Abstract
We have recently developed a fragment based selection strategy for targeting kinases, where a small molecule warhead can be non-covalently tethered to a phage-displayed library of peptides. This approach was applied to the conversion of the promiscuous kinase inhibitor, staurosporine, into a potent bivalent ligand for cAMP-dependent protein kinase (PKA). Herein we report a systematic evaluation of this new bivalent ligand (BL); (a) Lineweaver-Burke analysis revealed that the BL, unlike substrate-based bivalent kinase inhibitors, displayed non-competitive inhibition with respect to the peptide substrate, suggesting an allosteric mechanism of action; (b) linker optimization of the BL, afforded one of the most potent, sub-nanomolar, inhibitors of PKA reported to date; (c) the BL was found to be modular, where attachment of active site targeted small molecule warheads in lieu of staurosporine could achieve similar gains in affinity; and (d) profiling studies of both the staurosporine derivative and the BL (amide isostere) against a panel of 90 kinases revealed almost unique enhancement in selectivity against PKA (>5-fold) compared to the starting staurosporine derivative. These combined results provide new insights for BL discovery, which has the potential to provide guidance towards the development of kinase selective reagents while uncovering new allosteric sites on kinases for therapeutic targeting.
1. Introduction
The greater than 500 human protein kinases are implicated in the regulation of almost all cellular processes, ranging from cell growth and homeostasis to cell death.1,2 Not surprisingly, the deregulation of kinase activity has been implicated in diseases such as cancer, inflammation, diabetes, as well as neurological and immunological diseases.3–6 Significant efforts have focused on developing agents to modulate kinase activity, both as drugs and as specific reagents for studying kinase biology. However, considering that the 500 human protein kinases all possess structurally homologous active sites, achieving selectivity continues to remain a central challenge.1,7 To date, most research has focused on generating inhibitors of kinase activity through direct targeting of the ATP binding active site.8 Methods for identifying specific inhibitors include screens of heterocyclic compounds aided by structure based design9 and more recently, fragment based approaches.10,11 In spite of the enormous effort directed towards developing selective kinase ligands, recent studies have demonstrated a lack of selectivity for many widely used active site directed inhibitors when tested against large panels of kinases.12–15 Though polypharmacology can be therapeutically useful,16 it potentially limits the future utility of these compounds as research tools for understanding biology and also thereby warrants the search for new approaches for modulating selectivity.18
To develop new and general strategies for selective kinase compounds, sites other than the ATP binding cleft have recently been targeted.17 These approaches include targeting of allosteric sites that stabilize inactive conformations18, protein substrate binding sites,19 and noncatalytic domains. The generation of dual binding or bivalent inhibitors was proposed when it was recognized that protein kinases bind two substrates.20 Building on this concept, covalently linked substrate analogs (bisubstrate inhibitors) have achieved significant success with several kinases.21–26 For example, Cole and coworkers provided an elegant mechanism based approach for targeting the insulin receptor protein tyrosine kinase,22 while Schepartz and coworkers have engineered kinase surface selectivity utilizing a substrate structure guided bivalent strategy.27 In general, most methods for generating new classes of selective inhibitors rely on structural information of the targeted kinase, protein substrate information, or domain-specific protein partners; potentially limiting the use of these methods for kinases for which such information is unavailable.
