Abstract
Cyclic GMP modulates gene expression in vascular smooth muscle cells (SMC) in part by stimulating cGMP-dependent protein kinase I (PKGI) and the phosphorylation of transcription factors. In some cells, cGMP increases nuclear translocation of PKGI and PKGI-dependent phosphorylation of transcription regulators; however, these observations have been variable and the mechanisms mediating nuclear PKGI translocation are incompletely understood. We tested the hypothesis that proteolytic cleavage of PKGI is required for cGMP-stimulated nuclear compartmentation of PKGI and phosphorylation of transcription factors. We detected an NH2-terminal PKGI fragment with leucine zipper domain immunoreactivity in the cytosol and endoplasmic reticulum of SMC, but only a COOH-terminal PKGI fragment containing the catalytic region (now termed PKGIγ), was observed in the Golgi apparatus (GA) and nucleoplasm. Posttranslational PKGI processing in the GA was critical for nuclear compartmentation of PKGIγ because GA disruption with nocodazol or brefeldin A inhibited PKGIγ nuclear localization. PKGIγ immunoreactivity was particularly abundant in the nucleolus of interphase SMC where its colocalization with the nucleolar dense fibrillar component protein fibrillarin closely matched the level of nucleolar assembly. Purified nucleolar PKGIγ enzyme activity was insensitive to cGMP stimulation, which is consistent with its lack of the NH2-terminal auto-inhibitory domain. Mutation of a putative proteolytic cleavage region in PKGI inhibited cGMP-mediated phosphorylation of cAMP response element-binding protein, cAMP response element-dependent transcription, and nuclear localization of PKGIγ. These observations suggest that post-translational modification of PKGI critically influences the nuclear translocation of PKGI and activities of cGMP in SMC.
Keywords: cGKI, guanylate cyclase, gene expression regulation, signal transduction, vascular disease
Cyclic GMP is a key regulator of vascular smooth muscle cell (SMC) cytoskeletal kinetics, proliferation, and differentiation and abnormalities in cGMP signaling have been associated with pulmonary and peripheral vascular disease.1 cGMP is synthesized by guanylyl cyclases, which are activated by nitric oxide, carbon monoxide, and natriuretic peptides, and is metabolized by phosphodiesterases. Through the interaction with cytoplasmic proteins, cGMP influences SMC shape and migration and decreases vascular tone. Recently, cGMP has also been observed to modulate the expression of genes that influence SMC phenotype and proliferation.2 For example, cGMP has been noted to regulate SMC gene expression by increasing the phosphorylation of transcription factors, such as cAMP response element-binding protein (CREB)3-5 and activating transcription factor-1 (ATF-1),5 altering the expression of transcription regulators such as activator protein-1 (AP-1)3 and the growth arrest-specific homeobox transcription factor (GAX),6 and regulating the activity of other nuclear factors, such as serum response factor (SRF).7
cGMP-dependent protein kinase I (PKGI) appears to modulate many of the nuclear activities of cGMP. Upon activation by cGMP, PKGI increases the phosphorylation of several nuclear transcription factors. PKGI is a threonine/serine kinase present in two isoforms in SMC as a result of alternate mRNA splicing. These isoforms, PKGIα and PKGIβ, have distinct NH2-terminal leucine zipper (LZ) domains.1,8 Through these LZ domains, the PKGI isoforms homodimerize and also interact with heterologous proteins, which anchor them in the cytosol and in proximity to their phosphorylation targets.9 The conserved COOH-terminal portion of PKGI contains two cGMP-binding domains and a catalytic region containing Mg2+/ATP-binding, kinase, and substrate recognition domains. The importance of PKGI in modulating transcription factor phosphorylation is supported by the observation that in cells lacking PKGI, such as baby hamster kidney (BHK cells), cGMP does not phosphorylate CREB or ATF-1. In contrast, in cells that express PKGI, such as some low passage SMC, 3T3 fibroblasts, and BHK cells transfected with PKGI-encoding plasmids, cGMP stimulates phosphorylation of CREB and ATF-1.3,5
Several studies suggest that nuclear translocation of PKGI is important for cGMP-mediated regulation of transcription factors in SMC. PKGI has been detected in nuclei in some SMC lines.3,10 Moreover, cGMP induces nuclear localization of recombinant PKGI in BHK cells, which do not express endogenous PKGI, where it mediates cGMP-stimulated phosphorylation of CREB.3 Importantly, this activity of cGMP in BHK cells is dependent on the activity of a putative monomeric nuclear localization sequence in PKGI that resides within the Mg2+/ATP-binding domain.10 In addition, the nuclear activities of cGMP have been observed to occur independently of the Ca2+, PKA, and MAPK signaling pathways.3 However, there is variability in the PKGI nuclear localization and activities reported in several studies. In some investigations, PKGI was not observed in the nucleus of primary SMC or SMC lines11-13 and did not modulate cGMP-dependent gene expression.11 One study reported that in cells that lack cGMP-modulated PKGI translocation, cGMP does not modulate CREB phosphorylation or cAMP response element (CRE)-dependent gene expression.11 This variability in cGMP-mediated nuclear PKGI localization and nuclear function suggests that additional mechanisms might influence PKGI-dependent cGMP nuclear signaling. In this study, we investigated the role of PKGI proteolysis in PKGI function and localization and established its importance in mediating cGMP-induced nuclear events in SMC.
Materials and Methods
Detailed information is provided in the Supplemental Information.
Antibodies and reagents
The anti-PKGI regulatory domain (PKGIREG) antibody was kindly provided by S. Janssens14 and the human anti-pyruvate dehydrogenase complex E2 subunit autoantibodies were provided by D. Bloch.15 The other antibodies and reagents were obtained from commercial sources.
Cell culture and transfection
Pulmonary artery smooth muscle cells (PASMC) were isolated from rats;16 all other cells were obtained from American Type Culture Collection. PASMC and RFL-6 cells were maintained in RPMI 1640 and the others in DMEM medium. PASMC medium was supplemented with 10% Nuserum and all other media contained 10% fetal bovine serum. Cells were transfected using a cationic lipid-based reagent. Luciferase activities were measured using a commercially available kit and a luminometer and normalized to the average luciferase activity observed in cGMP-treated wild type PKGIβ expressing cells.
Subcellular fractionation
SMC nuclei and nucleoli were purified as described in the Supplemental Information in the presence of protease inhibitors. Nuclear purity was confirmed using phase contrast microscopy; nucleolar purity was confirmed using phase contrast microscopy and Azure C and fluorescent RNA-binding dyes.
