Abstract
The paradigm of gene regulation was forever changed by the discovery that short RNA duplexes could directly regulate gene expression. Most regulatory roles attributed to non-coding RNA were often repressive. Recent observations are beginning to reveal that duplex RNA molecules can stimulate gene transcription. These RNA activators employ a wide array of mechanisms to up-regulate transcription of target genes, including functioning as DNA-tethered activation domains, as co-activators and modulators of general transcriptional machinery, and as regulators of other non-coding transcripts. The discoveries over the past few years defy “Moore’s law” in the breath-taking rapidity with which new roles for non-coding RNA in gene expression are being revealed. As gene regulatory networks are reconstructed to accommodate the influence of non-coding RNAs, their importance in maintenance of cellular health will become increasingly apparent. In fact, a new generation of therapeutic agents will focus on modulating the function of non-coding RNA.
Keywords: Riboactivators, RNA activation/activators (RNAa), RNA switches, antigene agRNA, transcription regulation, gene expression, RNAi
Introduction
The role of RNA in transmitting genomic information for the synthesis of cellular machines, in the form of proteins, is a central tenet of biology. The realization that RNA molecules have multiple roles within cells inspired intensive research efforts towards revealing those roles as well as harnessing these biopolymers for novel therapeutics. From the start, the plasticity of RNA was evident from the numerous conformations that it adopted (Patel et al., 2000). Furthermore, the functional versatility of RNA was compellingly demonstrated by directed evolution methods that could generate molecules with enzymatic and regulatory properties (Davidson et al., 2007; Doudna et al., 2002; Joyce, 2004; Wilson et al., 1999). Selection strategies yielded artificial RNA ‘aptamers’ that bind desired small molecules and biopolymers with surprising degrees of specificity and affinity (Patel and Suri, 2000; Serganov et al., 2007). One such aptamer was recently approved as a therapeutic agent for macular degeneration in humans while several others directed at cancer targets such as the cell surface protein nucleolin, tenascin C of the extracellular matrix, and the DNA repair protein Ku are in clinical trials (Bunka et al., 2006; Chu et al., 2007; Hicke et al., 2001; Ireson et al., 2006; Ng et al., 2006).
Nature itself uses RNA structural plasticity and aptamer-type RNA modules to sense the levels of metabolites within cells and to regulate gene expression. In the classical example of attenuation in prokaryotes, nascent RNA adopts different structures to either block or permit transcription of the tryptophan biosynthetic genes in response to the cellular levels of this amino acid (Yanofsky, 2007). In more recent examples, nascent RNA was found to bind directly to small molecule metabolites through an aptamer module and transmit this information to regulate mRNA translation (Henkin et al., 2006; Nudler, 2006; Winkler et al., 2005). How these “riboswitches” transmit information is only now coming into focus (Serganov and Patel, 2007). Regulatory riboswitches were first discovered in microbes and have since been found in plants, fungi and humans (Cheah et al., 2007; Ray et al., 2009; Wachter et al., 2007). While most riboswitches negatively regulate gene expression, an example of positive regulation by a riboswitch has also been described (Mandal et al., 2004).
The role of RNA in regulating gene expression is not limited to riboswitches or attenuators. An unexpectedly robust ability of short duplex RNA molecules to inhibit gene expression led to the discovery of a more general mechanism of regulation by RNA duplexes (Fire et al., 1998). The RNA interference (RNAi) mechanism uses short non-coding RNA duplexes along with cellular proteins to target and silence mRNA by degradation or by translational blockage (Hannon et al., 2004). Intensive scrutiny of this post-transcriptional mechanism of gene expression has led to rapid gains in our understanding of the physiological importance of non-coding RNA transcripts and the cellular machinery that processes and utilizes these transcripts to regulate gene expression (Farazi et al., 2008; Hannon et al., 2006). Typically, the processed RNA is incorporated into the silencing complex called RISC that targets specific mRNA molecules for degradation or translational inhibition (Farazi, Juranek, and Tuschl, 2008; Meister et al., 2004). In plants, fission yeast and Drosophila, short RNA duplexes were also shown to participate in the formation of repressive heterochromatin that blocks gene transcription (Bartel, 2004; Baulcombe, 2006; Bernstein et al., 2005; Du et al., 2007; Grewal et al., 2007; White et al., 2008; Zofall et al., 2006). The principles of RNAi are now sufficiently well understood to harness this cellular process to block the expression of desired genes by using synthetic RNA duplexes that bear sequence complementarity to the targeted mRNA (Hannon and Rossi, 2004). The design of such repressive short interfering RNAs (siRNAs) and their applications has been widely described in the literature.
In this review, the focus is on RNA molecules that positively regulate transcription, the first step in gene expression. The identification of non-natural RNA aptamers that serve as surrogates for protein-based transcription factors (gene-specific activators) leads to questions such as how these molecules function to stimulate transcription. Riboactivators might directly recruit the transcriptional machinery to gene promoters, or they may act in conjunction with protein-based transcriptional activators. In support of both these possibilities, examples of naturally encoded RNA molecules that interact with the transcriptional machinery, as well as those that interact with transcriptional activators to function as co-activators are described. A closer examination of the targets of activators is coupled with the realization that a central component of the transcriptional machinery requires an RNA co-factor to mediate its function. In the final act, the focus is on the astounding revelation that short RNA duplexes require the RNAi machinery to stimulate gene transcription. This discovery, like many in this field, emerged from engineering efforts that utilized synthetic RNAs as “antigene” transcription inhibitors and led to the identification of natural non-coding RNA that target gene promoters to stimulate transcription. The review ends with a discussion of the emerging global role for non-coding RNA in positive regulation of mRNA synthesis.
Can RNA positively regulate gene expression at the level of transcription?
