000; 2005; Schrammeijer et al., 2003). As shown for the T-DNA substrate, VirE2 also docks at the VirD4 receptor for translocation through this secretion channel (Atmakuri et al., 2003).
Conjugative plasmids often encode factors that inhibit the transfer of coresident plasmids (Olsen and Shipley, 1975; Yusoff and Stanisich, 1984). Such fertility inhibition factors appear to fall into two broad classes, those acting to repress expression of transfer (tra) genes (Gasson and Willetts, 1976; Tanimoto and Iino, 1983), and those functioning at a step in the translocation pathway subsequent to Mpf subunit synthesis and assembly of the conjugation apparatus (Yusoff and Stanisich, 1984; Winans and Walker, 1985). Fertility inhibition via control of tra gene regulation has been extensively characterized for the F plasmid finOP inhibition system (van Biesen and Frost, 1994; Penfold et al., 1996; Ghetu et al., 2000). In contrast, little is known about the mechanism(s) by which fertility inhibition factors act downstream of T4S machine assembly (Lawley et al., 2004). In A. tumefaciens, two factors have been shown to render wild-type (WT) cells nearly or completely avirulent, a phenomenon termed ‘oncogenic suppression’ (Close and Kado, 1991; Ward et al., 1991). One factor is the RSF1010 transfer intermediate composed of the MobA relaxase covalently bound to the 5′ end of the transferred ssDNA (R-strand) (Ward et al., 1991; Stahl et al., 1998). The second is the Osa protein encoded by the IncW plasmid pSa (Close and Kado, 1991). Osa does not affect accumulation or membrane association of VirB proteins (Chen and Kado, 1994; 1996). Moreover, Osa shares sequence similarities to FiwA, a fertility inhibition factor of IncPα plasmids that interferes with transfer of coresident IncQ and IncW plasmids (Chen and Kado, 1994; Fong and Stanisich, 1989). It can thus be postulated that Osa represents a class of fertility inhibition factors acting after T4S machine biogenesis to block one or more steps of translocation.
Studies exploring the inhibition mechanism(s) of RSF1010 and Osa have exploited the early finding that two avirulent strains, one deleted of the T-DNA and a second lacking a gene for a protein substrate, e.g. VirE2, incite tumour formation when coinoculated onto plant wound sites (Otten et al., 1984). The simplest explanation for these findings is that these strains translocate the requisite complement of tumour-inducing T-DNA and protein substrates into the same plant cell (Christie et al., 1988; Binns et al., 1995). Such mixed infection experiments with ΔT-DNA or virE2 mutant strains harbouring RSF1010 or a plasmid producing the Osa protein supplied evidence that both suppressor factors inhibit VirE2 translocation (Binns et al., 1995; Lee et al., 1999), whereas only RSF1010 inhibits T-DNA transfer (Binns et al., 1995; Lee and Gelvin, 2004). Osa does, however, strongly inhibit intercellular transfer of other protein substrates, VirE3 and VirF (Schrammeijer et al., 2003), as shown by a Cre recombinase (CRAfT) assay (Vergunst et al., 2000).
In this study, we sought to define the mechanistic bases underlying ‘oncogenic suppression’. We present evidence that the RSF1010 substrate follows the same translocation pathway through the VirB/D4 secretion channel as recently described for the T-DNA substrate (Cascales and Christie, 2004a). We further report that the RSF1010 transfer intermediate and Osa suppress A. tumefaciens oncogenesis specifically by interfering with T-DNA and VirE2 substrate binding to the VirD4 receptor. The promiscuous plasmid RSF1010 and the pSa-encoded Osa fertility inhibition factor thus act on a common target, the T4S receptor, to inhibit substrate transfer through T4S machines.
Results
Plasmid RSF1010 (IncQ) follows the same translocation route as T-DNA through the VirB/D4 T4S system
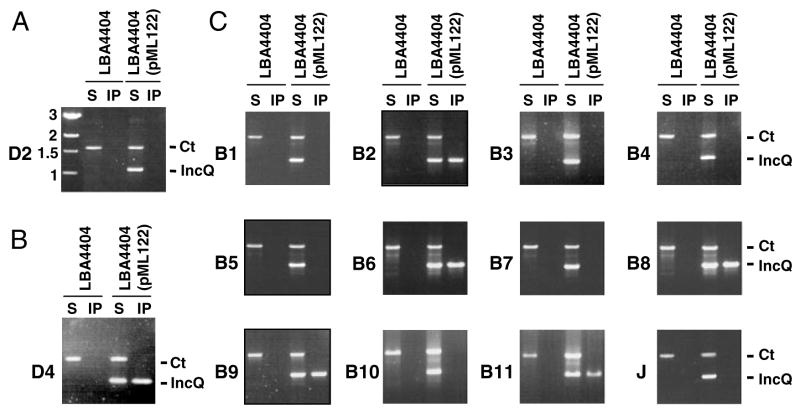
The T-DNA transfer pathway has been depicted as a series of spatially and temporally ordered close contacts with six VirB/D4 channel subunits – VirD4, VirB11, VirB6, VirB8, VirB2 and VirB9, as shown with the Transfer DNA ImmunoPrecipitation (TrIP) assay (Cascales and Christie, 2004a). We first tested whether a mobilizable RSF1010 (IncQ) derivative, pML122 (Fullner, 1998), establishes similar channel subunit contacts during translocation through the VirB/D4 T4S system. To avoid possible competitive effects of a coresident T-DNA substrate, TrIP assays were carried out with A. tumefaciens strain LBA4404 (ΔT-DNA) harbouring pML122. Intact cells were treated with formaldehyde (FA) and detergent solubilized, and then material precipitated with antibodies to the Vir proteins was subjected to polymerase chain reaction (PCR) amplification to detect coprecipitation of cross-linked channel subunit–pML122 substrate complexes. As a non-substrate DNA control, we included primers for amplification of the repAB locus from pTiAch5 in the PCR amplification reaction. We further confirmed that antibodies to the VirD2 relaxase, which is not required for RSF1010 transfer (Buchanan-Wollaston et al., 1987; Fullner, 1998), did not precipitate detectable levels of the pML122 or pTiAch5 plasmid fragments from LBA4404 or LBA4404(pML122) cell extracts (Fig. 1A).
Fig. 1. An RSF1010 derivative forms close contacts with six VirB/D4 subunits during translocation. Strain LBA4404 (ΔT-DNA) with or without pML122 was assayed for formation of pML122 contacts with VirB/D4 channel subunits by TrIP.
A. Control experiment showing PCR amplification of pTiAch5 repAB (Ct) and pML122 mobA (IncQ) fragments in the soluble fraction (S) but not material immunoprecipitated (IP) with anti-VirD2 antibodies from LBA4404 extracts.
B and C. Antibodies to the VirD4 receptor (panel B) and the VirB/VirJ subunits listed on the left (panel C) were used to immunoprecipitate material from extracts of FA-treated LBA4404 and LBA4404(pML122) cells. Both the repAB control (Ct) and pML122 mobA (IncQ) fragments were detected in solubilized material (S) by PCR amplification. The mobA fragment was detected only in the material precipitated with antibodies to VirD4, VirB2, VirB6, VirB8, VirB9 and VirB11.
Antibodies to the six putative VirB/D4 channel subunits named above specifically precipitated the mobA fragment carried by the pML122 substrate from extracts of the FA-treated cells (Fig. 1B and C). This fragment was undetectable in material precipitated with antibodies to the remaining six VirB proteins – VirB1, VirB3, VirB4, VirB5, VirB7, VirB10 – or to another postulated machine component VirJ (Pantoja et al., 2002) (Fig. 1C). This profile of RSF1010 substrate–channel subunit contacts matched that obtained previously for the T-DNA substrate (Cascales and Christie, 2004a). Further TrIP studies established that individual virB and virD4 null mutations blocked translocation of the pML122 substrate at the same stages as determined previously for the T-DNA (Cascales and Christie, 2004a; data not shown). We therefore conclude that the T-DNA and IncQ plasmid transfer intermediates follow the same route during translocation through the VirB/D4 secretion channel.
The IncQ plasmid inhibits T-DNA binding to the VirD4 receptor
Wild-type A. tumefaciens strain A348 incites abundant tumour formation on Kalanchoe daigremontiana leaves, whereas the isogenic strain harbouring an RSF1010 replicon is almost avirulent (Fig. 2A; Ward et al. 1991; Binns et al., 1995). In view of the above evidence that T-DNA and pML122 substrates share the same VirB/D4 translocation pathway, we tested whether pML122 inhibits one or more steps of T-DNA translocation. These steps include: (i) T-DNA processing by the VirD2 relaxase, (ii) T-DNA docking at the VirD4 receptor and (iii) T-DNA transfer through the VirB/D4 secretion channel. Substrate processing and channel subunit contacts were monitored by TrIP and the modified QTrIP assay (Cascales and Christie, 2004a). This latter assay is based on radiolabelling of PCR products generated from immunoprecipitated material to quantify levels of recovered substrate (see Experimental procedures).