Towards providing alternative methods for targeting kinases, we have recently developed a non-covalent fragment based approach for the discovery of BLs utilizing phage display. This approach does not rely on explicit structural information regarding ligands that bind outside the ATP-binding site.28 Our approach begins with an ATP-competitive warhead that then acts as an anchor for the selection of a peptide ligand that binds nearby such that the two ligands may be covalently linked to generate a single BL (Figure 1). Specifically, an ATP-competitive small molecule directs a phage displayed six residue cyclic peptide library to bind an adjacent site on a targeted kinase through non-covalent tethering of the phage particle via a Fos/Jun coiled-coil interaction. This approach allows us to link high-affinity small molecule ligands to the large chemical space encompassed by biological libraries. In our proof-of-principle studies we successfully converted the promiscuous kinase inhibitor, staurosporine, into a potent inhibitor of the cAMP-dependent protein kinase (PKA) with an IC50 = 2.6 nM, a >60-fold and >21,000-fold relative increases when compared to the staurosporine derivative, 1a, (IC50 = 159 nM) and the selected cyclic peptide, 1b, (IC50 = 57 µM) alone.28 Very recently, Maly and coworkers have also utilized an elegant strategy to develop highly specific bivalent kinase inhibitors that target both the active site and an adjacent SH3 site in Abl- and Src-family kinases.29
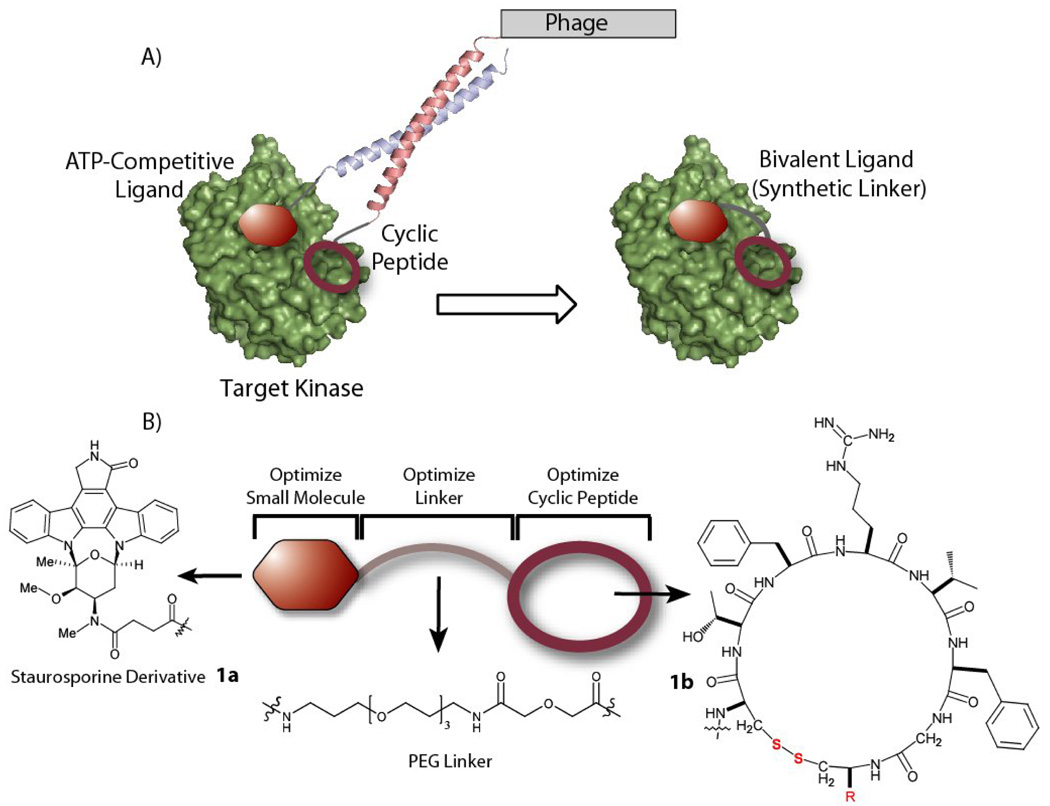
Figure 1. Bivalent Ligands of Protein Kinase A (PKA).
A) The general strategy for generating bivalent ligands for protein kinases is shown, where an ATP-competitive warhead directs the selection of a phage-displayed cyclic peptide through a coiled-coil interaction. B) The initial BL targeting PKA, 1, consists of the staurosporine derivative, 1a, a 30 Å PEG linker and the cyclic peptide with a C-terminal glycine (R = Gly), 1b.
Herein we focus our attention on this new class of BLs for kinases generated using our approach with an emphasis on addressing issues that are unique to this class of reagents: (a) Is the mode of binding competitive with respect to the kinase peptide binding site? (b) Can binding be enhanced by optimization of linker length? (c) Can the starting warhead, staurosporine, be replaced with another user-defined small molecule and still benefit from the affinity enhancement provided by the cyclic peptide? (d) Does the peptide contribute unique selectivity for PKA as compared to the promiscuous starting warhead staurosporine when interrogated against a large commercial kinase panel? These studies should help provide a framework for the further development of this new class of kinase selective bivalent ligands.
2. Results and discussion
2.1 Bivalent Ligand Optimization
The bivalent ligand (BL) for protein kinase A (PKA) is comprised of three modules: an ATP-competitive ligand, a selected cyclic peptide [cyclo(CTFRVFGC)G] ligand, and a linker tethering the two ligands (Figure 1). We have previously shown that covalent linkage of the small molecule to the cyclic peptide increased potency towards PKA by over 60-fold as compared to the staurosporine derivative alone.28 Herein we first explore the mode of inhibition with respect to the peptide substrate and the dependence of linker length upon activity.
