Abstract
Background and Aims
Dendrobium species show tremendous morphological diversity and have broad geographical distribution. As repetitive sequence analysis is a useful tool to investigate the evolution of chromosomes and genomes, the aim of the present study was the characterization of repetitive sequences from Dendrobium moschatum for comparative molecular and cytogenetic studies in the related species Dendrobium aphyllum, Dendrobium aggregatum and representatives from other orchid genera.
Methods
In order to isolate highly repetitive sequences, a c0t-1 DNA plasmid library was established. Repeats were sequenced and used as probes for Southern hybridization. Sequence divergence was analysed using bioinformatic tools. Repetitive sequences were localized along orchid chromosomes by fluorescence in situ hybridization (FISH).
Key Results
Characterization of the c0t-1 library resulted in the detection of repetitive sequences including the (GA)n dinucleotide DmoO11, numerous Arabidopsis-like telomeric repeats and the highly amplified dispersed repeat DmoF14. The DmoF14 repeat is conserved in six Dendrobium species but diversified in representative species of three other orchid genera. FISH analyses showed the genome-wide distribution of DmoF14 in D. moschatum, D. aphyllum and D. aggregatum. Hybridization with the telomeric repeats demonstrated Arabidopsis-like telomeres at the chromosome ends of Dendrobium species. However, FISH using the telomeric probe revealed two pairs of chromosomes with strong intercalary signals in D. aphyllum. FISH showed the terminal position of 5S and 18S–5·8S–25S rRNA genes and a characteristic number of rDNA sites in the three Dendrobium species.
Conclusions
The repeated sequences isolated from D. moschatum c0t-1 DNA constitute major DNA families of the D. moschatum, D. aphyllum and D. aggregatum genomes with DmoF14 representing an ancient component of orchid genomes. Large intercalary telomere-like arrays suggest chromosomal rearrangements in D. aphyllum while the number and localization of rRNA genes as well as the species-specific distribution pattern of an abundant microsatellite reflect the genomic diversity of the three Dendrobium species.
Keywords: Orchidaceae, Dendrobium moschatum, Dendrobium aphyllum, Dendrobium aggregatum, repetitive DNA, FISH, c0t-1 DNA
INTRODUCTION
Orchidaceae is the largest family among angiosperms, consisting of about 850 genera with 25 000 species (Dressler, 1993). Dendrobium is the third largest genus of Orchidaceae comprising, at the time of writing, of 1184 species (Leitch et al., 2009). Dendrobium species are characterized by a broad geographical distribution, a tremendous diversity in growth habits and the ability to produce a large number of interspecific hybrids with different morphology. The systematics of the subtribe Dendrobiinae was extensively studied on the basis of morphological key characters (Dressler, 1981), and on the basis of chloroplast DNA sequences (Yukawa et al., 1996, 2000). However, the classification of many Dendrobium species remains ambiguous (Clements, 2003).
For Bangladesh, 16 Dendrobium species have been described (Hossain, 2002), some of which either have economical value as ornamental plants or are regarded as endangered species. In cytogenetic analyses, karyotypes of some Dendrobium species from Bangladesh have been compared (Begum and Alam, 2004, 2005). However, unequivocal species differentiations are hampered by almost similar chromosome numbers (2n = 38–40) and only few differences in chromosome morphology.
Repetitive sequence families are major components of plant genomes (Heslop-Harrison, 2000). According to genomic organization, repeats are divided into tandemly arranged and dispersed sequences (Schmidt and Heslop-Harrison, 1998). Tandem repeats are divided into satellite DNA, micro- and minisatellites, telomeric repeats and ribosomal genes. Typical plant satellite DNA repeats range in size between 160–180 and 320–360 bp and are organized in tandem arrays with up to 105 copies per haploid genome (Hemleben et al., 2000). Microsatellites are simple sequence repeats representing a unique type of tandemly repeated genomic sequences. They are abundantly distributed in small arrays across the genome and show high levels of polymorphism in sequence and copy number. Some tandem repeats are also functional. The ends of eukaryotic chromosomes form a unique chromatin domain that comprises the telomeres and the adjacent subtelomeric regions, often consisting of degenerated telomere sequences. In most plants, telomeres are tandem arrays of the repeat unit 5′-TTTAGGG-3′, which is widely distributed among the majority of plant species (Richards and Ausubel, 1988; Ganal et al., 1991). The most conserved tandemly arranged sequences are ribosomal RNA genes in eukaryotes comprising 18S–5·8S–25S rRNA repeating units forming long arrays.
The rapid evolution of both tandemly arranged and dispersed repetitive DNA often results in species-specific repeat variants and the generation of novel sequence families. Therefore, comparative studies of plant repetitive sequences are useful to investigate the evolutionary relationships between plant species (Kamm et al., 1995) and suggest that comparative study of repetitive sequences would be useful for investigation of the relationships among orchid species.