Plasmid construction and mutagenesis
The plasmids encoding wild type and mutant PKGI were constructed using standard techniques17 and a plasmid encoding murine PKGIβ (provided by M. Uhler).11 FLAG-PKGIβ and PKGIβ-FLAG represent NH2-terminal and COOH-terminal FLAG epitope-tagged PKGIβ, respectively.
Purification and analysis of PKGI fragments
The purified NH2-terminal fragment of PKGIα was identified following trypsic digestion using LC-MS/MS and data base interrogation. To assess the NH2-terminal end of PKGIγ, nuclei of 8-Br-cGMP-treated A7r5 cells were purified and homogenized in lysis buffer and PKGIγ was collected using immobilized anti-PKGCR antibody. After SDS-PAGE, purified PKGIγ was subjected to protein microsequencing using Edman degradation.
Immunodetection of proteins
Cells and purified nuclei and nucleoli were fixed with 4% paraformaldehyde, exposed to 100% methanol, and blocked with 1% serum in PBS. After incubation with primary antibody or pre-immune serum, biotin-conjugated secondary antibodies and fluorochrome-linked strepavidin or fluorescent probe-labeled secondary antibodies were employed and subsequently detected using epifluorescent microscopy. For Western blotting, protein fractions were resolved using SDS-PAGE, transferred to polyvinylidene fluoride membranes, exposed to indicated primary antibodies and detected using enzyme-conjugated secondary antibodies and chemiluminescence.
PKGI enzyme activity
PKG enzyme activity was measured using a PKGI-specific peptide substrate as previously described.16
Quantification of PKGIγ nuclear localization
Cells transfected with plasmids encoding wild type and mutant PKGIβ-FLAG and green fluorescent protein with a nuclear localization sequence (GFPNLS) were treated with 8-Br-cGMP, exposed to digitonin in PBS containing protease inhibitors, and then fixed as described above. PKGIβ-FLAG, reacted with biotinylated anti-FLAG antibody and Alexa Fluor 610-conjugated streptavidin, and GFPNLS were detected using epifluorescence. Identically registered epifluorescent images of six randomly-oriented, non-overlapping 1.0 mm2-fields were obtained and analyzed using ImageJ.18
Statistical analysis
Data are represented as mean ± SD. Data were analyzed using a randomized complete block design and, because no differences were found between the groups, the results were analyzed using one-way ANOVA. When significant differences were detected, a Scheffe test was used post hoc. Significance was determined at P<0.05.
Results
Nuclear PKGI lacks NH2-terminal LZ domain immunoreactivity
We reasoned that the reported variability in nuclear localization of PKGI could be due to a post-translational modification of PKGI that modifies its immunologic detection. Therefore, several polyclonal antibodies (PKGI epitopes are schematically defined in Figure 1A) were used to localize PKGI functional domains to SMC compartments. Although the nuclei of many pulmonary artery SMC and cells of SMC and myofibroblast cell lines were observed to harbor PKGICR immunoreactivity, none exhibited PKGIβ LZ domain reactivity, although it was detected in the cytosol (Figure 1B). However, nuclear PKGICR staining was not detected in all SMC. For example, some later SMC and A7r5 cell passages did not exhibit nuclear PKGICR immunoreactivity despite the detection of PKGI LZ domain and COOH-terminal immunoreactivity in the cytosol. Moreover, in individual cultures that exhibited nuclear PKGICR immunoreactivity, expression was non-homogeneous. For example, in areas where SMC were confluent, abundant cytosolic and nuclear PKGICR immunoreactivity was often detected. In contrast, in sub-confluent areas of the same culture, less cytosolic anti-PKGICR reactivity and little or no nuclear PKGICR immunoreactivity were evident. These results suggest the importance of using an objective sampling method when quantifying nuclear compartmentation of PKGI in cultured cells.
Figure 1.
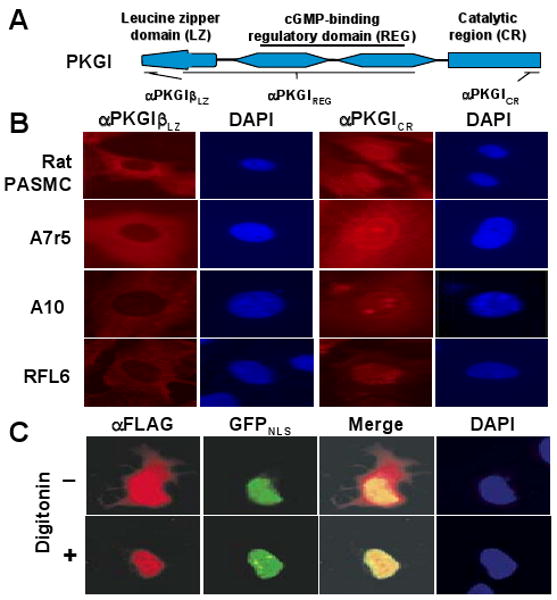
PKGI domain nuclear immunoreactivity in SMC. (A) PKGI has a functional leucine zipper (LZ) domain, cGMP-binding regulatory domain (REG), and catalytic region (CR), which were examined using the indicated antibodies. (B) In the nucleus of rat pulmonary artery smooth muscle cells (PASMC) and indicated SMC lines, PKGICR immunoreactivity was observed although PKGILZ reactivity was not detected. (C) Recombinant PKGI was also localized in SMC nuclei. RFL6 cells expressing PKGIβ-FLAG and GFP with a nuclear localization sequence (GFPNLS) were treated with digitonin, which permits soluble cytosolic PKGI to diffuse from the cell. PKGIβ-FLAG was detected in the nucleus with an anti-FLAG antibody (αFLAG) and co-localized with the GFPNLS epifluorescence.
Although PKGI expressed from transfected plasmids has been detected in nuclei of some cells,10,11 it is not known whether their functional domains are differentially localized. We examined whether the COOH-terminal portion of PKGI over-expressed in RFL6 cells localizes to the nucleus as the endogenous protein does. To improve detection of nuclear PKGI, after RFL6 cells were transiently transfected with plasmids encoding PKGI-FLAG, digitonin was used to permeabilize the plasma membrane and permit the egress of soluble cytosolic PKGI while nuclear PKGI was retained. To aid in nuclear PKGI-FLAG localization, cells were co-transfected with a plasmid encoding green fluorescent protein with a nuclear localization sequence. Immunohistochemistry with anti-FLAG revealed the COOH-terminal portion of PKGIβ co-localized with GFP in the nucleus (Figure 1C) but no nuclear PKGIβ LZ domain immunoreactivity (data not shown).