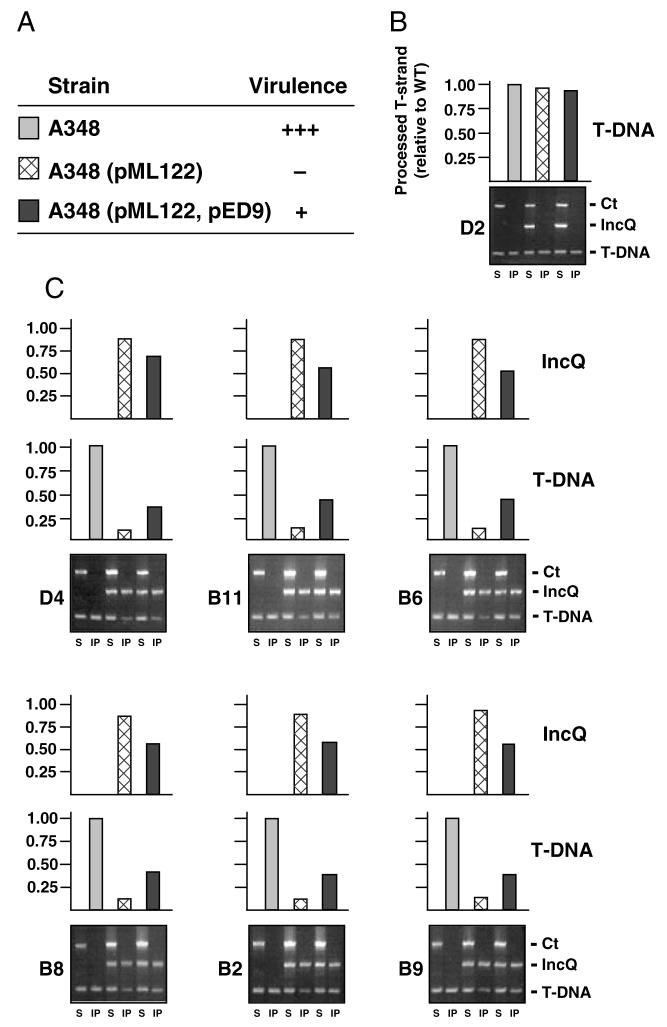
Fig. 2. The pML122 IncQ plasmid substrate blocks formation of T-DNA close contacts with VirB/D4 channel subunits.
A. Inoculated Kalanchoe daigremontiana leaves were scored for tumour formation on a scale of no tumours (−) to WT tumours (+++) 5 weeks after inoculation with WT A348, A348(pML122), or A348(pML122, pED9). The latter strain overproduces VirB9, VirB10 and VirB11 from an IncP replicon.
B. Effects of pML122 and VirB9, VirB10 and VirB11 overproduction on formation of the T-DNA transfer intermediate. Antibodies to VirD2 relaxase precipitated equivalent amounts of the T-DNA substrate from extracts of A348 strains listed in A, as shown by TrIP (bottom) and QTrIP (top). TrIP: The ophDC control (Ct), pML122 mobA (IncQ) and gene 7 (T-DNA) fragments were detected by PCR amplification from soluble fractions (S) and immunoprecipitates (IP) of A348, A348(pML122) and A348(pML122, pED9). QTrIP: Levels of processed T-DNA (T-strand) in the immunoprecipitates were quantitated as described previously (Cascales and Christie, 2004a). Results are presented as a fraction of the T-DNA detected in the immunoprecipitates from WT A348 (normalized to 1.0).
C. Antibodies to the VirD4 receptor and the five putative VirB channel subunits presented at the left of each TrIP panel were used to immunoprecipitate material from extracts of strains listed in A. TrIP: Detection of PCR amplification products corresponding to ophDC (Ct), pML122 and T-DNA fragments from soluble fractions (S) and immunoprecipitates (IP) recovered from the three strains listed in A. QTrIP: Levels of processed T-DNA (middle histograms) and pML122 (top histograms) in the immunoprecipitates, presented as described in B.
Anti-VirD2 antibodies precipitated similar levels of the T-DNA transfer intermediate from A348 and A348(pML122) cells, showing that the IncQ plasmid does not affect VirD2 processing at T-DNA border repeat sequences (Fig. 2B). In striking contrast, the anti-VirD4 antibodies precipitated a very low level of T-DNA substrate from extracts of A348(pML122) compared with A348. The level of T-DNA precipitated with the VirD4 receptor in the presence of the IncQ plasmid was approximately one-tenth that in its absence, as determined by QTrIP (Fig. 2C). Of further interest, the anti-VirD4 antibodies precipitated an abundant amount of pML122 from A348(pML122), approximately nine times that of precipitated T-DNA (Fig. 2C).
The above findings suggest that the pML122 substrate outcompetes the T-DNA substrate for binding to the VirD4 receptor (Fig. 2C). Next, we sought to determine whether pML122 additionally disrupts passage of the T-DNA through the VirB/D4 secretion channel. However, antibodies to VirB11, VirB6, VirB8, VirB2 and VirB9 precipitated T-DNA and pML122 at approximately the same 1 to 9 ratio as obtained with antibodies to VirD4 (Fig. 2C). The IncQ plasmid thus appears to specifically disrupt T-DNA substrate-receptor binding without further downstream effects on T-DNA translocation. Supporting genetic evidence that the inhibitory effect of RSF1010 is restricted to substrate docking at the T4S receptor is presented below.
VirB9, VirB10 and VirB11 overproduction confer altered substrate selection
Ward et al. (1991) reported that the suppressive effects of RSF1010 can be partially reversed by the co-overproduction of VirB9, VirB10 and VirB11 (Fig. 2A). In agreement with this finding, antibodies to VirD4 and each of the five presumptive VirB channel subunits precipitated T-DNA at appreciably higher levels from extracts of the VirB9-VirB11 overproducing strain A348(pML122, pED9) than from A348(pML122) (Fig. 2C). Of further interest, these antibodies precipitated the pML122 substrate from A348(pML122, pED9) at diminished levels compared with A348(pML122). In Fig. 2C, the amount of T-DNA or RSF1010 substrate recovered from each strain is presented as a fraction of total T-DNA recovered with the same antibody from WT A348. Whereas the amount of total substrate (T-DNA plus RSF1010) recovered from A348(pML122) approximated the level of T-DNA from A348, the amount of total substrate recovered from A348(pML122, pED9) was reproducibly approximately 5% higher than from the WT strain (data not shown). This finding is consistent with an earlier proposal that VirB9, VirB10 and VirB11 are rate limiting for assembly of functional VirB/D4 T4S machines (Ward et al., 1991). Yet, this small increase in substrate binding does not account for the pronounced approximately threefold increase in T-DNA binding and corresponding decrease in pML122 binding to the VirD4 and VirB channel subunits upon VirB overproduction (Fig. 2C). These findings and results of mutational analyses (see below) support a proposal that VirB proteins act in conjunction with VirD4 to modulate substrate selection and trafficking through the secretion channel (see Discussion).
The IncQ transfer intermediate inhibits T-DNA binding to the VirD4 receptor
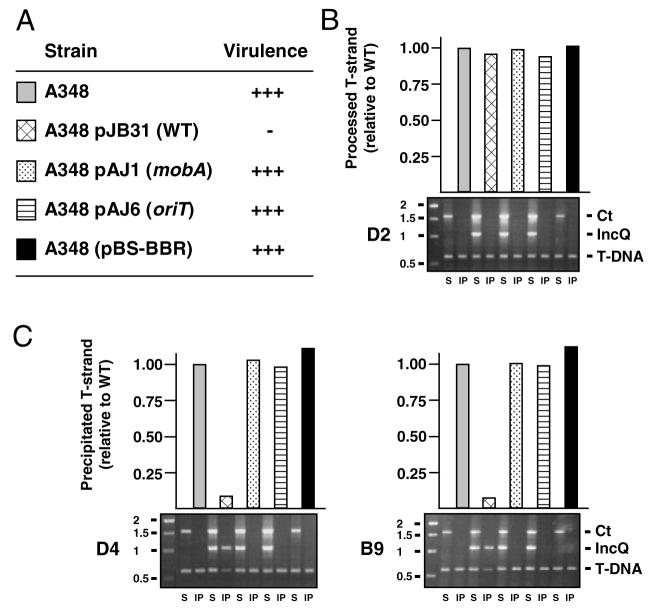
Processing at the oriT sequence by the MobA relaxase of RSF1010 is required for suppression of A. tumefaciens virulence (Fig. 3A; Stahl et al. 1998). In A348 cells harbouring the RSF1010 derivative pJB31, the T-DNA transfer intermediate accumulates at WT levels, establishing that the IncQ plasmid processing reaction does not disrupt processing by VirD2 relaxase at the T-DNA border sequences (Fig. 3B). In these cells, however, the T-DNA substrate binds VirD4 at only 5–10% of WT levels, indicative of a nearly complete block in substrate-receptor binding (Fig. 3C). In A348 strains harbouring the isogenic IncQ plasmids pAJ1 and pAJ6 bearing mutations in oriT and mobA, respectively (Stahl et al., 1998), the T-DNA bound VirD4 at WT levels (Fig. 3C). These findings establish that formation of the MobA-R-strand transfer intermediate is essential for the observed block in T-DNA substrate access to the VirD4 receptor.