2.1 Mode of Inhibition
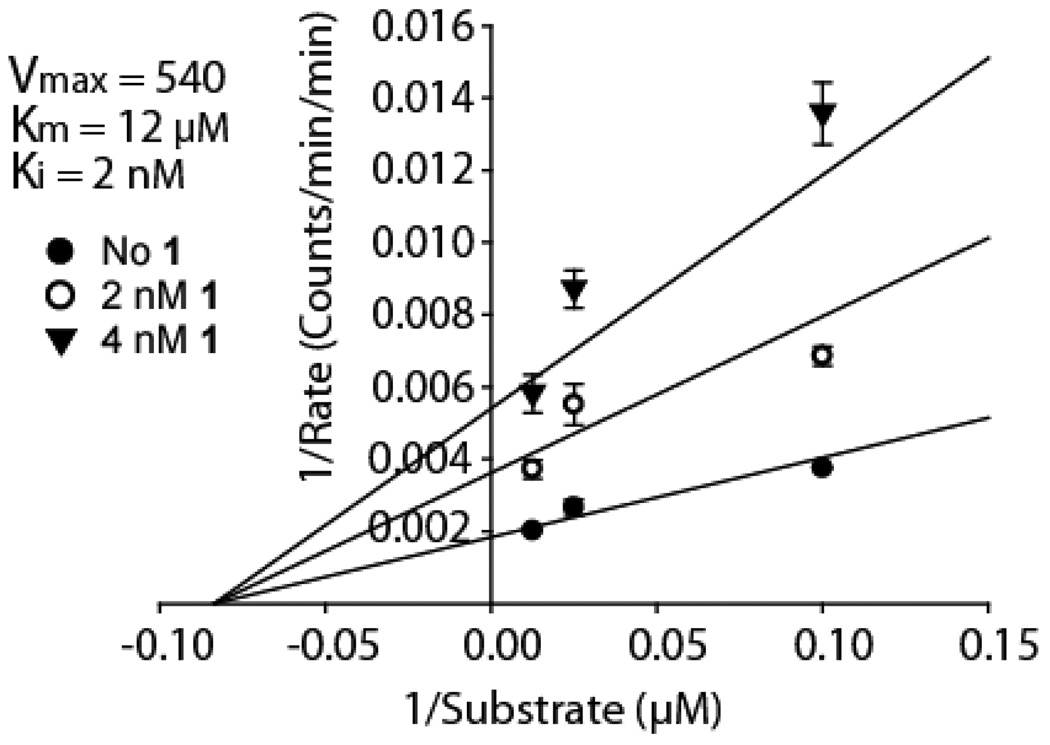
Though the staurosporine warhead in the BL binds to the ATP site, the mode of inhibition with respect to protein substrate cannot be inferred a priori. Our bivalent selection strategy (Figure 1) in principle allows for the discovery of any peptide that binds adjacent to the ATP binding site but not necessarily to the substrate site. Therefore, the mode of inhibition must be determined empirically, and this analysis would be warranted for any ligand discovered with our approach. Kinetic experiments were performed with PKA to determine the mode of inhibition of the BL with respect to a peptide substrate (Kemptide = LRRASLG). The first generation BL containing a PEG linker, 1, (IC50 = 2.6 nM) was used in these kinetic experiments. The resulting data for the BL was fit to competitive, noncompetitive, uncompetitive and mixed inhibition models (Supporting Information, Figure S1). A noncompetitive inhibition model was found to best fit the observed data (Figure 3), with a calculated Ki of 2 nM and Km of 12 µM for the PKA substrate, Kemptide, which is in good agreement with the IC50 value 2.6 nM of the inhibitor28 and reported Km 21. Furthermore, kinetic experiments with the cyclic peptide, 1b, alone were also best fit to a noncompetitive inhibition model (Supporting Information, Figure S2). Noncompetitive inhibition with respect to the peptide substrate implies that the cyclic peptide does not bind to the protein substrate binding site of PKA. This is consistent with the optimal linker being significantly longer (vide infra) than that required for binding to the substrate site, which is directly adjacent to the ATP binding pocket. If the cyclic peptide binds a remote site as suggested, an allosteric effect may contribute to the BL’s potency in addition to the ATP competitive binding by the staurosporine derivative. These results also suggest that the relatively unbiased search for a BL has the potential to uncover new allosteric sites on kinases that may result in new modalities for kinase inhibition.
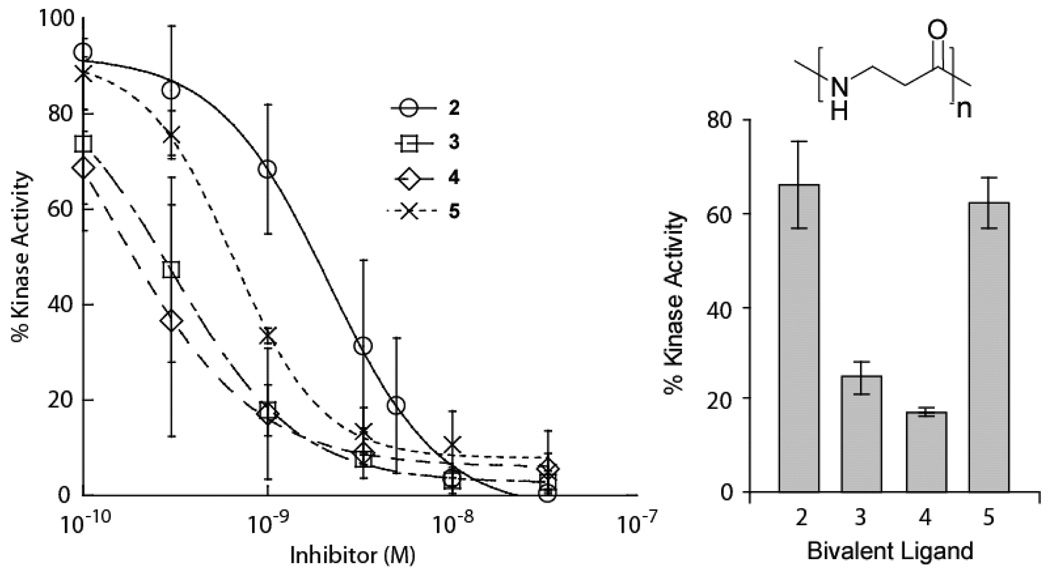
Figure 3. Bivalent Ligand Linker Length Optimization.
(a) PKA assays of the BLs (2–5) of varying linker length were performed at 0.5 nM PKA. Since full IC50 curves could not be obtained, the BLs were reevaluated at a single concentration of 3.3 nM against PKA (0.5 nM) to ascertain the relative activities.
2.2 Optimization of Bivalent Ligand Spacers
For a BL containing tethered components that bind two distinct sites on a protein, the length of the adjoining linker has the capacity to significantly influence binding affinity. A short linker will prevent simultaneous binding of both ligands, whereas a long linker would be entropically costly30,31. This is evident in previous work concerning designed bisubstrate inhibitors targeting two domains of protein kinases22,32. In these examples, the optimal linker lengths were determined empirically, differing from the initial design21,33, even when the distance between the binding sites of both ligands was known. In contrast the peptide binding site is unknown in our selection approach, thus the optimal linker length must be determined empirically. In principle the linker length will at most approximate the separation between the warhead and cyclic peptide when attached to the Fos/Jun dimer during the selection process (Figure 1). This corresponds to a radius of 10 – 42 Å from the ATP binding site. The initial ~30 Å PEG linker was chosen to span an intermediate length within the range of possible distances28.