PKGI is cleaved in vascular SMC
PKGI cleavage might account for the differential compartmentation of PKGI epitopes. We therefore evaluated purified nuclear proteins for evidence of PKGI fragmentation. Plasma membrane disruption and isopycnic density centrifugation permitted the purification of SMC nuclei and effective separation of nuclear and cytosolic proteins (Figure 2A). Immunoblotting using equal amounts of nuclear and cytosolic proteins revealed that SMC nuclear protein fractions did not contain full-length PKGI with NH2-terminal and COOH-terminal epitope immunoreactivity. However, an ∼18-kDa PKGI fragment with LZ domain immunoreactivity (Figure 2B; arrowhead), and a 60-kDa fragment (PKGIγ with PKGIREG and PKGICR immunoreactivity were detected in nuclear proteins. Because we previously found no nuclear immunoreactivity for the PKGIβ LZ domain in intact SMC (Figure 1B), we performed immunofluorescence studies using purified nuclei to clarify the PKGI LZ domain localization. PKGILZ immunoreactivity mapped to asymmetric perinuclear structures in purified SMC nuclei (Figure 2C), co-localizing with molecules restricted to the Golgi apparatus (Online Figure I). These data suggested that PKGI fragmentation occurs outside the nuclear envelope and that only the COOH-terminal fragment of PKGI enters the nucleus.
Figure 2.
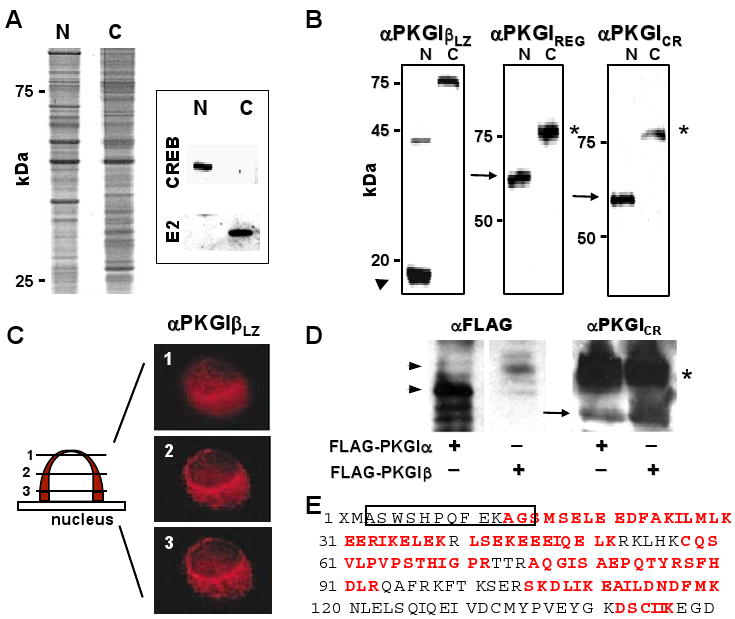
PKGI proteolytic cleavage in SMC. (A) Hypotonic cell shock and isopycnic density centrifugation yielded nuclear (N) and cytoplasmic (C) SMC protein fractions with differential protein abundance and CREB and pyruvate dehydrogenase E2 subunit (E2) immunoreactivity. (B) Purified A7r5 cell nuclear proteins contained a ∼18-kDa protein fragment (arrowhead) with PKGILZ immunoreactivity and a 60-kDa protein (arrow) with PKGIREG and PKGICR immunoreactivity. Although these PKGI fragments were not identified in the cytosolic fraction, full-length ∼78-kDa PKGI in the cytosol exhibited all of these immunoreactivities. (C) PKGIβ LZ domain immunoreactivity was identified in perinuclear regions in Z-dimension optical sections of A7r5 nuclei. (D) and (E) PKGIα was also fragmented in intact cells. NH2-terminal (arrowheads) and COOH-terminal fragments (arrow) of PKGIα and PKGIβ and full-length PKGI isoforms (*) were identified in lysates of BHK cells expressing FLAG-PKGIα and FLAG-PKGIβ. The apparent masses of the NH2-terminal fragments of the PKGI isoforms are consistent with the different sizes of the their LZ domains. Mass spectroscopy identified peptide portions (red letters) of the LZ domain of PKGIα and the precipitating SBP2 epitope (box).
We assessed whether PKGIα is similarly cleaved and compartmented in SMC. Because no antibody detecting the PKGIα LZ domain was available, BHK cells were transfected with a plasmid that encodes NH2-terminal FLAG-tagged PKGIα. The NH2-terminal fragment of PKGIα was detected with anti-FLAG antibody and the COOH-terminal PKGIγ fragment with anti-PKGILZ antibody. Immunoblotting revealed NH2-terminal fragments (Figure 2D, arrow heads) consistent with the lengths of PKGIα and PKGIβ LZ domains and a PKGIγ fragment (arrow). Mass spectroscopy confirmed the identity of the purified NH2-terminal fragment (Figure 2E). The observed cleavage of both PKGI isoforms, the generation of PKGIγ of an identical size upon fragmentation of either PKGIα or PKGIβ (Figure 2D; arrow), and the 60-kDa size of the PKGIγ fragment suggested that the PKGI cleavage site resides in a conserved cGMP-binding domain of the PKGI isoforms.
cGMP increases PKGIγ nuclear localization
Previous studies revealed that cGMP increased PKGI immunoreactivity in the nucleus of BHK and some SMC lines.10 Because only PKGIγ was identified in SMC nuclei, we examined the effect of cGMP on nuclear PKGIγ levels. cGMP was found to increase nuclear PKGIγ abundance in BHK and RFL-6 cells expressing PKGIβ-FLAG (Figure 3A). Moreover, because PKGI expression is increased in post-confluent SMC,19 the influence of SMC density on PKGIγ levels in purified nuclei was examined. cGMP increased nuclear PKGIγ levels in sparsely plated A7r5 cells, but as SMC density increased, so did the level of nuclear PKGIγ immunoreactivity in the absence of cGMP stimulation (Figure 3B). This suggests that as SMC become more confluent and PKGI expression increases, PKGI cleavage and nuclear PKGIγ translocation increases independent of cGMP exposure.