Early work explored the possibility that RNA aptamers could be selected to positively regulate gene expression. Three groups identified RNA activators (riboactivators) that could stimulate gene transcription with increasing efficiency and the final class of riboactivators was engineered to function in a ligand-responsive manner.
a) Genome-encoded RNA activators
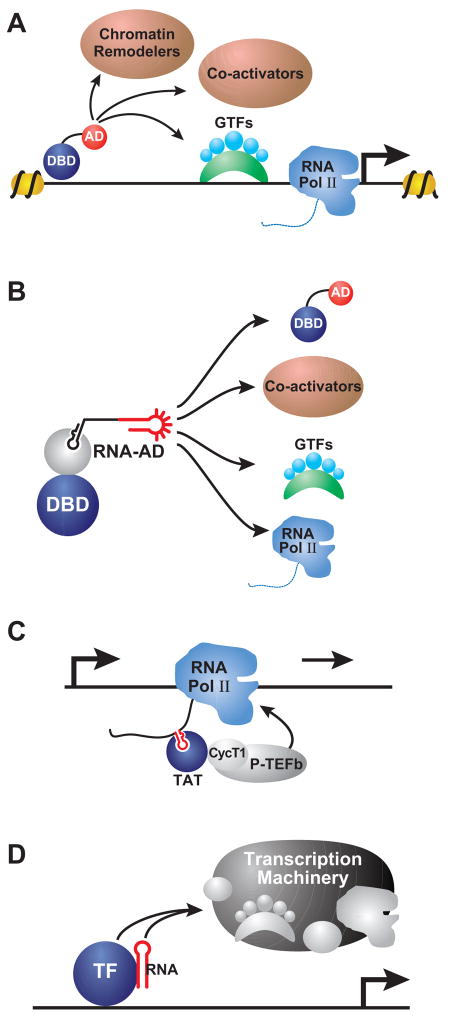
In a study designed to identify endogenous targets of an RNA binding protein it was found that some RNA molecules themselves could stimulate transcription when tethered upstream of a reporter gene. The study relied on an elegant modification of the widely used two-hybrid assay (Fields et al., 1989) (Figure 1A). This assay relies on the modular nature of eukaryotic transcription factors wherein the DNA binding domain (DBD) can be separated from the activation domain (AD) (Ptashne et al., 2002). If separated, the modules do not assemble into a functional unit and fail to stimulate transcription (Figure 1A). However, if each domain is fused to proteins that interact non-covalently then the interactions between the fused partner proteins tether the activation domain to the DNA bound domain (Ma et al., 1988). The resulting “two-hybrid” complex reconstitutes a functional activator that can recruit the transcriptional machinery and stimulate the expression of proximal genes (Figure 1A) (Fields and Song, 1989). The ability to non-covalently tether activation domains to a DNA bound protein was used to create a “three-hybrid” assay (SenGupta et al., 1996; Zhang et al., 2000). Here the DNA binding domain and the activation domain are fused to heterologous RNA binding proteins (Figure 1A). The bridging molecule is a bi-functional RNA where one part interacts with the protein fused to the DBD and the other part interacts with the protein fused to the AD.
Figure 1.
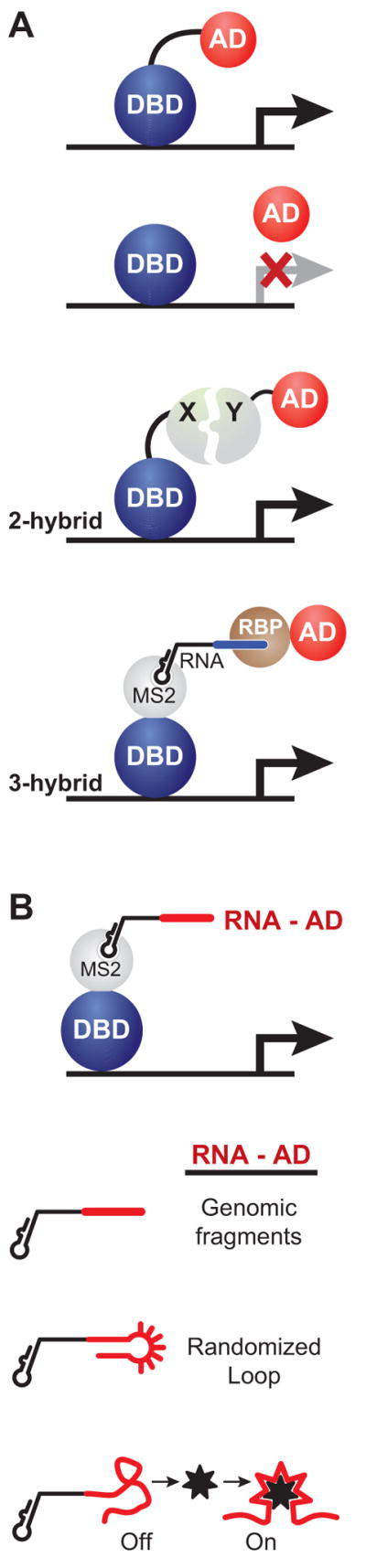
Modular architecture of transcription activators. (A) Eukaryotic transcription factors typically contain a DNA binding domain (DBD) and an activation domain (AD). Separating the two domains eliminate transcriptional activation of target genes. However, tethering the AD to a DBD, by non-covalent protein interactions (X and Y) suffices to stimulate transcription. This principle is the basis of the 2-hybrid assay. In the 3-hybrid assay, a third bridging molecule tethers the AD to the DBD. In this case the bridging molecule is a bifunctional RNA molecule that bears and MS2-binding hairpin and a region that interacts with an RNA binding protein (RBP). (B) The DNA bound MS2 protein is used to tether RNA molecules bearing an MS2-binding hairpin and a putative RNA-Activation Domain (in red) upstream of a reporter gene. Three types of riboactivators are illustrated below.
To identify RNA molecules that interact with Snp1, a yeast protein that interacts with U1 spliceosomal RNA, the authors fused RNA fragments from the S. cerevisiae genome to a RNA module that interacts with the bacteriophage MS2 coat protein (Figure 1B). The DNA-bound MS2 coat protein tethers the fused genomic RNA sequence upstream of a reporter plasmid (Sengupta et al., 1999). These RNA molecules were not expected to stimulate expression without the third “hybrid” partner, namely the Snp1 protein fused to an activator domain. While the most robust activation of the reporter gene resulted from interaction of Snp1 with the U1 RNA, a significant fraction of RNA molecules were able to weakly stimulate the reporter gene in the absence of Snp1 (Sengupta, Wickens, and Fields, 1999). These RNA activators show no sequence consensus and it remains unclear how they stimulate gene expression. Moreover, because these RNA activators were obtained from genomic sequences, it is possible that they function as bona fide activators at certain genes. Nevertheless, these serendipitously identified riboactivators were the first examples of non-coding RNAs that could stimulate transcription of proximal genes.
b) In vivo selection of non-natural riboactivators
Ptashne and co-workers instead were actively seeking RNA surrogates for transcription activators (Saha et al., 2003). The activation domains (AD, in Figures 1A and 2A) of eukaryotic transcription factors are thought to interact with components of the transcription machinery and the chromatin remodeling enzymes (Figure 2A) (Ptashne and Gann, 2002). These domains are often negatively charged with a few interspersed hydrophobic residues (Ansari et al., 1998; Cress et al., 1991; Drysdale et al., 1995). Importantly, robust non-natural octapeptide activation domains could be readily isolated from in vivo selections (Lu et al., 2000; Lu et al., 2005). However, it was unclear if non-peptic molecules could function as activation domains. To examine the importance of retaining the peptidic nature of activation domains, Ptashne and co-workers tested if RNA aptamers would function as activation domains (Saha et al., 2003). The premise was based on the chemical likeness of the RNA with peptidic activation domain (negative charge of the RNA phosphodiester backbone and the hydrophobicity of the nucleobases) as well as ability of RNA aptamers to target a wide array of ligands, from small molecules to complex biopolymers. If an aptamer, tethered to DNA upstream of a gene, could target a component of the machinery it would stimulate transcription.