To determine the optimal linker length between the staurosporine derivative and cyclic peptide, we varied the linker composition (3, 5, 7, and 9 β-alanines; compounds 2–5 respectively) corresponding to approximate maximum lengths of 12 Å to 37 Å. These new BLs were initially evaluated in kinase assays with PKA (0.5 nM), which indicated that the IC50 values of the most potent compounds were approximately half of the enzyme concentration, thus precluding the accurate determination of IC50 values (Figure 3). However, the relative potencies of the inhibitors were easily distinguishable at a single inhibitor concentration (3.3 nM) chosen to be 6-fold greater than the enzyme concentration (0.5 nM). From the relative differences in their ability to inhibit PKA, the optimal linker was determined to consist of 7 β-alanine residues (Figure 3), which can theoretically span a maximum distance of ~28 Å. The relatively long linker suggests the peptide does not bind at the protein substrate site, which is consistent with the observed noncompetitive mode of inhibition (vide supra). Simultaneous binding of a remote site would explain the increased potency of the BL, 4, (IC50 < 0.52 nM) compared to the cyclic peptide alone, 1b, (IC50 = 57 µM) or ATP-competitive warhead, 1a, (IC50 = 159 nM). The observed increase in binding of the optimized BL is >300-fold when compared to the starting staurosporine derivative. Thus this optimized inhibitor represents one of the most potent ligands for PKA reported to date.34
2.3. Modularity of the Active Site Directed Ligand
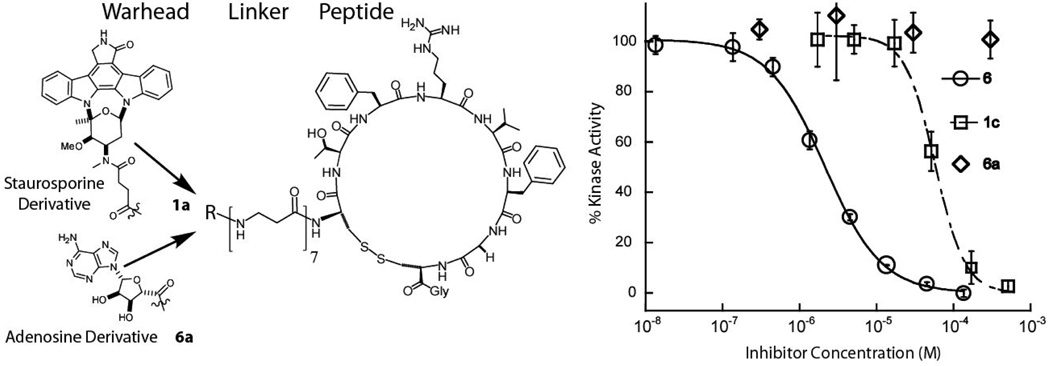
In our selection strategy a non-specific active site directed warhead, staurosporine, was initially chosen,28 eventually resulting in the identification of a potent and selective BL. However, the modularity of the BL was not known, that is could another ATP-cleft directed small molecule ligand replace staurosporine post-selection in the context of the optimized BL, 4? To address the modularity of the small molecule warhead, an adenosine derivative35 was substituted as the ATP-competitive ligand (Figure 4) conjugated to the cyclic peptide via the optimal 7 β-alanine linker, 6. The IC50 of the adenosine derived BL was found to be 2.9 µM, which is a >300-fold improvement from the adenosine derivative, 6a, (IC50 > 1 mM). These results suggest that the bivalent selection strategy can in principle provide a peptide ligand for a kinase juxtaposed to the ATP cleft facilitating rapid modification of new ATP-site binding small molecules to rapidly increase affinity in a fragment based fashion. Thus this method potentially affords access to more selective BLs, since a promiscuous warhead can be readily exchanged with a user-defined selective warhead.
Figure 4. Testing the Modularity of the Warhead.
The original warhead, staurosporine, was replaced in the BL with another weak ATP-competitive ligand, an adenosine derivative, 6a. The significant improvement of the BL, 6, (IC50 = 3 µM) versus the starting warhead, 6a, (IC50 > 1000 µM) demonstrates that attachment of the peptide module can confer significant improvement in affinity in an warhead independent fashion.
2.4. Selectivity Profiling of the Bivalent Ligands
Our earlier results demonstrated that the cyclic peptide module of the BL selectively increased affinity for PKA when compared against four other kinases that have similar affinity for staurosporine28. However, the presence of the disulfide moiety precluded a more extensive profiling of the BL, since the disulfide is susceptible to the reducing conditions (7.5 mM DTT) of large kinase panel screens and loses substantial activity when assayed in these conditions (Supporting Information, Figure S6). Thus to profile the cyclic peptide, an amide isostere, 7, that replaced the disulfide moiety in the BL was synthesized utilizing orthogonal solid phase peptide synthesis.36 The amide BL, 7, IC50 was determined to be 12.6 nM, maintaining a 12-fold improvement as compared to the staurosporine derivative, 1a. Similar decreases in overall activity have been previously observed for disulfide replacements in cyclic peptides37–39. However, importantly the amide compound was insensitive to reducing conditions as determined by kinase assays in the presence of 7.5 mM DTT (Supporting Information, Figure S4). Thus with the amide compound, 7, in hand, the selectivity of the BL could be addressed in the Ambit kinase screen which evaluates a compound’s kinase binding profile in a competitive binding assay.13 As our reference compound in the screen, we also evaluated staurosporine derivatized with a 3 β-alanine linker, 1a, which has an IC50 of 159 nM (Supporting Information, Figure S5). To ensure similar levels of compound binding for PKA in the 96 kinase panel screen, a 10 fold higher concentration of the 3-β-alanine staurosporine derivative, 1a, (500 nM) was utilized as compared to the amide isostere BL, 7, (50 nM).