Figure 3.
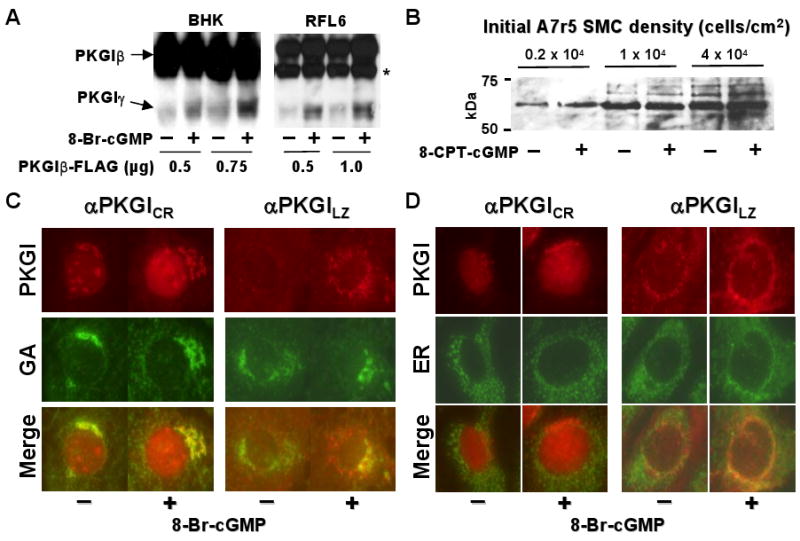
cGMP increased PKGIγ generation and nuclear localization. (A) 8-Br-cGMP increased PKGIγ levels in the lysates of cells transfected with the indicated amounts of a plasmid encoding PKGIβ-FLAG. A non-specific biotin-binding protein was also detected in RFL-6 (*) that did not have anti-PKGICR antibody reactivity. (B) With increased SMC density, PKGIγ levels in nuclei increased and became independent of cGMP treatment. A7r5 cells seeded at the indicated density were exposed to 8-CPT-cGMP and PKGIγ was detected in lysed purified nuclei using an anti-PKGICR antibody. (C) and (D) cGMP increased detection of PKGIγ in the GA and nucleoplasm and the NH2-terminal PKGIβ fragment in the ER. RFL-6 cells were treated with 8-Br-cGMP, exposed to digitonin, and fixed; PKGIγ was detected with an anti-PKGICR antibody, GA α-D-galactoside was visualized using lectin, and ER protein disulfide isomerase with an anti-antibody.
PKGI fragments were detected in the Golgi apparatus (GA; Figure 2C and Online Figure I) and cGMP appeared to increase PKGI fractionation, thus we investigated whether cGMP increases localization of PKGI to the GA. Exposure of RFL-6 cells to cGMP increased PKGI immunoreactivity in the GA and endoplasmic reticulum (ER) (Figures 3C and 3D). PKGICR domain immunoreactivity co-localized with the GA while PKGILZ immunoreactivity also co-localized with the ER. These results suggest that cGMP increases PKGI mobilization to the ER and GA, where cleavage might increase PKGIγ accumulation in the GA and its transfer to the nucleoplasm.
Intact Golgi apparatus is required for nuclear trafficking of PKGIγ
The GA contains endopeptidases that process some signaling proteins and thereby regulate their nuclear trafficking.20 We examined the role of the GA in regulating PKGIγ nuclear compartmentation. The effects of both Nz and BFA on PKGI nuclear localization were tested because they dissociate the GA through different mechanisms: whereas Nz causes microtubular depolymerization and relocation of GA fragments into cytosolic islands,21,22 BFA inhibits protein transport from the ER to the GA by inhibiting GTP-exchange factors23 and disassembles the cis/middle- and trans-Golgi complexes, causing them to fuse with the ER.24 PKGIγ nuclear localization was inhibited in cGMP-treated RFL-6 cells exposed to either Nz or BFA. Following cGMP exposure, PKGICR immunoreactivity continued to be associated with the Nz- and BFA-disrupted GA, however PKGIγ nuclear transport decreased (Figure 4A). Objective quantitative analysis revealed that GA disruption with Nz completely blocked cGMP-stimulated PKGIγ nuclear localization while BFA partially inhibited it (Figure 4B). GA disruption did not inhibit PKGI fractionation: immunoblotting revealed abundant PKGIγ in cGMP-exposed RFL-6 cells treated with these compounds (data not shown), indicating that the effect of GA disruption appears to be on the nuclear translocation of PKGIγ, and not on proteolysis of PKGI.
Figure 4.
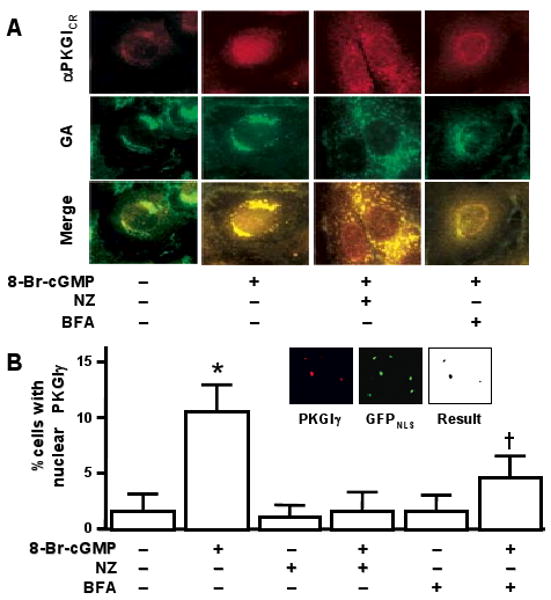
An intact GA was important for PKGIγ nuclear localization. (A) Nz and BFA caused GA disassociation and decreased PKGIγ nuclear localization in 8-Br-cGMP-treated RFL-6 cells. (B) cGMP-induced nuclear PKGIγ localization was abolished by Nz exposure and inhibited by BFA treatment. Nuclear colocalization of PKGIβ-FLAG and GFPNLS, shown in the inset, was objectively quantified in digitonin-treated RFL-6 cells as detailed in the text. Results are expressed as means±SD, n=6 per group, and typical of three independent experiments. * and †P<0.05, indicated treatment vs. the other treatment groups.