Figure 2.
Targets of protein and RNA activation domains. (A) Protein based transcription factors recruit several different protein complexes to gene promoters. These include several different chromatin modifying and remodeling complexes, co-activators, general transcription factors (GTFs), and the RNA polymerase II (Pol II). Nucleosomes are indicated as yellow spheres and the transcription start site denoted by a right-angled arrow. (B) An artificial riboactivator may interact with components of the transcriptional machinery, protein-based transcription factors and chromatin remodeling complexes. (C) The nascent transcript from the HIV-1 long terminal repeat (TAR) interacts with a viral protein transcription factor (TAT) and together they recruit pTEFb to a stalled polymerase. The pTEFb-associated kinase phosphorylates Pol II and promotes transcript elongation. (D) Developmental transcription factors (TF) like Dlx2 and NRSF/REST interact with their RNA partners to recruit the transcriptional machinery to their target genes.
The in vivo selection relied on three-hybrid system to tether a bi-functional RNA molecule upstream of a reporter (Saha et al., 2003). The aptamer was placed within a stem-loop scaffold that was known to be stable in vitro and in vivo (Figure 1B) (Tsai et al., 1992; van Venrooij et al., 1990). Ten positions in the loop were randomized, yielding a library of nearly a million non-identical sequence variants. The whole collection of molecules was screened to identify RNA aptamers that functioned as transcriptional activators. Unlike similar screens for non-natural peptide activators in which robust activators were obtained with high frequency (10−1), the screen for riboactivators yielded activators at significantly lower frequency (10−6). Nevertheless, this was the first demonstration of an engineered RNA aptamer functioning as a transcriptional activator. These results greatly strengthened the notion that RNA molecules could directly communicate with the promoter-associated proteins to stimulate gene transcription. The riboactivators were significantly more active than the genomic sequences (Sengupta, Wickens, and Fields, 1999). The riboactivators identified by Saha et al. were comparable in the activation potential to typical yeast transcription factors such as Gcn4 but were about 10% as active as the strongest activators, such as Gal4. Within the stem-loop scaffold the molecules that activated transcription showed a surprising consensus in their sequence (5′UGCNGGNUC3′). Based on the small library complexity (106 sequences), the scaffold constraints and the appearance of a clear consensus motif, the authors predicted that a library with more sequence variants might yield stronger riboactivators (Saha et al., 2003). Indeed, a later study utilizing significantly longer RNA molecules with greater sequence complexity identified stronger riboactivators (Buskirk et al., 2003).
c) Ligand-regulated riboactivators
Inspired by the design of natural riboswitches, where a ligand-responsive module is coupled to a regulatory module, Liu and co-workers integrated a small molecule aptamer within a non-essential element of their most robust riboactivator (Buskirk et al., 2004). Several rounds of selection and enrichment led to ligand-responsive riboactivators. The selection yielded molecules wherein ligand binding stabilized structural elements and enhanced the function of the riboactivator (Figure 1B). This study elegantly incorporated the principles of natural riboswitches to generate artificial riboactivators whose activity could be regulated by a cell-permeable small molecule ligand. While these molecules still require a DNA bound protein to localize them to the target gene, they provide a first step toward generating riboactivators that may include an additional aptamer component that targets transcription factors of choice at one end and delivers a ligand-responsive activation module at the other.
How do riboactivators mediate their function?
The mechanism by which riboactivators stimulate transcription is an important issue that remains unresolved. The manner, by which they were identified, as DNA-tethered molecules, suggests that riboactivators would function akin to natural peptide activators. While the bona fide targets of protein transcription activators remain an area of debate and investigation (Mapp and Ansari, 2007), it is clear that most protein activation domains facilitate the assembly of the transcriptional machinery at gene promoters (Figure 2A) (Ptashne and Gann, 2002). In addition to acting at early steps, protein activators may also act at later stages to facilitate efficient transcript elongation.
a) Targeting components of the transcriptional machinery
Riboactivators might function by recruiting limiting components of the transcriptional machinery and thereby nucleate the assembly of the multi-protein complex at gene promoters (Figure 2B). In support of this model, Saha and co-workers found that their short riboactivator interacted with the TATA box binding protein, TBP (Saha et al., 2003). This well studied protein is part of at least three complexes that bind to the promoters of the three RNA polymerases in eukaryotes. In the case of protein-coding genes, RNA polymerase II binds gene promoters at a step subsequent to the binding of TBP and its associated proteins (Bryant et al., 2003; Buratowski, 2000; Buratowski et al., 1989). At several promoters binding of TBP, as a subunit of the larger TFIID complex, is thought to be rate limiting. Recruitment of TBP/TFIID by protein activators or by direct tethering to the promoter proximal sites stimulates gene expression (Chatterjee et al., 1995; Ptashne and Gann, 2002; Roeder, 1996; Verrijzer et al., 1996). In addition, transcription factors often interact with other protein complexes such as Mediator proteins that interact with RNA polymerase or chromatin remodeling complexes that act on chromatin templates to make the underlying DNA elements accessible to the transcriptional machinery (Kornberg, 2005; Lee et al., 2000; Malik et al., 2005; Ptashne and Gann, 2002; Smith et al., 2005; Taatjes et al., 2004; Workman, 2006). Similarly, riboactivators might interact with the Mediator as well as other protein complexes that stimulate gene transcription. In a more direct recruitment event, riboactivators may interact with the polymerase and recruit it to the promoter. The C-terminal domain (CTD) of the largest subunit of RNA polymerase II is known to interact directly with RNA as well as RNA-binding proteins (Kaneko et al., 2005; Phatnani et al., 2006). It is possible that different riboactivators target specific components of the transcriptional machinery or that riboactivators interact with multiple targets and facilitate the recruitment of different rate-limiting components at different promoters. The outstanding mechanistic questions are only now beginning to be addressed.