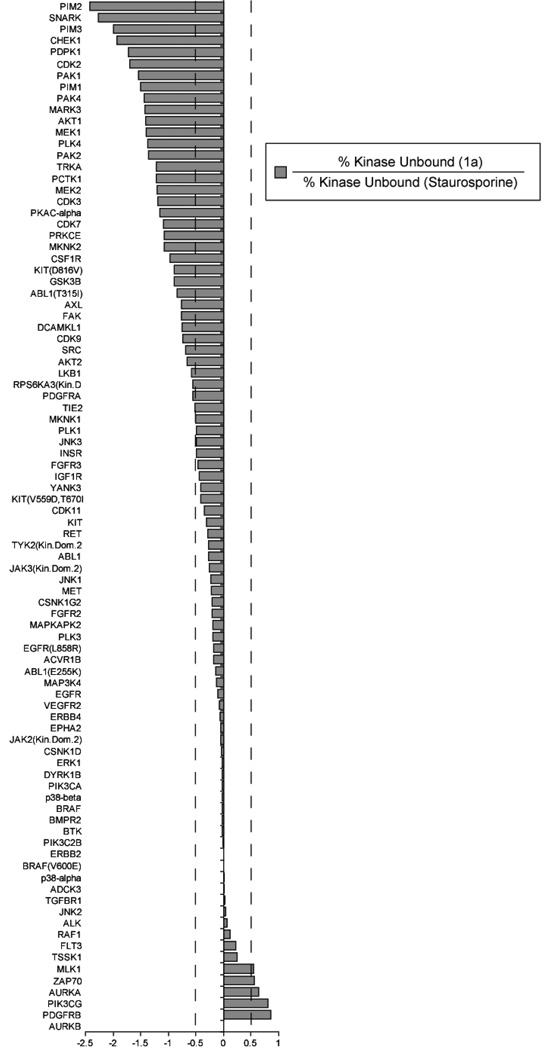
We first chose to evaluate changes in staurosporine affinity for the kinase panel attributable to the addition of the 3-β-alanine linker. The results (Figure 5) revealed a significantly altered binding profile, when compared to the reported binding profile for the parent warhead, staurosporine. Of the kinases tested, 39 kinases did not show significantly altered affinity for the staurosporine derivative (within 2-fold), 45 kinases displayed a >2 fold decrease in affinity, while 6 kinases gained >2 fold in affinity. Co-crystal structures of staurosporine with kinases have shown that the secondary amine of staurosporine can potentially form hydrogen bonds to the active site residues of several kinases.40–42 It has been proposed that the affinity of staurosporine for other kinases is also modulated by these hydrogen bonding networks.41,43,44 To understand if hydrogen bonding was responsible for the reduction in activity for the 39 kinases, we analyzed the 10 available co-crystal structures amongst the 90 kinases screened (Table II, Supplementary Information). Amongst these kinases, hydrogen bonding interactions with the aminomethyl group of staurosporine was clearly observed in several cases (PKA,40 CHEK1,43 PDPK1,45 CDK2,46 PIM1,42 JAK3,47 ZAP70,48 PIK3CG44). For kinases that make 2 hydrogen bonding interactions where at least one residue is an aspartic or glutamic acid, the staurosporine derivative showed decreased affinity ranging from 7 to > 80-fold lower than the parent, staurosporine. An exception was PIK3CG, which has two potential hydrogen bonding residues, T886 and D964, but binds compound 1a with a 6-fold higher affinity. For GSK3B there are no observable hydrogen bonding interactions with the aminomethyl group of staurosporine, and the resulting change in affinity (7-fold worse) may possibly arise from unfavorable sterics. For JAK3 and ZAP70, the interacting residues are arginine residues which may perhaps interact favorably with the amidated aminomethyl group. It is worth noting that when grouped according to kinase subfamily, the amidated staurosporine does not appear to favor any particular kinase subfamily (AGC, CAMK, CMGC, CK, STE, TK, TKL, or lipid) any more than does staurosporine.
Figure 5. The Affinity Profile of the Modified Staurosporine Compound, 1a, Relative to Staurosporine.
1a (500 nM) was screened against a panel of 90 kinases (Ambit Biosciences) and the resulting binding data was evaluated for the difference in binding relative to published binding values for staurosporine (500 nM). The data shows that the derivitization of staurosporine at its secondary amine results in a decrease in affinity for most kinases in the panel, though there are a few for which enhanced binding is observed.