Fig. 3. The IncQ plasmid transfer intermediate inhibits T-DNA transfer.
A. Kalanchoe daigremontiana leaves were scored for tumour formation on a scale of no tumours (−) to WT tumours (+++) 5 weeks after inoculation with WT A348 strain carrying RSF1010 plasmid pJB31 or the isogenic derivative plasmids pAJ1 (mobA) or pAJ6 (oriT), or control plasmid pBS-BBR.
B. Effects of the IncQ plasmid and derivatives on T-DNA processing, as monitored by precipitation of the VirD2-T-strand substrate with antibodies to VirD2 from extracts of the strains listed in A. Bottom: TrIP data showing amplification of ophDC (Ct), pML122 mobA (IncQ) and gene 7 (T-DNA) fragments from soluble fractions (S) and immunoprecipitates (IP). DNA size markers (kb) are at the left. Top: QTrIP data showing levels of T-DNA recovered from the various strains as a fraction of T-DNA substrate from WT A348 (normalized to 1.0).
C. Effects of the IncQ plasmid and derivatives on T-DNA transfer. Lower panel: TrIP data showing amplification of pTiAch5 repAB (Ct), pML122 mobA (IncQ) or T-DNA. DNA size markers (kb) are at the left. Upper panel: QTrIP data showing levels of T-DNA in the immunoprecipitates recovered with antibodies to VirD4 (left panels) and VirB9 (right panels).
Osa blocks T-DNA access to the VirD4 receptor
As mentioned above, the Osa protein encoded by plasmid pSa (IncW) also functions as an oncogenic suppressor (Chen and Kado, 1994; Lee et al., 1999). By use of the mixed infection assay, it was reported that Osa does not inhibit delivery of T-DNA to plant cells (Lee et al., 1999; Lee and Gelvin, 2004). However, in parallel studies in both of our laboratories, we have reproducibly detected strong suppressive effects of Osa on T-DNA transfer upon inoculation of K. daigremontiana leaf wounds as well as tobacco leaf discs. As summarized in Fig. 4A and Table 1, A348(pSa) expressing osa from its native promoter or A348(pUCD3960) expressing osa from an nptI promoter displayed highly attenuated virulence. In the mixed infection studies, coinoculation of LBA4404 (ΔT-DNA) with three independently derived virE2 null mutants [At12516, virE2::carb and Mx358 (data not shown)] yielded tumour formation on both K. daigremontiana and tobacco leaf discs. In striking contrast, coinoculation of LBA4404 with these virE2 mutant strains carrying the osa-expressing pSa or pUCD3960 plasmids produced no tumours or only a few small tumours. In these assays, the virE2 mutant strains represent the only source for the oncogenic T-DNA, indicating that the Osa protein at least partially blocks translocation of the T-DNA. Results of our studies examining the effects of Osa on VirE2 translocation are presented below.
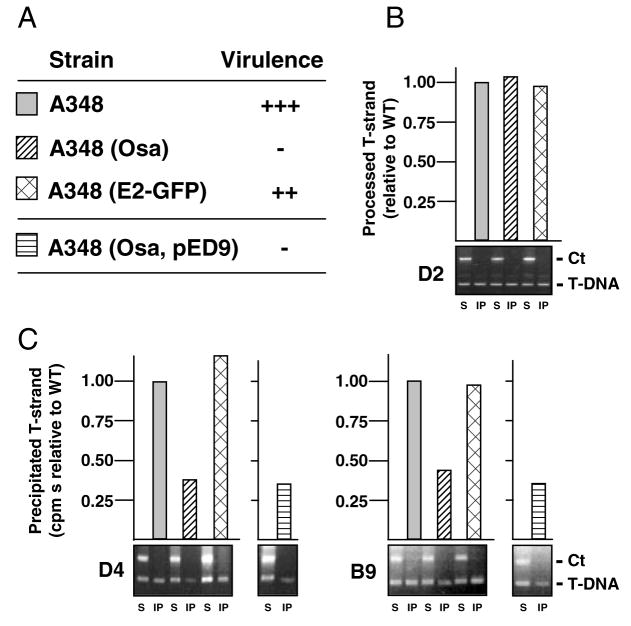
Fig. 4. The pSa Osa protein blocks VirD4 receptor access to the T-DNA.
A. Kalanchoe daigremontiana leaves were scored for tumour formation on a scale of no tumours (−) to WT tumours (+++) 5 weeks after inoculation with WT A348, or A348 derivatives producing Osa from plasmid pUCD3960 (Osa) or VirE2-GFP (E2-GFP) from plasmid pXZ66, or overproducing VirB9, VirB10 and VirB11 from pED9.
B. Effects of Osa and VirE2-GFP production on T-DNA processing, as monitored by precipitation of the T-DNA substrate with antibodies to VirD2 from extracts of the strains listed in A. For the QTrIP data presented in the upper histogram, the amounts of T-DNA recovered from the various strains are presented as a fraction of T-DNA recovered from WT A348 (normalized to 1.0).
C. Effects of Osa and VirE2-GFP production on T-DNA transfer. Lower panels: TrIP data showing amplification of ophDC (Ct) and gene 7 (T-DNA) fragments from soluble fractions (S) and immunoprecipitates (IP) with antibodies to VirD4 (left) or VirB9 (right). Upper panels: QTrIP data showing levels of T-DNA recovered from the various strains.
Table 1.
Effects of pML122 plasmid and Osa production on T-DNA and VirE2 translocation assessed by plant mixed infection assays.a
| T-strand donorb | VirE2 donorc | K. daigremontiana leaves | Tobacco leaf discs |
|---|---|---|---|
| virE2 | – | – | 0 |
| – | ΔT-DNA | – | 0 |
| virE2 | ΔT-DNA | +++ | 4.1 ± 0.6 |
| virE2 (pML122)d | – | – | 0.2 ± 0 |
| – | ΔT-DNA (pML122) | – | 0 |
| virE2 (pML122) | ΔT-DNA | + | 0 |
| virE2::carb (pML122) | ΔT-DNA | + | 0.2 ± 0.1 |
| virE2 | ΔT-DNA (pML122) | – | 0.1 ± 0 |
| virE2 (pUCD3960)e | – | – | 0 |
| – | ΔT-DNA (pUCD3960) | – | 0 |
| virE2 (pUCD3960) | ΔT-DNA | + | 0 |
| virE2::carb (pUCD3960) | ΔT-DNA | + | 0.2 ± 0.1 |
| virE2 | ΔT-DNA (pUCD3960) | – | 0 |
| virE2 (pSa)f | – | – | 0 |
| – | ΔT-DNA (pSa) | – | 0 |
| virE2 (pSa) | ΔT-DNA | + | 0 |
| virE2::carb (pSa) | ΔT-DNA | + | 0.9 ± 0.3 |
| virE2 | ΔT-DNA (pSa) | – | 0 |
T-DNA and VirE2 donor strains were coinoculated on wounded K. daigremontiana leaves or tobacco leaf discs as described in the Experimental procedures. Results are expressed as sizes of tumours on K. daigremontiana leaves 5 weeks after inoculation [on a scale of no tumour (−) to WT tumour (+++)], or as an average number of tumours arising along the edges of tobacco 16 leaf pieces.
virE2 – strain At12516 (Fullner, 1998) or virE2::carb (this study).
ΔT-DNA – strain LBA4404 (Ooms et al., 1982).
pML122, a derivative of RSF1010 (IncQ) (Fullner, 1998).
pUCD3960, produces Osa from a constitutive nptI promoter (Chen and Kado, 1996).
pSa, produces Osa from its native promoter (Close and Kado, 1991).
By use of the QTrIP assay, we determined that A348(pUCD3960) cells accumulate WT levels of the T-DNA transfer intermediate (Fig. 4B), but appreciably lower amounts of the putative T-DNA substrate–VirD4 receptor complex (Fig. 4C). In this Osa-producing strain, the T-DNA bound the receptor at 30–40% of WT levels (Fig. 4C). Similarly, in this strain the T-DNA substrate bound to VirB11, VirB6, VirB8, VirB2 and VirB9 at 30–40% of WT levels (Fig. 4C and data not shown). It is noteworthy that, in contrast to pML122 (Fig. 3C), Osa does not completely block formation of these T-DNA–channel subunit contacts, yet Osa-producing cells are nearly avirulent (Fig. 4A). Conceivably, Osa disrupts not only the T-DNA-receptor binding reaction, but also interferes with DNA transfer kinetics, a dynamic reaction that cannot be measured with the QTrIP assay. Additionally or alternatively, Osa’s strong ‘oncogenic suppression’ phenotype could be due to its ability to block VirE2 protein substrate binding to the VirD4 receptor (see below).
Overproduction of VirB9, VirB10 and VirB11 does not reverse Osa-mediated oncogenic suppression (Fig. 4A; Lee et al. 1999). Correspondingly, VirB9-VirB11 overproduction did not correlate with an increase in T-DNA binding to VirD4 or the VirB channel subunits in the presence of Osa, as shown by QTrIP (Fig. 4C and data not shown). These findings and additional data (see below), suggest that the pML122 transfer intermediate and Osa exert their effects on a common target, the VirD4 receptor, but most probably through different mechanisms.