b) Targeting protein-based transcription factors
The most celebrated example of a non-coding RNA modulating gene transcription is that of trans-activation response (TAR) element of the AIDS-causing virus HIV. In this example, RNA polymerase II transcribes a short region of the HIV-1 gene but fails to elongate the transcript. The TAR segment of the nascent transcript folds into a stem-loop structure and associates with a virally-encoded transcriptional activator protein called TAT (Figure 2C) (Rosen et al., 1985; Selby et al., 1989). The non-coding TAR segment and the associated TAT protein specifically interact with the cellular cyclinT1 (Garber et al., 1998). The cyclinT1-dependent kinase, Cdk9 in the pTEFb complex is thereby also recruited to the stalled polymerase (Figure 2C). This kinase phosphorylates the CTD of the polymerase and triggers the release of negative regulators and association of complexes that facilitate transcriptional elongation (Bres et al., 2008). The TAR segment thus up-regulates transcription by interacting with a transcription factor and a component of the transcriptional elongation machinery. It is important to note, that this RNA module only works in cis at the viral gene and is not known to stimulate transcription by stalled polymerases across the host genome.
Non-coding RNAs also interact with DNA-bound transcription factors to stimulate gene expression (Figure 2D). Two recent examples include the Evf-2 and NRSE non-coding transcripts. Evf-2 interacts with the homeodomain transcription factor, Dlx2 and augments the activation strength of that developmental activator (Feng et al., 2006). This non-coding RNA functions only at genes that are bound and regulated by Dlx2. The authors suggest that Evf-2 transcript acts together with Dlx2 as a “co-transcription factor” to robustly activate target genes and coordinate the induction of neuronal and craniofacial developmental programs. At this early stage many questions on the nature of Evf-2 function and generality of RNA co-transcription factors remain unanswered. Intriguingly, Evf-2 sequence is transcribed from an “ultraconserved” sequence overlapping the enhancer region of genes for Dlx5/6 developmental transcription factors (TF). Across the genome, several hundred such ultraconserved non-coding regions, whose transcription are linked to developmental transcription factors, have been bioinformatically identified (Feng et al., 2006). The possible existence of hundreds of evolutionarily conserved RNA transcription factor pairs imply an unexpectedly broad role for non-coding RNA as co-transcription factors.
In another example, a short non-coding RNA duplex associates with a neuron-restrictive silencing factor (NRSF/REST) and converts it into a transcriptional activator (Kuwabara et al., 2004). NRSF binds and represses the expression of genes that dictate stem cell differentiation into neurons. The ability of this RNA duplex (NRSE) to reverse the repressive state of NRSF led to hypothesis that the RNA displaces NRSF from its DNA binding sites. Removal of the repressor would lead to the expression of the cell-fate defining genes. The short RNA duplex shows sequence homology to DNA sites bound by NRSF and might function by strand-invasion and R-Loop formation at the DNA binding sites. The resulting RNA-DNA hybrid disallows NRSF binding to its regulatory sites in the genome. Alternatively, the DNA binding domain of NRSF could potentially bind to the RNA itself, despite the significantly different helical structure of RNA duplexes. Astonishingly, the RNA duplex does not act by disrupting the NRSF-DNA complex, instead it binds to DNA associated NRSF and likely blocks the ability of this transcription factor to interact with co-repressors (Kuwabara et al., 2004). The resulting complex stimulates the expression of the target genes that induce differentiation of stem cells into neurons (Figure 2D). It remains to be seen if the non-coding RNA, tethered to gene promoters by NRSF, actively stimulates gene expression by directly interacting with the transcriptional machinery or the chromatin remodeling machinery.
RNA as an adaptor between Transcription Factors and the transcriptional machinery
With increasing reports of transcription factor-RNA interactions at gene promoters, it becomes pertinent to explore the possibility that such RNA molecules participate as co-activators or molecular bridges between DNA-bound TFs and the transcriptional machinery. Two examples of natural RNA molecules that associate with the transcriptional machinery and facilitate gene activation are discussed below (Figure 3).
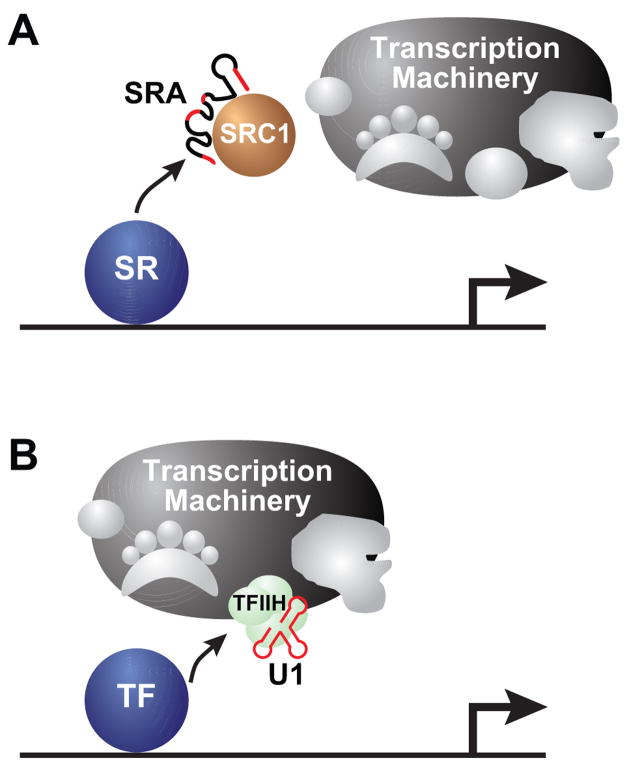
Figure 3.
RNA co-activators. (A) Upon binding their ligand, steroid receptors (SR) interact with several co-activators. A non-coding RNA (SRA) in complex with a protein (SRC-1) functions as a co-activator for steroid receptor class of transcription factors. Sub-elements of the RNA (in red) are important for SR mediated transcription activation. The co-activators interface with the transcriptional machinery and recruit it to the target gene promoter. (B) A general transcription factor complex (TFIIH) is often targeted by protein-based transcription factors, including steroid receptor class of TFs. TFIIH is found to be associated with the U1 snRNA. The RNA molecule is critical for the kinase activity and it strongly facilitates transcription reinitiation by Pol II.