The present paper describes the molecular characterization of repetitive sequences including dispersed, telomeric and microsatellite repeats in three Dendrobium species. The genomic organization of repetitive DNA sequences was elucidated by comparative Southern hybridization in Dendrobium species and species of distantly related orchid genera. Physical mapping by fluorescence in situ hybridization (FISH) was used to show the distribution of the different types of repeat families as well as 18S–5·8S–25S and 5S rRNA genes along chromosomes.
MATERIALS AND METHODS
Plant material and genomic DNA extraction
Nine different orchid species were used, six of which belong to the genus Dendrobium and the remaining three to different genera. Dendrobium moschatum Sw., Dendrobium aphyllum Roxb. and Dendrobium aggregatum Roxb. were collected in Bangladesh. Dendrobium anosmum L., Dendrobium fimbriatum var. oculatum Hook.f., Dendrobium palpebrae L. and other orchid species Nageliella angustifolia (Booth ex Lindl.) Ames et Correl., Oncidium sphacelatum L. and Phalaenopsis lueddemanniana var. purpurea (Rchb.f.) Sweet. were obtained from the Dresden Botanical Garden, Germany.
Genomic DNA from each species was isolated from young leaves using the DNeasy Maxi Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Chromosome preparation
Primary roots were collected and washed in running tap water for 4–5 min. After removal of excess water, roots were incubated in 8-hydroxyquinoline (0·002 m) for 5 h at 18 °C, fixed in 45 % acetic acid for 15 min at 4 °C and stored in 70 % ethanol at 4 °C. Fixed roots were washed in enzyme buffer (0·01 m citric acid–sodium citrate, pH 4·6) to remove the fixative and digested at 37 °C for 2 h in enzyme solution consisting of 2·5 % pectinase, 2·5 % cellulose, 2·5 % pectolyase and 1·0 % cytohelicase in enzyme buffer. The dropping method was applied for the preparation of mitotic metaphase chromosomes according to Schwarzacher and Heslop-Harrison (2000).
Fluorescence in situ hybridization
FISH probes were labelled with biotin-16-dUTP or digoxigenin-11-dUTP by PCR or nick translation, respectively. Sites of hybridization were detected immunologically by antibodies coupled to fluorochromes. The clone pTa71 from Triticum aestivum (Gerlach and Bedbrook, 1979) consisting of a large part of the 18S–5·8S–25S rRNA genes was labelled with digoxigenin-11-dUTP by nick translation, while the clone pXV1 (Schmidt et al., 1994) containing the 5S rRNA gene from Beta vulgaris was labelled with biotin-16-dUTP using PCR. Probes of D. moschatum were labelled with biotin-16-dUTP (clone DmoF14) and digoxigenin-11-dUTP (clone DmoO11). The telomeric sequence (clone DmoB22) was labelled with digoxigenin-11-dUTP using PCR.
FISH was performed according to Heslop-Harrison et al. (1991). Chromosome spreads were pre-treated with 100 µg mL−1 RNase A in 2× sodium saline citrate (SSC) for 1 h at 37 °C and washed twice in 2× SSC. After incubation with 10 µg mL−1 pepsin in 0·01 mm HCl for 20 min at 37 °C, preparations were stabilized in freshly de-polymerized 4 % (w/v) paraformaldehyde in water for 10 min, dehydrated in a graded ethanol series and air dried. The hybridization mixture, consisting of 50–150 ng μL−1 of DNA probe, 50 % (v/v) formamide, 10 % (w/v) dextran sulfate, 0·1 % sodium dodecyl sulfate (SDS) and 300 ng μL−1 of sheared salmon sperm DNA in 2× SSC, was incubated for 10 min at 70 °C and chilled on ice. Then, 30 µL of the hybridization mixture was added to the chromosome preparations and covered with a plastic coverslip. The hybridization mixture and the chromosomal DNA were denatured at 70 °C for 5 min and the temperature was gradually decreased to 37 °C using a Hybaid Omnislide temperature cycler (Thermo, Waltham, MA, USA). Hybridization was carried out overnight at 37 °C. Following hybridization, the slides were given a stringent wash in 20 % (v/v) formamide in 0·1× SSC at 42 °C to remove mismatched or unhybridized probe molecules. For the detection of digoxigenin- or biotin-labelled probes, slides were equilibrated in 4× SSC/0·1 % (v/v) Tween 20 and blocked in 5 % (w/v) bovine serum albumin in 4× SSC/0·1 % (v/v) Tween 20 for 5 min. Slides were incubated with a final concentration of 2 µg mL−1 of sheep anti-digoxigenin antibody conjugated with fluorescein isothiocyanate (FITC) or streptavidin-Cy3 in a moist chamber at 37 °C for 1 h. Excess antibody was removed by washing the slides in 4× SSC/0·1 % (v/v) Tween 20 three times for 5 min. After counterstaining with DAPI (4′,6-diamidino-2-phenylindole; 2 µg mL−1), the slides were mounted in antifade solution (AF1, Citifluor). Examination of slides was carried out with a Zeiss Axioplan2 fluorescence microscope equipped with filters 09 (FITC), 15 (Cy3) and 01 (DAPI). Images were acquired directly with Applied Spectral Imaging v. 3·3 software, coupled with a high-resolution ASI BV300-20A CCD camera, and printed from Adobe Photoshop after contrast optimization using only functions affecting the whole image equally.