Previously, we reported that virE2 fused at its 3′ end to the gene for green fluorescent protein (GFP) exerts negative dominance when expressed in A348 cells (Fig. 4A) (Zhou and Christie, 1999). In mixed infection experiments, virE2-GFP expression also was reported to suppress both T-DNA and VirE2 translocation (Zhou and Christie, 1999). However, TrIP and QTrIP studies did not reveal any effects of VirE2-GFP on T-DNA binding to the VirD4 receptor or VirB channel subunits (Fig. 4C and data not shown). We also have attempted, without success, to suppress dominance of virE2-GFP by overproduction of the VirB/D4 channel subunits, VirE2, or VirE1, a secretion chaperone for VirE2 (Zhao et al., 2001). On the basis of these findings, we suggest that VirE2-GFP acts very early in the translocation pathway, even prior to substrate-receptor docking, to disrupt translocation of VirE2.
The IncQ conjugative intermediate and Osa inhibit VirE2 access to the VirD4 receptor
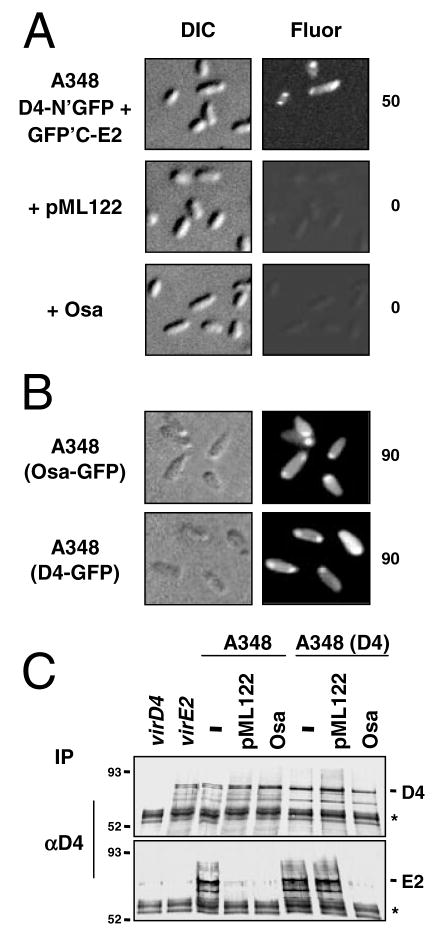
The RSF1010 transfer intermediate and Osa also block translocation of the VirE2 effector to plant cells (Table 1; Stahl et al., 1998; Lee et al., 1999). We next tested whether these suppressor factors interfere with the VirE2 substrate–VirD4 receptor contact. These studies made use of the bimolecular fluorescence complementation (BiFC) assay (Hu et al., 2002) adapted in our laboratory for in vivo studies of the VirE2–VirD4 docking reaction (Atmakuri et al., 2003). In this assay, VirE2 and VirD4 are tagged with half-length GFP molecules, such that the VirE2–VirD4 interaction serves to align the half-GFP’s for reconstitution of the active fluorescent protein (Atmakuri et al., 2003). Cells coproducing VirD4-N′GFP and C′GFP-VirE2 fluoresce at the cell poles (Fig. 5A), in agreement with positional information reported previously for VirD4 (Kumar and Das, 2002) and the VirE2–VirD4 complex (Atmakuri et al., 2003). In striking contrast, the isogenic cells carrying pML122 or osa-expressing pUCD3960 failed to display any detectable BiFC (Fig. 5A). As shown above for the T-DNA substrate, the suppressor factors similarly block access of the VirE2 protein substrate to the VirD4 receptor.
Fig. 5. The IncQ plasmid substrate and Osa disrupt VirE2 substrate–VirD4 receptor binding.
A. A348 strains coproduced VirD4-N′-GFP and GFP′C-VirE2 from plasmids pKA64 and pZD89, respectively, and harboured pML122 (IncQ plasmid) or pUCD3960 for Osa production as denoted at the left. Cells were examined at 12 h after vir gene induction by Nomarski (DIC) microscopy and fluorescence (Fluor) microscopy for detection of bimolecular fluorescence complementation (BiFC). Number to the right of each panel represents the percentage of cells with polar fluorescence among 104 cells examined.
B. Colocalization of Osa-GFP and VirD4-GFP at the cell poles. A348 (WT) cells producing Osa-GFP from pKA167 (this study) or VirD4-GFP from pKA62 (Atmakuri et al., 2003) were photographed 12 h after vir gene induction by Nomarski (DIC) and fluorescence (Fluor) microscopy. Number to the right of each panel represents the percentage of cells with polar fluorescence among 104 cells examined.
C. Coimmunoprecipitation of VirE2–VirD4. Isolated membranes were cross-linked with DSP and detergent-solubilized complexes were subjected to immunoprecipitation with antibodies to VirD4. Immunoblots were developed with antibodies listed at the right of the top two panels. Strains: Mx355 (virD4), At12516 (virE2), WT A348 (−), A348(D4) VirD4 is overproduced from pKA21. The latter strains also harboured pML122 (IncQ plasmid) or pUCD3960 for Osa production. Molecular weight markers are listed at the left of each panel. Asterisk, crossreactive IgG heavy chain. Except for the virE2 mutant, the A348 derivatives accumulated similar steady-state levels of VirE2 (data not shown); VirD4 overproduction in A348(pKA21) has been documented previously (Atmakuri et al., 2003).
These findings prompted a cytological test for colocalization of Osa and VirD4. As shown previously, VirD4 and VirD4-GFP localize at A. tumefaciens cell poles (Fig. 5B) (Kumar and Das, 2002; Atmakuri et al., 2003). We fused Osa at its C terminus to GFP, and determined that the fusion protein is stable in A. tumefaciens and phenotypically resembles the native protein by suppressing virulence, interfering with T-DNA and VirE2 binding to VirD4, and blocking pML122 transfer (data not shown). Very interestingly, Osa-GFP targeted to the A. tumefaciens poles, forming fluorescence patterns very similar to those of VirD4-GFP (Fig. 5B). Osa-GFP also targeted to poles of mutant strains deleted of VirD4, the VirB subunits, or the VirD2 relaxase, suggesting that Osa positioning is independent of T-DNA processing or polar localization of the VirB/D4 machine (data not shown). Moreover, native Osa did not affect polar targeting of VirD4-GFP (data not shown), excluding a mechanism whereby Osa interferes with targeting of the receptor to its site of action.
We next assayed for effects of Osa and pML122 on precipitation of a VirD4–VirE2 complex from A. tumefaciens cell extracts (Atmakuri et al., 2003). As shown in Fig. 5C, the anti-VirD4 antibodies precipitated abundant levels of VirD4 and VirE2 from extracts of WT A348 cells, but only a barely detectable level of VirE2 from extracts of isogenic strains carrying either pML122 or pUCD3960. These findings comprise further evidence that the suppressor factors act by inhibiting the VirE2–VirD4 binding reaction.
We also asked whether overproduction of the VirD4 receptor could reverse the inhibitory effects of pML122 or Osa. Not surprisingly, elevated receptor synthesis in the VirD4-overproducing strain A348(pKA21) was correlated with a ~5–10% increase in the amount of T-DNA precipitated with anti-VirD4 antibodies as compared with WT A348 cells. However, the QTrIP studies revealed no corresponding increase in T-DNA–VirB channel subunit contacts, in agreement with evidence presented above (Fig. 2) that one or more VirB subunits, not VirD4, is rate-limiting for assembly of the secretory apparatus. The VirD4 antibodies also coprecipitated VirD4 and VirE2 from extracts of A348(pKA21, pML122) (Fig. 5C), establishing that VirD4 overproduction also enabled detection of VirE2-receptor binding in the presence of the IncQ plasmid. In striking contrast, VirD4 overproduction in strain A348(pKA21, pUCD3960) did not reverse the inhibitory effect of Osa on VirE2–VirD4 complex formation (Fig. 5C). VirD4 overproduction therefore reversed the inhibitory effects of pML122 but not Osa on VirE2–VirD4 receptor binding.