a) RNA co-activators
In a two-hybrid assay designed to fetch targets of the progesterone receptor (PR), an unusual cDNA clone, riddled with stop codons and lacking a reasonable protein-coding region, was isolated (Lanz et al., 1999). Several elegant experiments indicated that the RNA transcript itself, rather than any potential protein encoded by it, was required for gene activation. This non-coding RNA molecule enhanced ligand-responsive transcription by several steroid receptors (SR) but did not alter the strength of other classes of transcription factors. The ability of this RNA molecule to discriminate between different classes of transcription factors yet function with several members of one class suggested that it was a class-specific co-activator molecule. The steroid receptor RNA activator (SRA) did not act alone, but was found to be associated with a protein (SRC-1) known to be a specific co-activator of this class of hormone receptors (Figure 3A). SRA can also interact with other proteins that selectively function with estrogen receptor alpha (Shi et al., 2001; Watanabe et al., 2001). The ribonucleoprotein complex interacts with steroid receptors and components of the transcriptional machinery (Figure 3A). Within the complex, SRA is thought to confer specificity for steroid receptors, and sub-elements within the RNA that are required for this function have been identified (Lanz et al., 2002). It remains to be seen how these elements modulate the interaction of steroid receptors with the co-activator complex and the transcriptional machinery.
b) RNA within the transcriptional machinery
In biochemical or genetic dissections of the transcriptional machinery, attention is rarely paid to non-protein components. So it came as a surprise that TFIIH, a well-studied protein complex that is a target of transcription factors and is required for transcription, associates with a non-coding RNA (Figure 3B) (Kwek et al., 2002). The TFIIH complex has 10 protein subunits, many of which have enzymatic functions that are essential for transcription by RNA polymerase II (Pol II). The association of a non-coding RNA greatly enhances those functions, especially the kinase and helicase activities that are vital for transcription initiation, promoter release and early steps of transcript elongation (O’Gorman et al., 2005). Loss of the associated RNA strongly diminishes the ability of the TFIIH complex to promote re-initiation of transcription in cell-free systems. The RNA in question is none other than the spliceosomal U1 snRNA. Moreover, genes with promoter-proximal splice sites are particularly responsive to the presence of U1 in the TFIIH complex. The non-coding U1 snRNA associates with TFIIH via the cyclinH subunit of the Cdk7 kinase. This is reminiscent of the TAR-cyclinT1 interaction that facilitates later stages of transcript elongation. Coincidentally, both TFIIH and pTEFb act on the repetitive heptapeptide motif within the CTD of Pol II. TFIIH-associated Cdk7 kinase phosphorylates Ser5 whereas pTEFb-associated Cdk9 phosphorylates Ser2 of the YS2PTS5PS heptapeptide of the CTD (Phatnani and Greenleaf, 2006). The phospho-Ser5 mark facilitates transcriptional initiation whereas the phospho-Ser2 mark stimulates transcription elongation. Thus, non-coding RNA via TFIIH or pTEFb overcome two rate limiting steps of mRNA synthesis.
As in the case of SRA, sub-elements of U1 that interact with TFIIH have been identified however it is unclear how this association modulates TFIIH function. Moreover, the precise role this RNA plays in global transcription by Pol II in vivo remains poorly understood. In this context, it is noteworthy that a sub-element of U1 that interacts with TFIIH, was used as a scaffold for riboactivators that were described in the earlier section. While the U1 sub-element lacking the artificial riboactivator loop sequence does not elicit transcription, it is possible that it contributes to the overall activity of the riboactivator.
In addition to general factors and apart from the CTD-RNA interactions, a different region of Pol II is targeted by a non-coding RNA that is expressed when cells experience stress (Allen et al., 2004; Espinoza et al., 2004; Goodrich et al., 2006). This non-coding B2 RNA, transcribed by Pol III under stress conditions, associates with Pol II and represses its function. A similar RNA-based mechanism inhibits eubacterial RNA polymerase in stationary phase (Wassarman et al., 2000). Non-natural aptamers that inhibit eukaryotic Pol II have also been isolated (Thomas et al., 1997) and their Pol II-bound structure determined by X-ray crystallography (Kettenberger et al., 2006). Together these studies define a non-coding RNA binding site on Pol II and it is possible that riboactivators bind this pocket and stimulate transcription.
Similarly, several studies have defined non-coding RNA that block transcription by inducing the formation of heterochromatin at target genomic loci (Bartel, 2004; Baulcombe, 2006; Bernstein and Allis, 2005; Du and Zamore, 2007; Grewal and Elgin, 2007; White and Allshire, 2008; Zofall and Grewal, 2006). As we learn more about the role of non-coding RNAs in transcription regulation it is likely that molecules that stimulate transcription by interacting with the chromatin machineries will also be discovered.
Several antigene-RNAs function as RNA activators (RNAa)
While RNAi-based approaches have proven to be extremely powerful, additional methods are being developed to inhibit gene expression at the level of transcription initiation. One such approach relies on RNA duplexes that share sequence homology with gene promoters rather than the protein-coding regions. These “antigene” (agRNA) molecules would hybridize with critical promoter elements and prohibit the binding of transcription factors and the transcriptional machinery (Janowski et al., 2005). In the process of developing such agRNA molecules, two groups serendipitously discovered that certain RNA duplexes could stimulate transcription of the targeted genes (Janowski et al., 2007; Li et al., 2006). This recent discovery of RNA activation (RNAa) surprised even the community that redefined the traditional notions of gene regulation by discovering RNAi. Astonishingly, RNAa duplexes that stimulate transcription do so with the help of the exact same cellular machinery that is needed for RNAi mediated inhibition of protein translation, the final step of gene expression (Janowski et al., 2006; Li et al., 2006; Place et al., 2008; Schwartz et al., 2008). RNAa does not appear to function by binding transcription factors or displacing repressors from promoters nor does it mediate its function by indirect perturbation of inhibitory loops within gene regulatory networks.