PKGIγ is associated with the nucleoli of interphase SMC
We further explored the apparent association between PKGIγ and SMC nucleoli (Figures 1B and 3C). PKGICR immunoreactivity co-localized with SMC nucleoli in situ and with SMC nucleoli purified by ultracentrifugation (Figures 5A and 5B). Moreover, isolated SMC nucleoli exhibited cGMP-independent PKGI enzyme activity at a level similar to that detected in cGMP-treated A7r5 whole cell lysates (Figure 5C). This suggests that removal of the NH2-terminal auto-inhibitory pseudosubstrate site of PKGI through proteolytic cleavage releases inhibition of kinase activity, revealing high constitutive PKGIγ activity. This observation is consistent with in vitro studies where the NH2-terminal region of PKGI was removed with purified proteases.25,26
Figure 5.
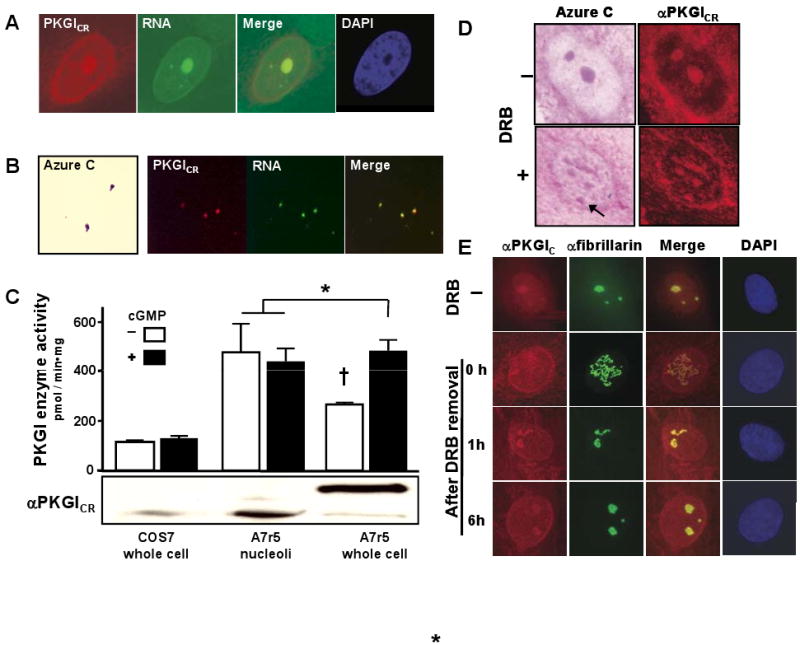
PKGIγ immunoreactivity and enzyme activity were localized in SMC nucleoli. (A) PKGIγ immunofluorescence co-localized with A7r5 cell nucleoli, which were identified by their characteristic interaction with RNA-binding dyes and exclusion of DAPI. (B) PKGIγ identified by PKGICR immunoreactivity also associated with purified A7r5 cell nucleoli, which exhibited Azure C and RNA-binding dye staining. (C) In contrast with whole cell lysates from COS7 cells, lysates of A7r5 cells and their nucleoli had abundant PKGI enzyme activity and immunoreactivity. Results are expressed as means±SD; n=3 each group, and typical of three independent experiments. * and †P<0.05, indicated groups vs. other treatment groups. (D) DRB caused nucleolar disassembly and disrupted nucleolar PKGICR immunoreactivity. PKGIγ was primarily observed in the beaded structures of the DRB-treated nucleoli detected by Azure C staining (arrow). (E) Following removal of DRB, PKGIγ localized in the reassembling nucleolus in a similar manner as fibrillarin, a nucleolar dense fibrillar component protein. The time in hours after DRB removal is indicated.
Transcription and processing of precursor rRNA and packaging of rRNA into ribosomal particles occur within structurally distinct areas of nucleoli. Specifically, the fibrillar centers are thought to harbor rDNA, the dense fibrillar component (DFC) is where pre-rRNA is transcribed and matured and pre-ribosome formation commences, and the granular compartment is where the pre-ribosome is assembled.27 DRB is an adenosine analogue that inhibits rRNA production and permits evaluation of nucleolar assembly and activity.28,29 DRB exposure causes the nucleoli to reversely unravel into intranuclear structures resembling a string of beads, which are comprised of RNA polymerase I and are thought to be single units of rDNA transcription, and a strand of non-transcribed DNA spacer regions.28-30 To investigate whether PKGIγ actively associates with the nucleolar subcomponents, SMC were treated with DRB and the nucleolar association of PKGIγ was assessed. DRB disassembled A7r5 cell nucleoli into beaded strands that retained PKGIγ immunoreactivity (Figure 5D; arrow). During DRB-mediated nucleolar disassembly and reassembly, PKGIγ localized in the nucleolus in a similar manner as fibrillarin (Figure 5E), a DFC protein that processes precursor rRNA,31 indicating that PKGIγ has a dynamic relationship with SMC nucleoli.
Regulation of gene expression by cGMP requires nuclear PKGIγ
Previous studies indicate that nuclear PKGI modulates gene expression in part by phosphorylating CREB.2-4 Because cGMP induces nuclear localization that appears dependent on PKGI cleavage, we examined whether mutation of a putative cleavage site inhibits cGMP-dependent CREB phosphorylation and activity and PKGIγ nuclear localization. To identify the PKGI cleavage site, the NH2-terminal end of immunopurified PKGIγ was detailed using Edman-based amino acid sequencing. Accounting for a protein fragment contributed by bovine serum albumin, peptides commencing with serine138 and glutamic acid153 of PKGIα were identified (detailed in Online Figure III). We generated plasmids with alanine substitutions in this region (mapped in Figure 6A and Online Figure III) and assessed their ability to express catalytically active PKGI and support cGMP-dependent CREB phosphorylation and CRE-dependent gene expression in BHK cells. Transfection conditions in these experiments were optimized so that wild type and mutant PKGI had equivalent immunoreactivity levels. The wild type and mutant PKGI forms exhibited similar cGMP-stimulated cytosolic kinase activity, as shown by the ability of cGMP to stimulate PKGI-dependent VASP serine239 phosphorylation (Figure 6B). However, Mut3 and Mut4 exhibited decreased cGMP-dependent CREB phosphorylation. Of interest, these two mutants had amino acid substitutions associated with the amino acid sequence KVEVTK, which has similarity to a putative proprotein convertase motif.32 Moreover, the amino acids in this area have a high level of species homology in PKGI (Online Figure III). Previous studies found that nuclear localization of PKGI critical for cGMP-dependent stimulation of CRE-mediated gene expression.10 We examined the ability of PKGI Mut3 to confer cGMP-dependent CRE-promoter activity. cGMP increased CRE-dependent promoter activity in BHK cells expressing wild-type PKGIβ but not in cells expressing PKGI Mut3 (Figure 6C). Additional studies revealed that this PKGI mutant exhibited less cGMP-stimulated PKGIγ nuclear localization (Figure 6D). These studies suggest that proteolytic cleavage of PKGI within the cGMP-binding domain is critical for the nuclear activities of cGMP.