Osa inhibits IncQ plasmid access to the VirD4 receptor
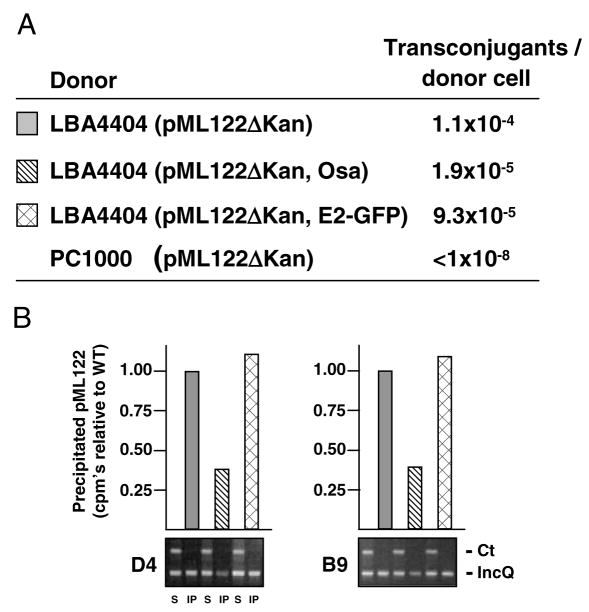
Osa also inhibits conjugative transfer of pML122 to agro-bacterial recipients (Fig. 6A; Lee and Gelvin, 2004). As assessed by QTrIP, Osa interfered with pML122 binding to the VirD4 receptor without further negative effects on substrate binding to the VirB subunits, as illustrated for VirB9 (Fig. 6B and data not shown). The magnitude of the Osa inhibitory effect – approximately 1 log reduction in conjugative transfer frequency and 65% reduction in pML122 binding to the VirD4 receptor as shown by QTrIP – correlate with each other and with the observed effect of Osa on the T-DNA substrate-receptor binding reaction (see Fig. 4B). Also in agreement with data presented above for the T-DNA substrate, VirE2-GFP did not affect pML122 transfer or VirD4 receptor binding (Fig. 6A and B). It is noteworthy that the strains characterized in this study carry two T4S systems in addition to the VirB/D4 T4S system (Hamilton et al., 2000; Chen et al., 2002). However, these T4S machines are not synthesized or functional under conditions required for vir gene expression and VirB/D4-mediated translocation, as shown previously and here by lack of detectable pML122 transfer by a PC1000 (ΔvirB operon) donor strain (Fig. 6A; Fullner, 1998; Lee and Gelvin, 2004). Thus, Osa inhibits conjugative transfer of pML122 specifically by interfering with substrate binding to the VirD4 receptor.
Fig. 6. The pSa Osa protein blocks VirD4 receptor access to the pML122.
A. Conjugative transfer frequencies of LBA4404 (ΔT-DNA) donor strains carrying IncQ plasmid pML122ΔKan without or with plasmids producing Osa protein (pUCD3960) or VirE2-GFP (pXZ66). Donors were mated with either WT A348 carrying plasmid pBBR1MCS (Strr). PC1000 (ΔvirB operon) carrying pML122ΔKan served as a donor in a control mating. Data are presented for a representative experiment.
B. Effects of the IncQ plasmid and derivatives on T-DNA transfer. Lower panel: TrIP data showing amplification of pTiAch5 repAB (Ct) and pML122 mobA (IncQ). Upper panel: QTrIP data showing levels of pML122 in the immunoprecipitates recovered with antibodies to VirD4 (left panels) and VirB9 (right panels).
The IncQ transfer intermediate and Osa do not affect VirD4-independent, VirB-dependent pCloDF13 transfer
Agrobacterium tumefaciens mobilizes the transfer of another plasmid, pCloDF13, to recipient cells (Escudero et al., 2003). pCloDF13 encodes its own VirD4 homologue, MobB, but lacks genes for the VirB Mpf subunits, and therefore transfer is dependent on assembly of a chimeric T4S system composed of the MobB receptor and VirB channel subunits. To determine whether RSF1010 and Osa act specifically to inhibit VirD4 receptor activity, we assayed for effects of these suppressor factors on pCloDF13 mobilization.
Table 2 shows that WT A348 and the virD4 null mutant Mx355 transferred the pCloDF13 derivative pJClo (See Experimental procedures) to agrobacterial recipients. Mx355 with or without pJClo failed to transfer pML122, showing that VirD4 is required for mobilization of the IncQ plasmid (Table 2; Fullner, 1998). Very interestingly, both A348 and Mx355 donor cells carrying pJClo and either pML122 or pUCD3960 (Osa producer) transferred pJClo at frequencies comparable to cells lacking the oncogenic suppressors. These findings show that the IncQ plasmid and Osa specifically inhibit VirD4 receptor activity without affecting activities of the MobB receptor or the VirB channel subunits.
Table 2.
Mobilization of pCloDF13 and RSF1010 derivatives between Agrobacterium strains.a
| Transfer of: |
||||
|---|---|---|---|---|
| RSF1010 |
CloDF13 |
|||
| Donorb | C58C1RSc | UIA143c | C58C1RS | UIA143 |
| A348 (pML122d) | 2.3 × 10−4 | 1.7 × 10−6 | NA | NA |
| A348 (pJClo) | NA | NA | 6.4 × 10−5 | 4.1 × 10−7 |
| A348 (pJClo, pML122d) | 8.9 × 10−5 | 5.8 × 10−7 | 2.1 × 10−5 | 1.3 × 10−7 |
| A348 (pJClo, pUCD3960f) | NA | NA | 7.9 × 10−5 | 4.9 × 10−7 |
| Mx355 (pML122e) | < 10−8 | < 10−8 | NA | NA |
| Mx355 (pJClo) | NA | NA | 0.9 × 10−4 | 6.8 × 10−7 |
| KA2000 (pML122d) | < 10−8 | < 10−8 | NA | NA |
| KA2000 (pJClo) | NA | NA | 8.1 × 10−5 | 6.0 × 10−7 |
| KA2000 (pJClo pML122e) | < 10−8 | < 10−8 | 7.7 × 10−5 | 5.4 × 10−7 |
| KA2000 (pJClo, pUCD3960f) | NA | NA | 7.4 × 10−5 | 5.7 × 10−7 |
Matings were carried out on a solid surface for 4 days at 20°C, as described in the Experimental procedures. Results are expressed as transconjugants per donor (Tc’s/donor).
Donor cells: WT A348; Mx355 or KA2000 (two independent virD4 mutants).
Recipient cells: C58C1RS (carries Ti plasmid which stimulates plasmid DNA acquisition); UIA143 (Ti plasmidless). Mobilizable plasmids:
pML122ΔKan;
pML122ΔGen; pJClo, a pCloDF13 derivative.
pUCD3960, constitutively synthesizes Osa protein.
NA, not applicable.
Discussion
In this study, a combination of biochemical, cytological and genetic findings established that two ‘oncogenic suppressors’ act on a common target, the VirD4 receptor, to block passage of the T-DNA and the VirE2 effector protein through the A. tumefaciens VirB/D4 T4S system. One suppressor factor, RSF1010 (IncQ), is a promiscuous plasmid that parasitizes the VirB/D4 system as well as many other T4S systems of Gram-negative bacteria (Rawlings and Tietze, 2001). Studies have shown that TrIP and the semi-quantitative version of this assay permit refined mechanistic studies of T4S machines (Atmakuri et al., 2004; Cascales and Christie, 2004a,b; Jakubowski et al., 2005). Here, by TrIP we provided evidence that the RSF1010 substrate follows the same pathway through the VirB/D4 T4S system as previously shown for the T-DNA (Cascales and Christie, 2004a). Moreover, in strains producing both DNA substrates, the IncQ transfer intermediate formed close contacts with the channel subunits at levels eight to nine times higher than the T-DNA substrate. The copy numbers of RSF1010 and the Ti plasmid in A. tumefaciens are estimated at 10–20 and 1–2 respectively (Binns et al., 1995). This correlation between substrate binding and plasmid copy number suggests the RSF1010 substrate accumulates in stoichiometric excess and probably outcompetes the T-DNA for available receptors. By contrast, data presented here and elsewhere (see below) suggest that the second suppressor factor, the Osa protein of pSa (IncW), blocks VirD4 receptor activity not as a competing substrate but rather through a mechanism shared by related fertility inhibition factors encoded by IncP, W and N conjugative plasmids.
Although we favour a model in which RSF1010 competitively blocks T-DNA access to VirD4, our studies also have supplied evidence that the VirB channel subunits influence receptor activity as well as trafficking through the T4S system. Strikingly, cells engineered to overproduce VirB9, VirB10 and VirB11 displayed a pronounced shift in favour of T-DNA versus RSF1010 binding to VirD4 and the VirB channel subunits (Fig. 2). The VirB overproducing strain might assemble elevated numbers of channel complexes compared with the WT strain, as suggested by an overall increase in levels of substrate binding to channel subunits. However, this slight increase, estimated at approximately 5% by QTrIP, by itself cannot account for the shift in the ratio of T-DNA versus RSF1010 substrate binding. On the basis of these findings, we suggest that overproduction of these VirB proteins preferentially stimulates the VirD4–T-DNA binding reaction. Cells overproducing only one of these subunits, VirB9, VirB10 or VirB11, still display RSF1010 suppression of T-DNA transfer (Ward et al., 1991; data not shown), further indicating that these VirB proteins function together as a subcomplex to modulate receptor activity. Consistent with this view, elsewhere we have presented evidence for a network of VirD4, VirB9, VirB10 and VirB11 interactions in directing substrate translocation across the A. tumefaciens cell envelope. Specifically, ATP utilization by VirD4 and VirB11 induces a conformational switch in VirB10 that enables stable complex formation with the outer membrane-associated VirB9. In turn, the T-DNA substrate passes from the inner membrane to the periplasmic and outer membrane portions of the secretion channel (Cascales and Christie, 2004b). Such dynamic interactions between VirB9, VirB10, VirB11 and VirD4 conceivably also regulates early substrate recruitment and receptor binding reactions.