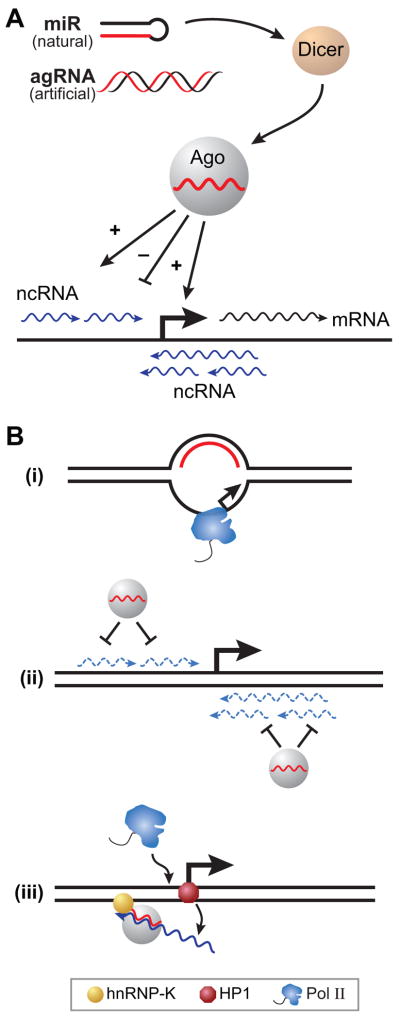
A computational search across the genome hinted that several natural non-coding RNA transcripts showed sequence homology to endogenous gene promoters (Place et al., 2008). Some of these transcripts due to a self-complementary sequence could form RNA hairpins. In studying RNAi it was discovered that short non-coding RNA hairpins, also called microRNA (miR), serve as substrates for Dicer, a protein that cleaves and processes these hairpins to generate RNA duplexes (Figure 4A). A single RNA strand of the duplex is loaded onto a member of the Argonaute (Ago) family of proteins. The single stranded RNA guides Ago2 and its partners to complementary mRNA where they mediate their repressive function. A recently discovered microRNA, miR-373, contains sequences that are homologous to a region several hundred base pairs upstream of the E-cadherin and CSD-C2 genes. Previously, miRs that did not target mRNA sequences were thought to be irrelevant since they would not function in RNAi. However, while investigating the function of miR-373 Dahiya and co-workers found that it required both Dicer and Ago2 to stimulate the transcription of the two target genes (Place et al., 2008). The result suggested that RNAa molecules, in association with a member of the Ago family, targeted intergenic transcripts rather than the underlying promoter elements. Indeed, RNAa molecules excised from miR-373 targeted non-coding transcripts at the E-cadherin promoter. In the absence of anti-sense transcripts that traversed the promoter, the RNAa molecule was incapable of stimulating mRNA synthesis of E-cadherin.
Figure 4.
RNA activation (RNAa) by natural or artificial RNA duplexes. (A) Naturally encoded RNA microRNAs (miRs) are processed into duplexes by Dicer and a single stranded ‘guide’ is loaded on to Ago2. The synthetic or artificial RNA duplexes, designed to target gene promoters also associate with Ago2. The RNA-Ago2 complex can stimulate (+) or repress (−) mRNA synthesis based on the precise sequence complementarity of its guide RNA. In addition to the bona fide mRNA (wavy black line) several non-coding sense and antisense transcripts are also generated at active promoters (wavy blue lines). (B) Three proposed models for the RNAa function. In the first, RNA hybridizes to the non-template strand leaving the template strand exposed for Pol II binding. The second model suggests that non-productive transcripts are eliminated by Ago-RNAa, thereby enhancing mRNA synthesis. The third model suggests that Ago-RNAa complex is transiently tethered to the PR gene promoter by hnRNP-K protein. This interaction facilitates the displacement of the repressive chromatin-binding protein HP-1γ from the promoter onto the antisense RNA. The promoter is then accessible to Pol II whereas the HP-1γ bearing RNA is likely released from the promoter.
A biochemical study showed that a positively acting agRNA duplex (not all agRNAs are stimulatory) was bound to Ago2 and to an antisense transcript that traversed the progesterone receptor (PR) gene (Schwartz et al., 2008). Tagging the “guide” strand of this agRNA/RNAa with biotin did not affect its biological function, thus providing a convenient handle to affinity-purify the agRNA/RNAa and the associated cellular machinery. The short RNA not only targeted the PR antisense transcript but also hnRNP-K, a protein that binds DNA and RNA. The PR promoter has hnRNP-K binding sites. Thus, the isolation of hnRNP-K bound to the antisense RNA offers a tantalizing clue as to how RNAa molecules may be targeted to gene promoters. Perhaps hnRNP-K serves as a protein bridge, binding both the promoter DNA and the RNAa associated antisense transcript. If so, this interaction must be transient or fairly unstable because a DNA-hnRNP RNAa ternary complex was not detected by standard experiments. A more baffling observation is that the positioning of the RNAa on the anti-sense transcript was critical because a few base pair offset could yield a molecule that inhibited rather than stimulated transcription (Figure 4A) (Janowski et al., 2007). The opposing regulatory functions of overlapping agRNAs are even more perplexing because both molecules require the antisense transcript to manifest their function.
Both genetic and biochemical studies have revealed an exciting and unexpected mode of RNA activation. The stimulatory role of RNA duplexes targeted at the transcription start site or significantly upstream has raised many interesting questions. For example, does RNAa act to dampen antisense transcription or does it utilize these transcripts to further tune mRNA synthesis? The requirement for prior transcription raises the critical issue of RNAa molecules functioning merely as “signal amplifiers” or “rheostats” rather than gene switches. In this scenario, protein factors function as key regulatory switches whereas RNAa may simply buffers gene expression from stochastic transcriptional variability. This question arises from the observation that stimulatory agRNA are most effective at lower levels of PR gene expression but reverse function to inhibit high degree of PR gene expression in the same cell line.
The initial exploration of RNAa action has also raised several mechanistic questions. A major enigma involves the rules that differentiate stimulatory from repressive agRNA duplexes. RNA molecules that bind overlapping sites on the antisense RNA utilize identical cellular machines (Dicer and Ago complexes) but have opposite regulatory effects on gene transcription. These inexplicable differences will have to be clarified for RNAa molecules to have general applications as research tools or therapeutics. More immediately, how RNAa molecules stimulate gene expression remains unresolved. Several models have been proposed to explain their stimulatory function (Figure 4B). An early model, positing that the RNA hybridizes with the non-template strand of the promoter to create an exposed single-stranded template has not been satisfactorily ruled out (Britten et al., 1969; Frenster, 1976). The formation of the DNA-RNA hybrid would generate a stable R-loop and the single-stranded template stand could enter the narrow cleft of RNA polymerase II to gain access to the active site (Armache et al., 2003; Bushnell et al., 2003). An alternative hypothesis is that RNAa molecules bound to the Ago complex dampen the expression of upstream transcripts (Figure 4B). In S. cerevisiae, such transcripts have been shown to regulate the transcription of genes by displacing transcription factors and machinery that binds to gene promoters (Martens et al., 2004). The Ago bound RNAa molecules may also dampen downstream antisense transcripts. Whether these “dampening” mechanisms require the degradation of upstream or downstream transcripts remains to be conclusively determined. Moreover, why bona fide protein-coding mRNA molecules would escape similar inhibition and instead be stimulated remains unclear, especially because in the case of the PR gene, the longer antisense transcript has been shown to be m7G capped, spliced, and polyadenylated (Schwartz et al., 2008). As noted above, a more elaborate model states that RNAa or RNAi molecules are localized to gene promoters via protein bridges that bind both RNA and promoter DNA. This act of promoter localization facilitates the transfer of chromatin-bound repressors (HP1γ) onto the antisense transcripts (Figure 4B). The experimental results support the last model and run counter to Occam’s razor of simplicity (Schwartz et al., 2008). In light of the unexpected ability of RNAa molecules to stimulate gene transcription, it should not be surprising if mechanisms as elaborate as those attributed to protein-based activators are involved in RNAa function.