Figure 6.
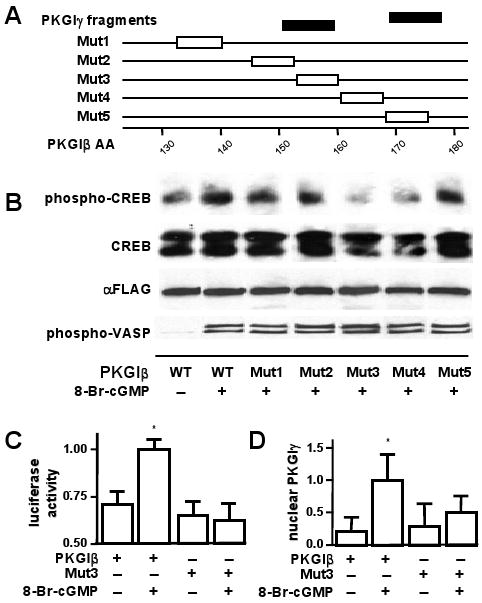
Mutation of the putative proteolytic cleavage area in PKGI inhibited cGMP-mediated gene expression and nuclear localization of PKGIγ. (A) PCR generated plasmids encoding mutant PKGIβ-FLAG (Mut1—5) with alanines (box) in a putative proteolysis area suggested by NH2-terminal amino acid sequence analysis of a fragment of immuno-purified A7r5 cell PKGIγ. (B) Mutation of the putative PKGI cleavage site inhibits cGMP-mediated CREB phosphorylation. Immunoblotting with anti-phospho-CREB antibodies revealed a decrease in phospho-CREB in lysates of cGMP-treated BHK cells expressing mutant PKGIβ-FLAG compared to those expressing wild type PKGI-FLAG. The cells had equivalent cGMP-stimulated cytosolic PKGI activity levels as reflected by phospho-VASP immunoreactivity. (C) PKGI cleavage site mutation inhibits cGMP-dependent CRE-activated gene transactivation. BHK cells transfected with a CRE-luciferase reporter plasmid and expressing Mut3 PKGI-FLAG did not have increased luciferase activity when exposed to 8-Br-cGMP. Results are expressed as means±SD, n=6 each group, and typical of three independent experiments. *P<0.05, indicated treatment vs. the other treatment groups. (D) PKGI cleavage site mutation inhibited PKGIγ nuclear localization. BHK cells expressing Mut3 or wild type PKGI-FLAG were treated with 8-Br-cGMP and examined for FLAG immunoreactivity. 8-Br-cGMP did not increase nuclear PKGIγ immunoreactivity in cells expressing Mut3. Results are expressed as means±SD, n=6 each group, and typical of three independent experiments. *P<0.05, indicated treatment vs. the other treatment groups.
Discussion
The principal objective of this investigation was to examine mechanisms that regulate the nuclear localization of PKGI and modulation of gene expression by cGMP. In SMC, cGMP exposure facilitated the cleavage of the NH2-terminal LZ domain from PKGI and the nuclear localization of PKGIγ, a constitutively active kinase fragment of PKGI. The Golgi apparatus appeared to have an important role in modulating the nuclear translocation of PKGI; Golgi apparatus disruption with either nocodazol or brefeldin A inhibited the cGMP-stimulated nuclear localization of PKGIγ. Additionally, PKGIγ nuclear localization appeared to be required for the regulation of gene expression by cGMP. Mutation of PKGI in the putative proteolysis site inhibited cGMP-mediated CREB-phosphorylation, CRE-dependent gene expression, and the nuclear localization of PKGIγ. cGMP regulates the expression of genes that modulate SMC proliferation and differentiation, therefore, these results support a central role for PKGI in mediating the influence of cGMP on cell phenotype. Moreover, they provide additional evidence that proteolytic pathways regulate some canonical signaling systems. Protein cleavage is also a critical step in the nuclear translocation and activities of sterol regulatory element binding protein (SREBP) and Notch.20 In these pathways, proteolytic removal of a membrane-binding domain releases a protein fragment that can enter the nucleus and modulate gene expression. Similarly, NH2-terminal cleavage of PKGI removes an LZ domain that tethers PKGI to cytosolic proteins. The resulting active kinase PKGI fragment enters the SMC nucleus and phosphorylates proteins that regulate gene expression.
Although the functional domains of PKGI and PKA are arranged similarly, the requirement for proteolytic processing to enable cyclic nucleotide-dependent nuclear signaling distinguishes PKGI from PKA. The PKA heterodimer consists of separate regulatory and catalytic subunits. Upon cAMP binding, the catalytic subunit is released from the regulatory unit which anchors it in the cytosol and enters the nucleus where it modulates gene expression.33 In PKGI, the anchoring (LZ), and regulatory and catalytic (PKGIγ) domains are on a single protein molecule that appear to require cleavage to allow PKGIγ nuclear translocation. Another difference between PKA and PKGI is the requirement for a nuclear localization sequence (NLS) for PKGI nuclear localization. Unlike the PKA catalytic subunit, which is small enough to passively diffuse into the nucleus,34 PKGIγ is too large to enter the nucleus without an NLS to facilitate docking to nuclear pore complexes.35 Gudi and coworkers identified a monomeric NLS in PKGI that facilitated nuclear PKGI localization in BHK cells upon cGMP treatment.3,10 This NLS is in the Mg2+/ATP binding domain of PKGI and is present in PKGIγ.
Studies suggest that cGMP-induced conformational changes in PKGI may expose a cryptic proteolytic site to endopeptidases, regulating PKGI cleavage. For example, in vitro studies indicate that cGMP-binding unfolds PKGI and increases the sensitivity of PKGI to endopeptidase-induced fractionation.36,37 In these experiments, PKGI was not cleaved in the cGMP-binding region, as we report here. This suggests that SMC contain an endopeptidase that cleave amino acid residues in the cGMP-binding region and is distinct from those investigated in the in vitro studies. It might also indicate that additional PKGI post-translational modifications or protein interactions are required to permit PKGI proteolysis within the cGMP-binding region.