Additional evidence that VirB subunits contribute to substrate selection derives from mutational studies. Small peptide insertions in both VirB9 and VirB11 were shown to confer a substrate discrimination phenotype, such that strains producing the mutant proteins translocate either but not both of the T-DNA or RSF1010 substrates (Sagulenko et al., 2001; Jakubowski et al., 2005; Cascales and Christie, unpublished data). In principle, such mutations could modulate substrate selection through effects on VirD4 receptor activity. However, at least for VirB9 the QTrIP studies instead supplied evidence that the substrate discrimination mutations do not affect the efficiencies of RSF1010 and T-DNA binding to VirD4 or early reactions leading to transfer across the inner membrane. Rather, the mutations selectively block passage of one but not the second DNA substrate through the distal portion of the channel comprised of VirB2 and VirB9. Consequently, VirB9 is postulated to function as a checkpoint to regulate delivery of DNA substrates through the periplasm to the cell surface. Given that these DNA transfer intermediates correspond to the VirD2 and MobA relaxases covalently bound to their respective ssDNA substrates, VirB9 might regulate substrate trafficking through recognition of novel translocation signals carried by the two relaxases (Jakubowski et al., 2005).
Finally, it is noteworthy that RSF1010 also blocks access of the VirE2 effector protein to the VirD4 receptor. This is in spite of the fact that WT A. tumefaciens cells synthesize VirE2 in far greater excess than the RSF1010 transfer intermediate (Binns et al., 1995; Zhou and Christie, 1999; Zhao et al., 2001). Conversely, abundant synthesis of native VirE2 or even of VirE2-GFP, a protein that could conceivably jam into the secretion channel due to rapid folding kinetics of GFP, does not affect transfer frequencies of either RSF1010 or T-DNA (Figs 4 and 6; Lee and Gelvin, 2004). These observations strongly suggest that RSF1010 inhibits VirE2 translocation by a mechanism other than simple competitive inhibition. It is also noteworthy that RSF1010 blocks access of both DNA and protein substrates to VirB/D4 channel complexes, whereas native substrates of this T4S system, e.g. T-DNA and VirE2, appear to have coevolved to minimize competition for available channels. Overall, these observations raise intriguing questions relating to substrate selection by individual VirB/D4 channel complexes. Is a given channel competent to translocation both DNA and protein substrates and, if so, what reactions temporally regulate recruitment and translocation of these substrates? Alternatively, is a channel competent to translocation one type of substrate – DNA or protein – and, if so, what factors govern the biogenesis and function of such dedicated systems? In future studies, it will be of interest to explore mechanistic differences underlying T4S-dependent trafficking of DNA versus protein substrates.
In contrast to RSF1010, pSa-encoded Osa most probably is not a secretion substrate of the VirB/D4 T4S system, as judged by the absence of a characteristic C-terminal translocation signal (Vergunst et al., 2005) and experimental evidence that Osa localizes at the inner membrane (Chen and Kado, 1996). Additionally, Osa does not affect T-DNA substrate processing (Fig. 4), or synthesis or subcellular localization of VirB subunits (Chen and Kado, 1994; 1996). Similar findings for the related FiwA fertility inhibition factor of plasmid RP1 strongly suggest that Osa and FiwA represent a class of plasmid-encoded factors that block trafficking by heterologous T4S systems at a stage subsequent to machine assembly (Olsen and Shipley, 1975; Fong and Stanisich, 1989; Chen and Kado, 1996). Here, we supplied evidence that Osa exerts its effects specifically on the substrate-receptor docking reaction (Fig. 4). The effect of Osa on DNA substrate-receptor binding is less pronounced, however, than that of RSF1010, and general comparisons of QTrIP and virulence data (Cascales and Christie, 2004a; Jakubowski et al., 2005; this study) suggest that this level of inhibition is not sufficient to account for the nearly avirulent phenotype of Osa-producing A. tumefaciens cells. Moreover, VirB9, VirB10 and VirB11 overproduction did not reverse Osa-mediated oncogenic suppression or confer elevated levels of T-DNA binding to VirD4 or the VirB channel subunits (Fig. 4). Together, these observations suggest, first, that Osa differs from RSF1010 in its mechanism of action, in agreement with a proposal derived from results of plant infection assays (Lee et al., 1999; Lee and Gelvin, 2004). Second, Osa likely suppresses virulence predominantly by inhibiting the binding of VirE2 and possibly other effector proteins to the VirD4 receptor. Very interestingly, Osa-GFP colocalized with VirD4 at the A. tumefaciens poles, constituting the first report of a cell positioning phenotype for a plasmid fertility inhibition factor. Yet, Osa targeting proceeds independently of VirD4 or the VirB channel subunits, and even occurs in a strain deleted of the Ti plasmid. Additionally, we have been unable to gain biochemical or two-hybrid evidence for complex formation between Osa and VirD4 (data not shown). Therefore, while enticing it remains a working model that Osa exerts its effects by modulating VirD4 receptor activity through a direct protein–protein interaction.
To complement our biochemical studies, we exploited a genetic screen originally devised in a characterization of the FipA and PifC fertility inhibition factors of plasmids pKM101 and F respectively (Santini and Stanisich, 1998). In that study, FipA and PifC were shown to block transfer of IncPα plasmid RP1 (equivalent to RP4) as well as RSF1010 through the RP1-encoded T4S system, but not the transfer of pCloDF13 by cells carrying a RP1 traG mutant. pCloDF13 encodes its own MobB receptor, which interacts with the RP1 Mpf proteins to yield a functional, chimeric T4S system. This specificity of PifC and FipA for RP1 TraG and not for pCloDF13 MobB constituted early genetic evidence that these factors inhibit plasmid transfer via effects on the RP1 TraG receptor (Santini and Stanisich, 1998). Similarly, we determined that Osa inhibits VirD4 but not CloDF13 MobB receptor activities. Thus, a combination of genetic, cytological, and biochemical findings (Santini and Stanisich, 1998; this study) strongly support a general proposal that a class of fertility inhibition factors represented by Osa (IncW), PifC (IncF), FipA (IncN) and FiwA (IncP) exert their effects by interfering with substrate docking at cognate VirD4/TraG-like receptors. The ability of MobB to interface with various T4S systems, together with its apparent immunity to inhibition factors, makes this an intriguing receptor for future structure–function studies exploring molecular details of substrate–receptor and receptor–Mpf channel subunit contacts.
The strong blocking action by Osa on an effector protein–T4S receptor binding reaction is of special interest because, as noted above, plasmid fertility inhibition has been considered a mechanism for preventing translocation of coresident plasmids. With a revised view that such factors also inhibit protein substrate trafficking through T4S systems, it is tempting to suggest that Osa or molecular mimics will prove effective in suppressing virulence of medically significant pathogens, e.g. Helicobacter pylori, Legionella pneumophila, and Bartonella and Burkholderia spp. whose infection cycles are dependent on T4S-mediated protein translocation (Segal and Shuman, 1998; Vogel et al., 1998; Cascales and Christie, 2003; Dehio, 2005). Additionally, there is increasing evidence that conjugation systems translocate protein substrates independently of DNA to bacterial recipients (Wilkins and Thomas, 2000; Luo and Isberg, 2004). Studies with fertility inhibition factors also should prove useful in evaluating the extent to which T4S-mediated protein trafficking represents a source of information flow in bacterial communities.
Experimental procedures
Strains, growth conditions and plasmids
Agrobacterium tumefaciens A348 is strain C58 bearing the octopine-type pTiA6NC plasmid (Zhu et al., 2000). A348 derivatives include Mx358 (virE2), Mx355 (virD4) (Stachel and Nester, 1986), At12516 (virE2) (Fullner et al., 1996), PC1000 (ΔvirB operon) (Fernandez et al., 1996) and LBA4404 (ΔT-DNA) (Ooms et al., 1982). C58C1RS (C58, Strr, Rifr) (Van Larebeke et al., 1975) and the Ti-plasmid-less strain UIA143 (C58 recA, Eryr) (Farrand et al., 1989) were used for conjugation experiments. Strain KA2000 (A348ΔvirD4) was constructed by deletion of the virD4 gene in A tumefaciens strain A348 by marker exchange-eviction mutagenesis (Berger and Christie, 1993) using plasmid pKA126 (Atmakuri et al., 2004).
Agrobacterium tumefaciens strains were grown in Luria–Bertani (LB) supplemented with mannitol and glutamate at 28°C (Zhou and Christie, 1999). Conditions for induction of the A. tumefaciens vir genes in AB inducing medium [ABIM, glucose-containing minimal medium (pH 5.5), 1 mM phosphate, 200 μM acetosyringone (AS)] have been described previously (Zhou and Christie, 1999). When necessary, medium was supplemented with antibiotics (in μg ml−1) as follows: gentamicin (100), kanamycin (100), tetracycline (5), carbenicillin (100 or 5 for strain LBA4404), spectinomycin (500), rifampicin (75), erythromycin (100), streptomycin (100).