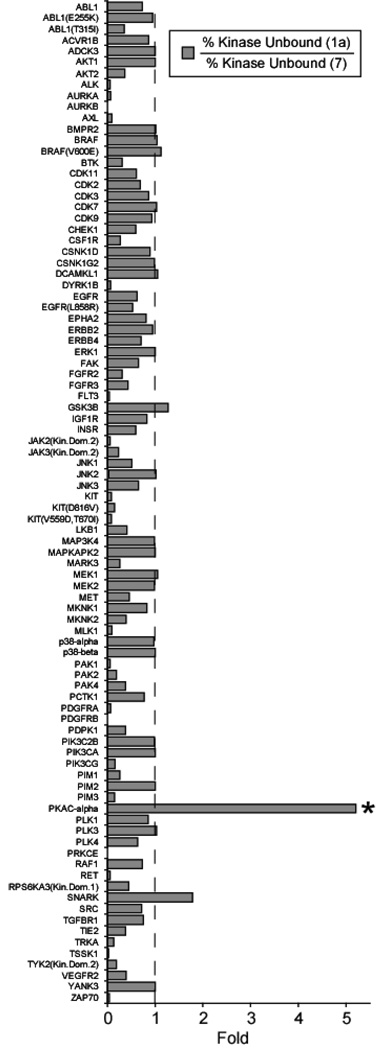
To determine the enhancement in selectivity brought about by the addition of the cyclic peptide module of the BL, we normalized for the relative potency of the starting staurosporine derivative, 1a, (500 nM) as compared to the amide isostere, 7, (50 nM). Thus, the profiling data is presented as a ratio of kinase binding values for the two inhibitors (Figure 6). In this analysis, a value of 1 ± 0.2 implies little change in affinity profiles (33 kinases); a value of >1.2 indicates a gain in affinity imparted by the cyclic peptide (2 kinases); and a value of <0.8 indicates that the attachment of the peptide, surprisingly, diminishes binding (55 kinases) (Figure 6). It is immediately clear from this analysis (Figure 6) that we only achieve a significant enhancement of selectivity (5-fold) for PKA. It is worth noting that the disulfide constrained BL under the same reducing conditions of the assay (7.5 mM DTT) demonstrated a significantly lower preference (2.7-fold) for PKA (Figure S6, Supporting Information). The only other kinase among the 90 analyzed that showed a small increase in selectivity (1.8-fold) by addition of the peptide was SNARK. It is interesting but difficult to explain why addition of the peptide module results in a drastically reduced binding (>2-fold) for many kinases (55 kinases), when compared to the starting staurosporine derivative. Thus overall, the profiling data goes towards directly confirming that the bivalent selection approach yields cyclic peptide modules that can confer significant selectivity, likely due to the low homology shared between kinase surfaces when compared to the ATP binding cleft.
Figure 6. Selectivity Profiling of the Amide BL, 7, isostere (50 nM) for Kinases Relative to the Modified Staurosporine Warhead, 1a (500 nM).
The greatest enhancement is observed for the target kinase: PKA-Cα, with an overall 5-fold enhancement in binding, while almost all other kinases in the panel are not targeted by the peptide with the exception of SNARK.
3. Conclusions
We have thus demonstrated that this approach for bivalent ligand discovery, selected by means of a fragment-based approach, has the potential to discover new and potential allosteric sites at the kinase surface. The cyclic peptide was shown to impart very high affinity (sub-nanomolar) as well as enhanced selectivity (>5-fold) for the targeted kinase when compared to a panel of 90 kinases. Furthermore the demonstrated modularity, where staurosporine was substituted for an adenosine analog, potentially implies that BLs derived from our selection methodology can be rapidly reconfigured to be more specific by replacement of the starting promiscuous inhibitor, staurosporine, with a user-defined kinase selective ligand. We believe that several of the considerations addressed herein (mode of binding, linker optimization, modularity, selectivity) are generally applicable to future ligands discovered by this fragment-based bivalent in vitro selection approach. Furthermore, a careful analysis of changes in affinity/selectivity brought about by modifications to the active-site directing warhead, in this case staurosporine, should also be an essential component for evaluating bivalent selection approaches.
4. Experimental
4.1 Compound Synthesis
The peptide portion of each BL was synthesized using standard Fmoc protection strategies in solid phase peptide synthesis on Rink Amide resin (substitution level = 0.3 mmol/g). Coupling conditions were PyBOP (3 eq), DIEA (6 eq) and 3 equivalents of the appropriate Fmoc protected amino acid in DMF. The synthesis of the succinic acid staurosporine derivative and the oxidized adenosine derivative were carried out as described previously28. Coupling of the staurosporine derivative (1.5 eq) was carried out with protection from light under standard Fmoc strategy conditions, as above. Coupling of the adenosine derivative was carried out as with Fmoc protected amino acid residues with no further precautions taken. BLs were cleaved from the resin with 94% TFA, 2.5% EDT, 2.5% water, and 1% TIPS for 2 hours. They were then precipitated in chilled ether. Oxidation to yield the cyclic peptide portion was achieved by dissolving the compound in 20% DMSO in PBS, pH = 7.4, at 37 °C for 36 hours and was monitored with Ellman’s Reagent49. Compounds were purified by HPLC (20 – 65% acetonitrile gradient in water with 0.1% TFA). Fractions containing the compounds were pooled and lyophilized. The compounds were characterized by MALDI mass spectrometry and amino acid analysis.