The association of PKGIγ with the nucleolus in SMC is a novel finding. The PKGIγ nucleolar compartmentation we report, particularly during nucleolar disassociation and reassembly, and presence of PKGI enzyme activity within purified nucleoli support the notion that PKGI is actively integrated within the nucleolus of SMC. These data suggest that cGMP signaling might influence nucleolar function. Recent proteomic screens indicate that the nucleolus contains not only proteins required for ribosomal biogenesis, but also kinases such as protein kinase C and PKA, which likely transduce cytoplasmic signals.38
The identification of PKGI proteolysis in SMC has important implications for vascular disease mechanisms and the development of novel therapies. Indirect evidence suggests that abnormalities in PKGI processing contributes to neointima formation in injured vessels.13,14,16,39 For example, Monks and coworkers observed that carotid artery injury in rats increases PKGI expression in neointimal SMC.39 However, because the increased PKGI was detected in the perinuclear region of SMC and not within the nucleus, PKGI proteolysis likely was diminished in the injured vessels.39 Decreased nuclear PKGI activity might contribute to SMC proliferation in injured carotid arteries. In early passaged SMC, in which we detected PKGI proteolysis and nuclear PKGIγ, PKGI decreases cell proliferation.16 In contrast, in murine SMC, in which PKGI does not localize in the nucleus,13 PKGI increases cell proliferation.40 Such an attenuation of PKGI proteolysis might inhibit the cytostatic effect of PKGI-based therapies. For example, Sinnaeve and coworkers found that although over-expression of PKGI does not decrease neointimal formation in the injured porcine aorta, transduction of an LZ domain-deficient mutant PKGI, which could passively diffuse into SMC nuclei, is protective.14 Thus, although PKGI expression is up-regulated in some injured vessels, deficient proteolysis could prevent PKGIγ generation and nuclear translocation, blocking cGMP-induced protection against neointimal cell proliferation. Our data indicate that PKGIγ-like molecules may be able to protect against vascular diseases in which PKGI proteolytic mechanisms are diminished.
In summary, we identified a novel mechanism that regulates cGMP nuclear signaling. The observation that PKGI proteolytic processing is a critical step has important implications for understanding how cyclic nucleotides regulate gene expression in health and disease.
Supplementary Material
Acknowledgments
We thank Michael Uhler (Univ. Michigan), Stefan Janssens (Univ. Leuven), Donald Bloch (Mass General Hospital) for generously providing reagents and Ashok Khatri (Mass General Hospital Protein Core Facility) for assisting in the amino acid microsequencing.
Sources of Funding: The National Institutes of Health grant HL080316 supported this work.
Footnotes
Disclosures: None.
References
- 1.Lincoln TM, Dey N, Sellak H. Invited review: cGMP-dependent protein kinase signaling mechanisms in smooth muscle: from the regulation of tone to gene expression. J Appl Physiol. 2001;91:1421–30. doi: 10.1152/jappl.2001.91.3.1421. [DOI] [PubMed] [Google Scholar]
- 2.Pilz RB, Broderick KE. Role of cyclic GMP in gene regulation. Front Biosci. 2005;10:1239–68. doi: 10.2741/1616. [DOI] [PubMed] [Google Scholar]
- 3.Gudi T, Casteel DE, Vinson C, Boss GR, Pilz RB. NO activation of fos promoter elements requires nuclear translocation of G-kinase I and CREB phosphorylation but is independent of MAP kinase activation. Oncogene. 2000;19:6324–33. doi: 10.1038/sj.onc.1204007. [DOI] [PubMed] [Google Scholar]
- 4.Gudi T, Huvar I, Meinecke M, Lohmann SM, Boss GR, Pilz RB. Regulation of gene expression by cGMP-dependent protein kinase. Transactivation of the c-fos promoter. J Biol Chem. 1996;271:4597–600. doi: 10.1074/jbc.271.9.4597. [DOI] [PubMed] [Google Scholar]
- 5.Sauzeau V, Rolli-Derkinderen M, Marionneau C, Loirand G, Pacaud P. RhoA expression is controlled by nitric oxide through cGMP-dependent protein kinase activation. J Biol Chem. 2003;278:9472–80. doi: 10.1074/jbc.M212776200. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita J, Itoh H, Ogawa Y, Tamura N, Takaya K, Igaki T, Doi K, Chun TH, Inoue M, Masatsugu K, Nakao K. Opposite regulation of Gax homeobox expression by angiotensin II and C-type natriuretic peptide. Hypertension. 1997;29:381–7. doi: 10.1161/01.hyp.29.1.381. [DOI] [PubMed] [Google Scholar]
- 7.Gudi T, Chen JC, Casteel DE, Seasholtz TM, Boss GR, Pilz RB. cGMP-dependent protein kinase inhibits serum-response element-dependent transcription by inhibiting rho activation and functions. J Biol Chem. 2002;277:37382–93. doi: 10.1074/jbc.M204491200. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann F. The biology of cyclic GMP-dependent protein kinases. J Biol Chem. 2005;280:1–4. doi: 10.1074/jbc.R400035200. [DOI] [PubMed] [Google Scholar]
- 9.Surks HK. cGMP-dependent protein kinase I and smooth muscle relaxation: a tale of two isoforms. Circ Res. 2007;101:1078–80. doi: 10.1161/CIRCRESAHA.107.165779. [DOI] [PubMed] [Google Scholar]
- 10.Gudi T, Lohmann SM, Pilz RB. Regulation of gene expression by cyclic GMP-dependent protein kinase requires nuclear translocation of the kinase: identification of a nuclear localization signal. Mol Cell Biol. 1997;17:5244–54. doi: 10.1128/mcb.17.9.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins SP, Uhler MD. Cyclic AMP- and cyclic GMP-dependent protein kinases differ in their regulation of cyclic AMP response element-dependent gene transcription. J Biol Chem. 1999;274:8391–404. doi: 10.1074/jbc.274.13.8391. [DOI] [PubMed] [Google Scholar]
- 12.Browning DD, Mc Shane M, Marty C, Ye RD. Functional analysis of type 1alpha cGMP-dependent protein kinase using green fluorescent fusion proteins. J Biol Chem. 2001;276:13039–48. doi: 10.1074/jbc.M009187200. [DOI] [PubMed] [Google Scholar]