RSF1010 plasmid derivatives used in this study are: pML122 (Genr, Kanr) and pML122ΔKan (Genr) (Fullner, 1998), pJB31 (Spcr) (Beaupre et al., 1997), pAJ1 (pJB31 mobA) and pAJ6 (pJB31 oriT) (Stahl et al., 1998). Additional plasmids include pSa (osa expressed from native promoter), pUCD3960 (Spcr, Crbr; osa constitutively expressed from nptI promoter) (Chen and Kado, 1994), pXZ66 (Kanr, PvirB-virE2-gfp) (Zhou and Christie, 1999), pED9 (Tetr, PvirB-virB9-10-11) (Ward et al., 1990), pJClo, a pBin19 derivative carrying the mob region of pCloDF13 (Kanr; gift from J. Escudero, A. Vergunst and P. J. J. Hooykaas) (Nijkamp et al., 1986), pBSBBR (Crbr, Kanr) constructed by ligation of pBluescript plasmid (Crbr) with the broad host range vector pBBR1MCS2 (Kanr) (Kovach et al., 1994), the IncP derivative pKA21 (PvirB-virD4) (Atmakuri et al., 2003), and ColE1 plasmids pKA64 (PvirB-virD4-N′GFP) and pKV38 (PvirB-GFP′C-virE2) (Atmakuri et al., 2003), respectively, ligated into the broad host range plasmids pBBRMCS-General (Genr) (Kovach et al., 1994) and pSW172 (Tetr) (Chen and Winans, 1991).
The osa gene was PCR-amplified with primers 5′-GCGCTAACGATGCATATGTTGCTACGGCGG-3′ and 5′-AAAGCGCAGGGACTCGAGTCACTATTCTAGAATCTTCCT GCATTG-3′ and pUCD3960 (Lee et al., 1999) as a template. Amplified DNA was cloned as a 0.57-kb NdeI/XhoI fragment into corresponding sites of pPC914KS+ (Berger and Christie, 1994) to obtain pKA165, which expresses PvirB-osa. Osa fused at its C terminus to GFP was constructed as follows. The MCS upstream of the virB promoter was destroyed by digesting pKA165 with SacII/EcoRV, blunt-ending and religating to obtain pKA166. A 0.73-kb XbaI/KpnI fragment containing GFP from pXZ63 (Rashkova et al., 2000) was ligated into the corresponding sites of pKA166 to obtain pKA167, which expresses PvirB-osa-gfp.
Virulence and extracellular complementation assays
Kalanchoe leaf assay. Agrobacterium tumefaciens strains were tested for virulence by inoculating wound sites of K. daigremontiana leaves as previously described (Berger and Christie, 1994). For extracellular complementation experiments, 0.5 ml of both strains at a similar optical density (OD600~0.8) were mixed in an Eppendorf tube and centrifuged. The pellet was resuspended in 100 μl and 25 μl was used to inoculate wounded K. daigremontiana leaves. WT A348 and avirulent mutant strains served as controls for the tumorigenesis assay. Virulence of strains or mixed infections was assayed at least three times on separate leaves.
Tobacco leaf square assay. Pieces (4 mm2) from surface sterilized mature young vegetative leaves of Nicotiana tabacum cv H425 plants were briefly soaked in a bacterial culture in LB (OD600~0.5) and then placed onto MS basal media plates supplemented with 300 μM AS. After 2 days at 25°C in the dark, pieces were washed with LB medium containing timentin (200 μg ml−1) and vancomycin (200 μg ml−1). Pieces were then placed onto MS basal medium plates supplemented with timentin and vancomycin. After 10 days at 25°C in the dark, tumours arising along the edges of each piece were counted. In each experiment, 16 leaf pieces were tested per bacterial strain (or mix) and each experiment was repeated in triplicate.
Conjugation assays
Agrobacterium tumefaciens donor strains carrying the RSF1010 derivative pML122ΔKm were mated with recipient strains A348 or At12516 (virE2 −) carrying plasmid pBBR1MCS (Strr). Briefly, mid-log phase (OD600~0.4) cells grown in MG l−1 broth were harvested and incubated in ABIM for 8 h at 22°C to induce vir gene expression. Cells were adjusted to an OD600~0.5, mixed in a donor to recipient ratio of 1 to 5 and spotted onto a nitrocellulose filter placed on an ABIM agar plate. A. tumefaciens donor cells carrying pJClo and UIA143 (Eryr) recipient cells were mixed in a ratio of 1 to 25 before inoculation on filter discs. The mating mixtures were incubated for 4 days at 18°C. Cells were recovered from the filters, serially diluted, and plated onto MG/l medium selective for transconjugant or donor cells. Frequencies of transfer were estimated as transconjugants recovered per donor. Experiments were repeated three times and results are reported for a representative experiment.
Coimmunoprecipitation
Coimmunoprecipitation was performed as described previously (Atmakuri et al., 2003). Briefly, 500 ml of induced A. tumefaciens cultures were harvested and cells were lysed by French press treatment. Total membranes were recovered by ultracentrifugation, cross-linked with 0.5 mg ml−1 of dithiobis(succidimidyl propionate; DSP), and solubilized with 1.5% N-Lauoryl sarcosine. The solubilized membranes were used as starting material for immunoprecipitation with anti-VirD4 antibodies coupled to Protein A-Sepharose CL-4B beads (Amersham Biosciences).
Bimolecular fluorescence complementation interaction assay
The BiFC assay was carried out as previously described (Atmakuri et al., 2003). Freshly transformed A. tumefaciens cells were incubated in ABIM and examined between 6 and 16 h post induction by fluorescence microscopy using an Olympus BX60 microscope equipped with a 100× oil immersion phase-contrast objective (Ding et al., 2002).
Transfer DNA ImmunoPrecipitation
Transfer DNA ImmunoPrecipitation and semi-quantitative TrIP (QTrIP) were carried out as previously described (Cascales and Christie, 2003). Primers for amplification were: 5′-GGGCGATTATGGCATCCAGAAAGCC and 5′-GTCGGC GGCCCACTTGGCACACAG for gene 7 present on the TL-DNA, 5′-CCTGCGGATGTCAGGGCTCTCGT and 5′-TGTC CGTGCTTGCCAATCCCCG for the ophDC locus on pTiA6NC (control), 5′-CTCAGTGGTTCAAGCGGTACA and 5′-TGATAGTTCTTCGGGCTGGTT for a fragment of mobA carried on RSF1010 derivatives used in this study, and 5′-GTGAGCAAAGCCGCTGCC and 5′-AGCCAATTGATCCT GCA for a fragment of the repAB loci on pTiAch5 (control).
Acknowledgments
The authors thank Lan-Ying Lee and Stan Gelvin for plasmids pUCD3960 and pSa and for helpful discussions, and members of the Christie and Binns labs for helpful discussions. We also thank J. Escudero, A. Vergunst and P. J. J. Hooykaas for the gift of pJClo. We thank S. Farrand for the gift of strain C58C1RS. We thank the Margolin lab for use of their microscopy facility. This study was supported by NIH Grant GM48746 to P.J.C. and NSF MCB0421885 to A.N.B.