4.2 Amide Isostere
The solid phase peptide synthesis was performed as with all other compounds. Allyl/Alloc amino acids: Fmoc-L-Dap(Alloc)-OH (AnaSpec, Inc.) and Fmoc-L-Asp(Allyl)-OH (Fluka) were coupled and orthogonally deprotected with 0.2 eq Pd(PPh3)4 and 20 eq PhSiH3 in Drisolv DCM for 1 hour36. The resin was extensively washed with DCM and the side chain cyclization was performed under standard amide coupling conditions. After cyclization on the resin, the linker and staurosporine derivative were coupled as with the other BLs, and was then cleaved and purified by HPLC.
4.3 Kinetic Assays
Kinetic assays were performed in duplicate. In a 60 µl final volume, [γ-32P]ATP (30 µM) initiated the reaction with 0.52 nM PKA and Kemptide (LRRASLG, 20, 40, and 80 µM) in the presence of the BL (0, 2 and 4 nM) in PKA Assay Buffer (40 mM Tris, 20 mM Mg Acetate, pH 7.4) with 0.01% BSA and 2.5% DMSO. At 7 minute intervals, 10 µl of the reaction mixture was spotted on P81 phosphocellulose paper. The samples were washed three times in 500 ml of 0.85% phosphoric acid and once in 500 ml of ethanol for 5 minutes each. The amount of 32P labeling of the peptide substrate was quantified using a Beckman LS 6000IC liquid scintillation counter.
PKA Inhibitor IC50 Determination
Kinase assays were performed in triplicate under similar conditions to those described above. In a 30 µl final volume, [γ-32P]ATP (30 µM) initiated the reaction with 2.6 nM PKA and Kemptide (LRRASLG, 30 µM) in PKA Assay Buffer (40 mM Tris, 20 mM Mg Acetate, pH 7.4) with 0.01% BSA and 2.5% DMSO. The reaction was quenched with 15 µl of 1.8% phosphoric acid, and 30 µl of the reaction mixture was spotted on P81 phosphocellulose paper. The samples were washed, and the extent of 32P labeling of the peptide substrate was quantified as described above. Data were normalized to reactions containing no inhibitors, which were run in triplicate.
4.4 Kinase Panel Screen
Stock solutions of each screened kinase inhibitor were provided to Ambit Biosciences (San Diego, CA) in DMSO at the appropriate concentrations. Inhibitors were screened at Ambit Biosciences against the scanEDGE kinase panel under reported conditions.12
Supplementary Material
Figure 2. The Mode of Inhibition of the Bivalent Ligand (BL), 1, and the Cyclic Peptide, 1b.
The BL with a 30 Å PEG linker was evaluated in kinetic assays in duplicate at 0, 2 nM, and 4 nM inhibitor, with 20, 40 and 80 µM Kemptide. A noncompetitive inhibition model provided the best fit with a Ki of 2 nM and a Km of 12 µM. These values are consistent with the observed IC50 of 2.6 nM and the Km of Kemptide, 16 µM.
Acknowledgement
We thank members of the Ghosh laboratory for helpful discussions and the NIH (R21CA141974 and R01AI068414) and the NSF (CHE-0548264) for partial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information
Details of synthesis, purification, characterization, and assays are available.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. Trends. Biochem. Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 3.Kyriakis JM, Avruch J. Physiol. Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 4.Martinez A, Castro A, Dorronsoro I, Alonso M. Med. Res. Rev. 2002;22:373–384. doi: 10.1002/med.10011. [DOI] [PubMed] [Google Scholar]
- 5.Billingsley ML, Kincaid RL. Biochem. J. 1997;323:577–591. doi: 10.1042/bj3230577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levitzki A, Gazit A. Science. 1995;267:1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 7.Vieth M, Higgs RE, Robertson DH, Shapiro M, Gragg EA, Hemmerle H. Biochim. Biophys. Acta Proteins Proteomics. 2004:243–257. doi: 10.1016/j.bbapap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 8.McInnes C, Fischer PM. Curr. Pharm. Design. 2005;11:1845–1863. doi: 10.2174/1381612053764850. [DOI] [PubMed] [Google Scholar]
- 9.Gray NS, Wodicka L, Thunnissen A, Norman TC, Kwon SJ, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, Kim SH, Lockhart DJ, Schultz PG. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 10.Chen JH, Zhang ZM, Stebbins JL, Zhang XY, Hoffman R, Moore A, Pellecchia M. ACS Chem. Biol. 2007;2:329–336. doi: 10.1021/cb700025j. [DOI] [PubMed] [Google Scholar]
- 11.Gill A. Mini Rev. Med. Chem. 2004;4:301–311. doi: 10.2174/1389557043487385. [DOI] [PubMed] [Google Scholar]
- 12.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 13.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 14.Davies SP, Reddy H, Caivano M, Cohen P. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bain J, McLauchlan H, Elliott M, Cohen P. Biochem. J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overington JP, Al-Lazikani B, Hopkins AL. Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 17.Knight ZA, Shokat KM. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Gray NS. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 19.Ye GF, Tiwari R, Parang K. Curr. Opin. Investig. Drugs. 2008;9:605–613. [PubMed] [Google Scholar]