Future Scope
It is increasingly plain that RNA can regulate gene expression by a plurality of mechanisms. Early pioneering work led to aptamers against transcription factors and components of the transcriptional machinery and these have proven to be valuable mechanistic tools (Fan et al., 2005; Sevilimedu et al., 2008; Shi et al., 1999; Thomas et al., 1997; Wurster et al., 2008; Zhai et al., 2001). The eventual goal is to use such aptamers as therapeutic agents that can act on aberrantly functioning transcriptional circuits. Rather than use aptamers to competitively perturb molecular interactions, three groups used aptamers tethered upstream of promoters to stimulate gene expression. In a somewhat similar manner, a pTEFb binding TAR “aptamer” is used by HIV-1 to recruit the machinery required to overcome a rate-limiting step of transcriptional elongation. As we explore further, a broader role for natural aptamers in regulating transcription elongation will come into focus.
The Evf-2 RNA activator also hints at a major role for non-coding RNA in regulating gene networks in animal development. The paradigm set by this example implies that ultraconserved non-coding regions that are linked with genes encoding developmental transcription factors (TF) may well generate RNA molecules that interact with the protein TFs and reconstitute potent RNA-TF activators. A bioinformatic search for similar examples reveals hundreds of potential RNA-TF pairs. That non-coding RNA sequences may evolve more easily than protein domains has interesting implications on coupling of animal development and evolution of new functions in transcriptional regulators.
The role of non-coding transcripts in modulating gene expression of β-globin genes surprised all who had studied the regulation of this gene for decades (Gribnau et al., 2000). The intergenic transcripts were important for placing chromatin marks that facilitate transcription. This is contrary to the accepted role of RNA molecules in formation of repressive heterochromatin and in facilitating repressive DNA methylation patterns at promoters. The direct role of RNA molecules in targeting chromatin modifying/remodeling enzymes for transcriptional stimulation has yet to be demonstrated. However, it is not difficult to imagine that such RNA molecules will soon be found.
Another role of intergenic RNA has emerged from the search for RNA duplexes that could target gene promoters and silence transcription of desired genes. Instead, several synthetic RNA duplexes stimulated the expression of targeted genes. This led to the identification of endogenous non-coding RNA that target gene promoters and regulate their transcriptional robustness. Computational analysis has implicated many potential non-coding transcripts in similar functions. Initial bioinformatic searches for miRs focused solely on sequences that share sequence homology with coding regions of the genome. Recent high throughput sequencing methods have not only begun to reveal the existence of miRs that could target regulatory regions but have also found antisense transcription from bona fide mRNA promoters and from within coding regions of genes (Kapranov et al., 2007). These anti-sense and cryptic transcripts are not restricted to a subset of genes but are prevalent across the genome (Amy et al., 2008; Core et al., 2008; He et al., 2008; Preker et al., 2008). More importantly, in cases where such transcripts have been eliminated, they were found to diminish mRNA synthesis in the sense direction (Schwartz et al., 2008). The prevalence of such transcripts suggests that this newly discovered mode of gene regulation by RNAa is likely a general modulator of transcription.
As these new and exciting mechanisms of gene stimulation become apparent, they will undoubtedly be harnessed to regulate the transcription of desired genes. The goal of engineering gene regulatory networks will inevitably have to account for the role of endogenous non-coding RNAs. Indeed, synthetic biological applications led to the discovery of riboactivators. As our appreciation for the role of RNA activation increases and the role of such molecules in maintenance of health and the onset of disease becomes apparent, a new class of therapeutic agents targeting such molecules will be developed. RNA-targeting drugs may well overcome the limitations of targeting only protein regulators of gene expression.
Each of the examples of RNA activation described here has far-reaching implications. Most of these stimulatory functions were only recently discovered and the rate of new discoveries in this area shows no sign of abating. A new RNA world awaits discovery and a slight modification of the scout’s motto “be prepared to be surprised” may serve us best as we go forward.
Acknowledgments
I would like to thank Mary Ozers and Leslie Donato for thoughtful comments on the manuscript. The work in my laboratory is supported by the NIH, NSF, USDA, March of Dimes Foundation and Shaw scholar award.
References
- Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- Amy C, Seila JMC, Levine Stuart S, Yeo Gene W, Rahl Peter B, Flynn Ryan A, Young Richard A, Sharp Phillip A. Divergent Transcription from Active Promoters. Science. 2008;322:1849. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari AZ, Reece RJ, Ptashne M. A transcriptional activating region with two contrasting modes of protein interaction. Proc Natl Acad Sci USA. 1998;95:13543–13548. doi: 10.1073/pnas.95.23.13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc Natl Acad Sci U S A. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC. Short silencing RNA: the dark matter of genetics? Cold Spring Harb Symp Quant Biol. 2006;71:13–20. doi: 10.1101/sqb.2006.71.052. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- Bunka DH, Stockley PG. Aptamers come of age -at last. Nat Rev Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Snapshots of RNA polymerase II transcription initiation. Curr Opin Cell Biol. 2000;12:320–325. doi: 10.1016/s0955-0674(00)00095-8. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc Natl Acad Sci U S A. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk AR, Kehayova PD, Landrigan A, Liu DR. In vivo evolution of an RNA-based transcriptional activator. Chem Biol. 2003;10:533–540. doi: 10.1016/s1074-5521(03)00109-1. [DOI] [PubMed] [Google Scholar]
- Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Chu T, Ebright J, Ellington AD. Using aptamers to identify and enter cells. Curr Opin Mol Ther. 2007;9:137–144. [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008;322:1845. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress WD, Triezenberg SJ. Critical Structural Elements of the Vp16 Transcriptional Activation Domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Davidson EA, Ellington AD. Synthetic RNA circuits. Nat Chem Biol. 2007;3:23–28. doi: 10.1038/nchembio846. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- Drysdale CM, Duenas E, Jackson BM, Reusser U, Braus GH, Hinnebusch AG. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. Beginning to understand microRNA function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Fan X, Shi H, Lis JT. Distinct transcriptional responses of RNA polymerases I, II and III to aptamers that bind TBP. Nucleic Acids Res. 2005;33:838–845. doi: 10.1093/nar/gki212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1204. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Frenster JH. Selective control of DNA helix openings during gene regulation. Cancer Res. 1976;36:3394–3398. [PubMed] [Google Scholar]