References
- Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol. 2003;49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaupre CE, Bohne J, Dale EM, Binns AN. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek A, Zettl M, Lorenzoni K, Schauer A, Manhart M, Koraimann G. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J Bacteriol. 2004;186:6999–7006. doi: 10.1128/JB.186.20.6999-7006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger BR, Christie PJ. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993;175:1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Biesen T, Frost LS. The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol Microbiol. 1994;14:427–436. doi: 10.1111/j.1365-2958.1994.tb02177.x. [DOI] [PubMed] [Google Scholar]
- Binns A, Beaupre C, Dale E. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J Bacteriol. 1995;177:4890–4899. doi: 10.1128/jb.177.17.4890-4899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Passiatore JE, Cannon F. The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature. 1987;328:172–175. [Google Scholar]
- Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–150. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004a;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA. 2004b;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Winans SC. Controlled expression of the transcriptional activator gene virG. Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J Bacteriol. 1991;173:1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Kado CI. Inhibition of Agrobacterium tumefaciens oncogenicity by the osa gene of pSa. J Bacteriol. 1994;176:5697–5703. doi: 10.1128/jb.176.18.5697-5703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Kado CI. Osa protein encoded by plasmid pSa is located at the inner membrane but does not inhibit membrane association of VirB and VirD virulence proteins in Agrobacterium tumefaciens. FEMS Microbiol Lett. 1996;135:85–92. doi: 10.1111/j.1574-6968.1996.tb07970.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Chen Y, Wood DW, Nester EW. A new type IV secretion system promotes conjugative transfer in Agrobacterium tumefaciens. J Bacteriol. 2002;184:4838–4845. doi: 10.1128/JB.184.17.4838-4845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Cascales E. Structural and dynamic properties of bacterial type IV secretion [Review] Mol Membr Biol. 2005;22:51–61. doi: 10.1080/09687860500063316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Ward JE, Winans SC, Nester EW. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close SM, Kado CI. The osa gene of pSa encodes a 21.1-kilodalton protein that suppresses Agrobacterium tumefaciens oncogenicity. J Bacteriol. 1991;173:5449–5456. doi: 10.1128/jb.173.17.5449-5456.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio C. Bartonella-host-cell interactions and vascular tumour formation. Nat Rev Microbiol. 2005;3:621–631. doi: 10.1038/nrmicro1209. [DOI] [PubMed] [Google Scholar]
- Ding Z, Zhao Z, Jakubowski SJ, Krishnamohan A, Margolin W, Christie PJ. A novel cytology-based, two-hybrid screen for bacteria applied to protein–protein interaction studies of a type IV secretion system. J Bacteriol. 2002;184:5572–5582. doi: 10.1128/JB.184.20.5572-5582.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Atmakuri K, Christie PJ. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 2003;11:527–535. doi: 10.1016/j.tim.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero J, Den Dulk-Ras A, Regensburg-Tuink TJ, Hooykaas PJ. VirD4-independent transformation by CloDF13 evidences an unknown factor required for the genetic colonization of plants via Agrobacterium. Mol Microbiol. 2003;47:891–901. doi: 10.1046/j.1365-2958.2003.03328.x. [DOI] [PubMed] [Google Scholar]
- Farrand SK, O’Morchoe SP, McCutchan J. Construction of an Agrobacterium tumefaciens C58 recA mutant. J Bacteriol. 1989;171:5314–5321. doi: 10.1128/jb.171.10.5314-5321.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D, Spudich GM, Zhou XR, Christie PJ. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong ST, Stanisich VA. Location and characterization of two functions on RP1 that inhibit the fertility of the IncW plasmid R388. J Gen Microbiol. 1989;135:499–502. doi: 10.1099/00221287-135-3-499. [DOI] [PubMed] [Google Scholar]
- Fullner KJ. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J Bacteriol. 1998;180:430–434. doi: 10.1128/jb.180.2.430-434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullner KJ, Lara JC, Nester EW. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- Gasson M, Willetts N. Transfer gene expression during fertility inhibition of the Escherichia coli K12 sex factor F by the I-like plasmid R62. Mol Gen Genet. 1976;149:329–333. doi: 10.1007/BF00268535. [DOI] [PubMed] [Google Scholar]
- Ghetu AF, Gubbins MJ, Frost LS, Glover JN. Crystal structure of the bacterial conjugation repressor finO. Nat Struct Biol. 2000;7:565–569. doi: 10.1038/76790. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Lee H, Li PL, Cook DM, Piper KR, von Bodman SB, et al. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugative transfer system of pTiC58. J Bacteriol. 2000;182:1541–1548. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Jakubowski SJ, Cascales E, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol. 2005;187:3486–3495. doi: 10.1128/JB.187.10.3486-3495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Phillips RW, Elzer PH, Roop RM, 2nd, Peterson KM. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- Kumar RB, Das A. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol Microbiol. 2002;43:1523–1532. doi: 10.1046/j.1365-2958.2002.02829.x. [DOI] [PubMed] [Google Scholar]
- Lawley T, Wilkins BM, Frost LS. Bacterial conjugation in Gram-negative bacteria. In: Funnell BE, Phillips GJ, editors. Plasmid Biology. Washington, DC: American Society for Microbiology Press; 2004. pp. 203–226. [Google Scholar]
- Lee LY, Gelvin SB. Osa protein constitutes a strong oncogenic suppression system that can block vir-dependent transfer of IncQ plasmids between Agrobacterium cells and the establishment of IncQ plasmids in plant cells. J Bacteriol. 2004;186:7254–7261. doi: 10.1128/JB.186.21.7254-7261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Gelvin SB, Kado CI. pSa causes oncogenic suppression of Agrobacterium by inhibiting VirE2 protein translocation. J Bacteriol. 1999;181:186–196. doi: 10.1128/jb.181.1.186-196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- Lessl M, Balzer D, Weyrauch K, Lanka E. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J Bacteriol. 1993;175:6415–6425. doi: 10.1128/jb.175.20.6415-6425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Zunzunegui S, De la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci USA. 2003;100:10465–10470. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by inter-bacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijkamp HJ, de Lang R, Stuitje AR, van den Elzen PJ, Veltkamp E, van Putten AJ. The complete nucleotide sequence of the bacteriocinogenic plasmid CloDF13. Plasmid. 1986;16:135–160. doi: 10.1016/0147-619x(86)90072-7. [DOI] [PubMed] [Google Scholar]
- Olsen RH, Shipley PL. RP1 properties and fertility inhibition among P, N, W, and X incompatibility group plasmids. J Bacteriol. 1975;123:28–35. doi: 10.1128/jb.123.1.28-35.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms G, Hooykaas PJJ, Van Veen RJM, Van Beelen P, Regensburg-Tunik R, Schilperoort RA. Octopine Ti-plasmid deletion mutants of Agrobacterium tumefaciens with emphasis on the right side of the T-region. Plasmid. 1982;7:15–19. doi: 10.1016/0147-619x(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Otten L, De Greve H, Leemans J, Hain R, Hooykaas P, Schell J. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol Gen Genet. 1984;195:159–163. [Google Scholar]
- Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Progr Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- Pantoja M, Chen L, Chen Y, Nester EW. Agrobacterium type IV secretion is a two-step process in which translocation substrates associate with the virulence protein VirJ in the periplasm. Mol Microbiol. 2002;45:1325–1335. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- Penfold SS, Simon J, Frost LS. Regulation of the expression of the traM gene of the F sex factor of Escherichia coli. Mol Microbiol. 1996;20:549–558. doi: 10.1046/j.1365-2958.1996.5361059.x. [DOI] [PubMed] [Google Scholar]
- Rashkova S, Zhou XR, Christie PJ. Self-assembly of the Agrobacterium tumefaciens VirB11 traffic ATPase. J Bacteriol. 2000;182:4137–4145. doi: 10.1128/jb.182.15.4137-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings DE, Tietze E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev. 2001;65:481–496. doi: 10.1128/MMBR.65.4.481-496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko Y, Sagulenko V, Chen J, Christie PJ. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol. 2001;183:5813–5825. doi: 10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini JM, Stanisich VA. Both the fipA gene of pKM101 and the pifC gene of F inhibit conjugative transfer of RP1 by an effect on TraG. J Bacteriol. 1998;180:4093–4101. doi: 10.1128/jb.180.16.4093-4101.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B, Dulk-Ras Ad A, Vergunst AC, Jurado Jacome E, Hooykaas PJ. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroder G, Lanka E. The mating pair formation system of conjugative plasmids – A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Schroder G, Krause J, Zechner EL, Traxler B, Yeo HJ, Lurz R, et al. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for translocationed substrates? J Bacteriol. 2002;184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Shuman HA. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugative components of IncQ plasmid RSF1010. Mol Microbiol. 1998;30:197–208. doi: 10.1046/j.1365-2958.1998.01054.x. [DOI] [PubMed] [Google Scholar]
- Simone M, McCullen CA, Stahl LE, Binns AN. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol Microbiol. 2001;41:1283–1293. doi: 10.1046/j.1365-2958.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Nester EW. The genetic and transcriptional organization of the vir region of the A6 Ti. EMBO J. 1986;5:1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl LE, Jacobs A, Binns AN. The conjugative intermediate of plasmid RSF1010 inhibits Agrobacterium tumefaciens virulence and VirB-dependent translocation of VirE2. J Bacteriol. 1998;180:3933–3939. doi: 10.1128/jb.180.15.3933-3939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K, Iino T. Transfer inhibition of RP4 by F factor. Mol Gen Genet. 1983;192:104–109. doi: 10.1007/BF00327654. [DOI] [PubMed] [Google Scholar]
- Van Larebeke N, Genetello C, Schell J, Schilperoort RA, Hermans AK, Van Montagu M, Hernalsteens JP. Acquisition of tumour-inducing ability by non-oncogenic agrobacteria as a result of plasmid transfer. Nature. 1975;255:742–743. doi: 10.1038/255742a0. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, van Lier MC, den Dulk-Ras A, Grosse Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci USA. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Andrew HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Ward JEJ, Dale EM, Christie PJ, Nester EW, Binns AN. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and virB11 are essential virulence genes. J Bacteriol. 1990;172:5187–5199. doi: 10.1128/jb.172.9.5187-5199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JE, Dale EM, Binns AN. Activity of the Agrobacterium T-DNA transfer machinery is affected by virB genes products. Proc Natl Acad Sci USA. 1991;88:9350–9354. doi: 10.1073/pnas.88.20.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins BM, Thomas AT. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol Microbiol. 2000;38:650–657. doi: 10.1046/j.1365-2958.2000.02164.x. [DOI] [PubMed] [Google Scholar]
- Winans SC, Walker GC. Fertility inhibition of RP1 by IncN plasmid pKM101. J Bacteriol. 1985;161:425–427. doi: 10.1128/jb.161.1.425-427.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusoff K, Stanisich VA. Location of a function on RP1 that fertility inhibits Inc W plasmids. Plasmid. 1984;11:178–181. doi: 10.1016/0147-619x(84)90022-2. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Sagulenko E, Ding Z, Christie PJ. Activities of virE1 and the VirE1 secretion chaperone in translocation of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J Bacteriol. 2001;183:3855–3865. doi: 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XR, Christie PJ. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self association and interaction with VirE1 and a permissive site for hybrid protein construction. J Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Oger PM, Schrammeijer B, Hooykaas PJ, Farrand SK, Winans SC. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]