- 20.Jencks WP. Proc. Natl. Acad. Sci. U. S. A. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hines AC, Cole PA. Bioorg. Med. Chem. Lett. 2004;14:2951–2954. doi: 10.1016/j.bmcl.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 22.Hines AC, Parang K, Kohanski RA, Hubbard SR, Cole PA. Bioorganic Chem. 2005;33:285–297. doi: 10.1016/j.bioorg.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Lee JH, Kumar S, Lawrence DS. ChemBioChem. 2008;9:507–509. doi: 10.1002/cbic.200700583. [DOI] [PubMed] [Google Scholar]
- 24.Loog M, Uri A, Raidaru G, Jarv J, Ek P. Bioorg. Med. Chem. Lett. 1999;9:1447–1452. doi: 10.1016/s0960-894x(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 25.Parang K, Cole PA. Pharmacol. Therapeut. 2002;93:145–157. doi: 10.1016/s0163-7258(02)00184-5. [DOI] [PubMed] [Google Scholar]
- 26.Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA. Nat. Struct. Biol. 2001;8:37–41. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]
- 27.Schneider TL, Mathew RS, Rice KP, Tamaki K, Wood JL, Schepartz A. Org. Lett. 2005;7:1695–1698. doi: 10.1021/ol050179o. [DOI] [PubMed] [Google Scholar]
- 28.Meyer SC, Shomin CD, Gaj T, Ghosh I. J Am Chem Soc. 2007;129:13812–13813. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
- 29.Hill ZB, Perera BGK, Maly DJ. J. Am. Chem. Soc. 2009;131:6686–6688. doi: 10.1021/ja900871y. [DOI] [PubMed] [Google Scholar]
- 30.Qin DH, Sullivan R, Berkowitz WF, Bittman R, Rotenberg SA. J. Med. Chem. 2000;43:1413–1417. doi: 10.1021/jm990340z. [DOI] [PubMed] [Google Scholar]
- 31.Weatherman RV, Mortell KI, Chervenak M, Kiessling LL, Toone EJ. Biochemistry. 1996;35:3619–3624. doi: 10.1021/bi951916z. [DOI] [PubMed] [Google Scholar]
- 32.Hines AC, Cole PA. Bioorg. Med. Chem. Lett. 2004;14:2951–2954. doi: 10.1016/j.bmcl.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 33.Profit AA, Lee TR, Lawrence DS. J. Am. Chem. Soc. 1999;121:280–283. [Google Scholar]
- 34.Murray AJ. Sci Signal. 2008;1 doi: 10.1126/scisignal.122re4. re4. [DOI] [PubMed] [Google Scholar]
- 35.Loog M, Uri A, Raidaru G, Jarv J, Ek P. Bioorg. Med. Chem. Lett. 1999;9:1447–1452. doi: 10.1016/s0960-894x(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 36.Grieco P, Gitu PM, Hruby VJ. J. Pept. Res. 2001;57:250–256. doi: 10.1111/j.1399-3011.2001.00816.x. [DOI] [PubMed] [Google Scholar]
- 37.Hargittai B, Sole NA, Groebe DR, Abramson SN, Barany G. J. Med. Chem. 2000;43:4787–4792. doi: 10.1021/jm990635c. [DOI] [PubMed] [Google Scholar]
- 38.Rajarathnam K, Sykes BD, Dewald B, Baggiolini M, Clark-Lewis I. Biochemistry. 1999;38:7653–7658. doi: 10.1021/bi990033v. [DOI] [PubMed] [Google Scholar]
- 39.Ram N, Weiss N, Texier-Nogues I, Aroui S, Andreotti N, Pirollet F, Ronjat M, Sabatier JM, Darbon H, Jacquemond V, De Waard M. J. Biol. Chem. 2008;283:27048–27056. doi: 10.1074/jbc.M804727200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prade L, Engh RA, Girod A, Kinzel V, Huber R, Bossemeyer D. Structure. 1997;5:1627–1637. doi: 10.1016/s0969-2126(97)00310-9. [DOI] [PubMed] [Google Scholar]
- 41.Lamers M, Antson AA, Hubbard RE, Scott RK, Williams DH. J. Mol. Biol. 1999;285:713–725. doi: 10.1006/jmbi.1998.2369. [DOI] [PubMed] [Google Scholar]
- 42.Jacobs MD, Black J, Futer O, Swenson L, Hare B, Fleming M, Saxena K. J. Biol. Chem. 2005;280:13728–13734. doi: 10.1074/jbc.M413155200. [DOI] [PubMed] [Google Scholar]
- 43.Zhao B, Bower MJ, McDevitt PJ, Zhao HZ, Davis ST, Johanson KO, Green SM, Concha NO, Zhou BBS. J. Biol. Chem. 2002;277:46609–46615. doi: 10.1074/jbc.M201233200. [DOI] [PubMed] [Google Scholar]
- 44.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 45.Komander D, Kular GS, Bain J, Elliott M, Alessi DR, van Aalten DMF. Biochem. J. 2003;375:255–262. doi: 10.1042/BJ20031119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawrie AM, Noble MEM, Tunnah P, Brown NR, Johnson LN, Endicott JA. Nat. Struct. Biol. 1997;4:796–801. doi: 10.1038/nsb1097-796. [DOI] [PubMed] [Google Scholar]
- 47.Boggon TJ, Li YQ, Manley PW, Eck MJ. Blood. 2005;106:996–1002. doi: 10.1182/blood-2005-02-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin YJ, Yun CL, Burakoff SJ. J. Biol. Chem. 1999;274:28301–28307. doi: 10.1074/jbc.274.40.28301. [DOI] [PubMed] [Google Scholar]
- 49.Meyer SC, Gaj T, Ghosh I. Chem. Biol. Drug Des. 2006;68:3–10. doi: 10.1111/j.1747-0285.2006.00401.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.