- Garber ME, Wei P, Jones KA. HIV-1 Tat interacts with cyclin T1 to direct the P-TEFb CTD kinase complex to TAR RNA. Cold Spring Harb Symp Quant Biol. 1998;63:371–380. doi: 10.1101/sqb.1998.63.371. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rivas FV, Murchison EP, Steitz JA. The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:551–564. doi: 10.1101/sqb.2006.71.064. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The Antisense Transcriptomes of Human Cells. Science. 2008;322:1855. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM, Grundy FJ. Sensing metabolic signals with nascent RNA transcripts: the T box and S box riboswitches as paradigms. Cold Spring Harb Symp Quant Biol. 2006;71:231–237. doi: 10.1101/sqb.2006.71.020. [DOI] [PubMed] [Google Scholar]
- Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, Schmidt PG, Warren S. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Corey DR. Inhibiting transcription of chromosomal DNA using antigene RNAs. Nucleic Acids Symp Ser (Oxf) 2005:367–368. doi: 10.1093/nass/49.1.367. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Younger ST, Hardy D, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells withpromoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- Joyce GF. Directed evolution of nucleic acid enzymes. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Manley JL. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3′ end formation. Mol Cell. 2005;20:91–103. doi: 10.1016/j.molcel.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Eisenfuhr A, Brueckner F, Theis M, Famulok M, Cramer P. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat Struct Mol Biol. 2006;13:44–48. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O’Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O’Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 comple. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lanz RB, Razani B, Goldberg AD, O’Malley BW. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA) Proc Natl Acad Sci U S A. 2002;99:16081–16086. doi: 10.1073/pnas.192571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ansari AZ, Ptashne M. An artificial transcriptional activating region with unusual properties. Proc Natl Acad Sci U S A. 2000;97:1988–1992. doi: 10.1073/pnas.040573197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Rowe S, Brennan BB, Davis S, Metzler RE, Nau JJ, Majmudar CY, Mapp AK, Ansari AZ. Unraveling the mechanism of a potent transcriptional activator. J Biol Chem. 2005;280:29689–29698. doi: 10.1074/jbc.M504895200. [DOI] [PubMed] [Google Scholar]
- Ma J, Ptashne M. Converting a Eukaryotic Transcriptional Inhibitor into an Activator. Cell. 1988;55:443–446. doi: 10.1016/0092-8674(88)90030-x. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Dynamic regulation of Pol II transcription by mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mapp AK, Ansari AZ. A TAD further: exogenous control of gene activation. ACS Chem Biol. 2007;2:62–75. doi: 10.1021/cb600463w. [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Ng EW, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- Nudler E. Flipping riboswitches. Cell. 2006;126:19–22. doi: 10.1016/j.cell.2006.06.024. [DOI] [PubMed] [Google Scholar]
- O’Gorman W, Thomas B, Kwek KY, Furger A, Akoulitchev A. Analysis of U1 small nuclear RNA interaction with cyclin H. J Biol Chem. 2005;280:36920–36925. doi: 10.1074/jbc.M505791200. [DOI] [PubMed] [Google Scholar]
- Patel DJ, Suri AK. Structure, recognition and discrimination in RNA aptamer complexes with cofactors, amino acids, drugs and aminoglycoside antibiotics. J Biotechnol. 2000;74:39–60. doi: 10.1016/s1389-0352(99)00003-3. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup M, Jensen TH. RNA Exosome Depletion Reveals Transcription Upstream of Active Human Promoters. Science. 2008;322:1851. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Genes and Signals. Cold Spring Harbor Laboratory Press; New York: 2002. [Google Scholar]
- Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009 doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Rosen CA, Sodroski JG, Haseltine WA. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985;41:813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- Saha S, Ansari AZ, Jarrell KA, Ptashne M. RNA sequences that work as transcriptional activating regions. Nucleic Acids Res. 2003;31:1565–1570. doi: 10.1093/nar/gkg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby MJ, Bain ES, Luciw PA, Peterlin BM. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989;3:547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Sengupta DJ, Wickens M, Fields S. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. Rna. 1999;5:596–601. doi: 10.1017/s1355838299002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci U S A. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilimedu A, Shi H, Lis JT. TFIIB aptamers inhibit transcription by perturbing PIC formation at distinct stages. Nucleic Acids Res. 2008;36:3118–3127. doi: 10.1093/nar/gkn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hoffman BE, Lis JT. RNA aptamers as effective protein antagonists in a multicellular organism. Proc Natl Acad Sci U S A. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat Rev Mol Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- Thomas M, Chedin S, Carles C, Riva M, Famulok M, Sentenac A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J Biol Chem. 1997;272:27980–27986. doi: 10.1074/jbc.272.44.27980. [DOI] [PubMed] [Google Scholar]
- Tsai DE, Kenan DJ, Keene JD. In vitro selection of an RNA epitope immunologically cross-reactive with a peptide. Proc Natl Acad Sci U S A. 1992;89:8864–8868. doi: 10.1073/pnas.89.19.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Venrooij WJ, Hoet R, Castrop J, Hageman B, Mattaj IW, van de Putte LB. Anti-(U1) small nuclear RNA antibodies in anti-small nuclear ribonucleoprotein sera from patients with connective tissue diseases. J Clin Invest. 1990;86:2154–2160. doi: 10.1172/JCI114954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:176–181. [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, Masuhiro Y, Kato S. A subfamily of RNA-binding DEAD-box proteins acts as anestrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. Embo J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- White SA, Allshire RC. RNAi-mediated chromatin silencing in fission yeast. Curr Top Microbiol Immunol. 2008;320:157–183. doi: 10.1007/978-3-540-75157-1_8. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- Wurster SE, Maher LJ. Selection and characterization of anti-NF-kappaB p65 RNA aptamers. RNA. 2008;14:1037–1047. doi: 10.1261/rna.878908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA. 2007;13:1141–1154. doi: 10.1261/rna.620507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai G, Iskandar M, Barilla K, Romaniuk PJ. Characterization of RNA aptamer binding by the Wilms’ tumor suppressor protein WT1. Biochemistry. 2001;40:2032–2040. doi: 10.1021/bi001941r. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kraemer B, SenGupta D, Fields S, Wickens M. Yeast three-hybrid system to detect and analyze RNA-protein interactions. Methods Enzymol. 2000;318:399–419. doi: 10.1016/s0076-6879(00)18066-8. [DOI] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2006;71:487–496. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]