- 13.Feil R, Gappa N, Rutz M, Schlossmann J, Rose CR, Konnerth A, Brummer S, Kuhbandner S, Hofmann F. Functional reconstitution of vascular smooth muscle cells with cGMP-dependent protein kinase I isoforms. Circ Res. 2002;90:1080–6. doi: 10.1161/01.res.0000019586.95768.40. [DOI] [PubMed] [Google Scholar]
- 14.Sinnaeve P, Chiche JD, Gillijns H, Van Pelt N, Wirthlin D, Van De Werf F, Collen D, Bloch KD, Janssens S. Overexpression of a constitutively active protein kinase G mutant reduces neointima formation and in-stent restenosis. Circulation. 2002;105:2911–6. doi: 10.1161/01.cir.0000018169.59205.ca. [DOI] [PubMed] [Google Scholar]
- 15.Van de Water J, Fregeau D, Davis P, Ansari A, Danner D, Leung P, Coppel R, Gershwin ME. Autoantibodies of primary biliary cirrhosis recognize dihydrolipoamide acetyltransferase and inhibit enzyme function. J Immunol. 1988;141:2321–4. [PubMed] [Google Scholar]
- 16.Chiche JD, Schlutsmeyer SM, Bloch DB, de la Monte SM, Roberts JD, Jr, Filippov G, Janssens SP, Rosenzweig A, Bloch KD. Adenovirus-mediated gene transfer of cGMP-dependent protein kinase increases the sensitivity of cultured vascular smooth muscle cells to the antiproliferative and pro-apoptotic effects of nitric oxide/cGMP. J Biol Chem. 1998;273:34263–71. doi: 10.1074/jbc.273.51.34263. [DOI] [PubMed] [Google Scholar]
- 17.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Somerset, NJ: John Wiley & Sons; 2007. [Google Scholar]
- 18.Rasband WS. ImageJ. Bethesda, Maryland USA: 19972007. [Google Scholar]
- 19.Lin G, Chow S, Lin J, Wang G, Lue TF, Lin CS. Effect of cell passage and density on protein kinase G expression and activation in vascular smooth muscle cells. J Cell Biochem. 2004;92:104–12. doi: 10.1002/jcb.20043. [DOI] [PubMed] [Google Scholar]
- 20.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–8. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 21.Thyberg J, Moskalewski S. Microtubules and the organization of the Golgi complex. Exp Cell Res. 1985;159:1–16. doi: 10.1016/s0014-4827(85)80032-x. [DOI] [PubMed] [Google Scholar]
- 22.Robbins E, Gonatas NK. Histochemical and Ultrastructural Studies on Hela Cell Cultures Exposed to Spindle Inhibitors with Special Reference to the Interphase Cell. J Histochem Cytochem. 1964;12:704–11. doi: 10.1177/12.9.704. [DOI] [PubMed] [Google Scholar]
- 23.Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–7. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 24.Alcalde J, Bonay P, Roa A, Vilaro S, Sandoval IV. Assembly and disassembly of the Golgi complex: two processes arranged in a cis-trans direction. J Cell Biol. 1992;116:69–83. doi: 10.1083/jcb.116.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue M, Kishimoto A, Takai Y, Nishizuka Y. Guanosine 3′:5′-monophosphate-dependent protein kinase from silkworm, properties of a catalytic fragment obtained by limited proteolysis. J Biol Chem. 1976;251:4476–8. [PubMed] [Google Scholar]
- 26.Lincoln TM, Flockhart DA, Corbin JD. Studies on the structure and mechanism of activation of the guanosine 3′:5′-monophosphate-dependent protein kinase. J Biol Chem. 1978;253:6002–9. [PubMed] [Google Scholar]
- 27.Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2000;2:E107–12. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 28.Granick D. Nucleolar necklaces in chick embryo fibroblast cells. I. Formation of necklaces by dichlororibobenzimidazole and other adenosine analogues that decrease RNA synthesis and degrade preribosomes. J Cell Biol. 1975;65:398–417. doi: 10.1083/jcb.65.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haaf T, Hayman DL, Schmid M. Quantitative determination of rDNA transcription units in vertebrate cells. Exp Cell Res. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-b. [DOI] [PubMed] [Google Scholar]
- 30.Le Panse S, Masson C, Heliot L, Chassery JM, Junera HR, Hernandez-Verdun D. 3-D organization of ribosomal transcription units after DRB inhibition of RNA polymerase II transcription. J Cell Sci. 1999;112:2145–54. doi: 10.1242/jcs.112.13.2145. [DOI] [PubMed] [Google Scholar]
- 31.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–57. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 32.Seidah NG, Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 33.Lalli E, Sassone-Corsi P. Signal transduction and gene regulation: the nuclear response to cAMP. J Biol Chem. 1994;269:17359–62. [PubMed] [Google Scholar]
- 34.Hagiwara M, Brindle P, Harootunian A, Armstrong R, Rivier J, Vale W, Tsien R, Montminy MR. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13:4852–9. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–8. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 36.Takio K, Smith SB, Walsh KA, Krebs EG, Titani K. Amino acid sequence around a hinge region and its autophosphorylation site in bovine Lung cGMP-dependent protein kinase. J Biol Chem. 1983;258:5531–6. [PubMed] [Google Scholar]
- 37.Ruth P, Pfeifer A, Kamm S, Klatt P, Dostmann WR, Hofmann F. Identification of the amino acid sequences responsible for high affinity activation of cGMP kinase Ialpha. J Biol Chem. 1997;272:10522–8. doi: 10.1074/jbc.272.16.10522. [DOI] [PubMed] [Google Scholar]
- 38.Jeong JS, Kim IH, Lee HJ, Choi YC. Nucleolus contains signal molecules that constitute membrane-nucleolus linked pathway. Exp Mol Med. 1998;30:205–13. doi: 10.1038/emm.1998.30. [DOI] [PubMed] [Google Scholar]
- 39.Monks D, Lange V, Silber RE, Markert T, Walter U, Nehls V. Expression of cGMP-dependent protein kinase I and its substrate VASP in neointimal cells of the injured rat carotid artery. Eur J Clin Invest. 1998;28:416–23. doi: 10.1046/j.1365-2362.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- 40.Wolfsgruber W, Feil S, Brummer S, Kuppinger O, Hofmann F, Feil R. A proatherogenic role for cGMP-dependent protein kinase in vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2003;100:13519–24. doi: 10.1073/pnas.1936024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