Construction of a c0t-1 DNA library
The c0t-1 DNA for isolation of repetitive sequences with high or moderate copy number was prepared according to the protocol from Zwick et al. (1997) with minor modifications. Genomic DNA (100 ng μL−1) from D. moschatum was sheared at 99 °C for 10 min followed by sonication at 80 °C for 3 min to generate fragments ranging in size predominantly between 0·5 and 1·0 kb. Renaturation of DNA fragments was carried out in a 0·3 m NaCl solution at 65 °C after initial denaturation at 92 °C for 10 min. The renaturation time was calculated according to Zwick et al. (1997). Following S1 nuclease (Promega, Madison, WI, USA) treatment to remove single-stranded DNA and single-strand overhangs on renaturated double-stranded DNA, enzyme inactivation was carried out with stop solution [3 m Tris, pH 8·0, 0·5 m ethylenediaminetetraacetic acid (EDTA)] according to Ostermeier et al. (1999) and incubation at 72 °C for 20 min. Blunt-ended c0t-1 DNA fragments were used for ligation into the SmaI site of the dephosphorylated pUC18 vector. After transformation in XL1Blue cells (Stratagene, La Jolla, CA, USA) by electroporation, positive clones were transferred to 384-well plates, grown in LB freezing medium and stored at –80 °C. For the identification of repetitive DNA sequences, clones were spotted onto nylon membranes (GE Healthcare, Chalfont, UK) and sequentially probed with radiolabelled genomic DNA of D. moschatum, D. aphyllum and D. aggregatum. The overnight hybridizations were performed at 60 °C in 5× SSPE (0·75 m NaCl, 50 mm NaH2PO4, 5 mm EDTA, pH 7.0) with 5× Denhardt's solution and 0·2 % SDS. Post-hybridization washings were performed twice at 60 °C in 1× SSC/0·1 % SDS for 10 min. The signals were detected by autoradiography.
PCR amplification of DmoF14 repeats
For amplification of the dispersed repeat DmoF14 from different orchid species, the primer pair 5′-CAGTTCACAAAGAGCTAATGC-3′ and 5′-TAGTCAGAGGTAGTCGACCCAACC-3′ was used for PCR with genomic DNA as template at an annealing temperature of 54 °C. PCR reactions with 50 ng template DNA and a final primer concentration of 0·5 µm were performed in a volume of 50 µL containing 0·2 mm dNTPs, 50 mm KCl, 1·5 mm MgCl2, 10 mm Tris–HCl (pH 9·0) and 1 unit of Taq DNA polymerase (GE Healthcare). Standard PCR conditions were 94 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 49–55 °C for 45 s and 72 °C for 3·5 min, and a final incubation at 72 °C for 10 min.
After gel electrophoresis, PCR fragments were purified with the QIAquick gel extraction kit (Qiagen) and cloned in the pGEM-T vector (Stratagene).
Sequence analyses
Plasmid clones from the c0t-1 DNA library and plasmid clones harbouring the dispersed repeat DmoF14 amplified from different orchid species were sequenced on a CEQ 8000 capillary sequencer (Beckman, Fullerton, CA, USA). Sequences from the c0t-1 DNA library were aligned via the MegAlign option within the Lasergene 6·0 software (DNAStar, Madison, WI, USA) by using CLUSTAL with default parameters. Divergence between DmoF14 in different orchid species was analysed by the neighbour-joining algorithm of the MEGA4·0 program (Tamura et al., 2007).
Southern hybridization
Southern hybridization to genomic DNA of different orchid species was performed using standard conditions with 32P-labelled probes (Sambrook et al., 1989). DNA from all nine species used was digested with different restriction endonucleases, separated on 1·1 % agarose gels and transferred to HybondN+ (GE Healthcare) membranes. After hybridization, filters were washed at 60 °C in 2× SSC/0·1 % SDS and 1× SSC/0·1 % SDS for 10 min each. Signals were detected by autoradiography.
RESULTS
Identification of repetitive DNA sequences in D. moschatum
To identify repetitive DNA sequences in Dendrobium species, the restriction endonucleases BamHI, XbaI, HindIII, AluI, RsaI, HaeIII, PstI, SacI, SmaI and KpnI were used to digest total genomic DNA of D. moschatum. After gel electrophoresis, distinct bands of highly abundant repetitive DNA families were not observed. Therefore, as an alternative approach, a c0t-1 plasmid library from D. moschatum was constructed.
Experimental parameters were chosen to enrich repetitive sequences ranging from 100 to 1000 bp. The c0t-1 plasmid library consists of 384 clones which were separately hybridized with genomic DNA of D. moschatum, D. aphyllum and D. aggregatum. Based on the strength of the signals, eight clones harbouring highly repetitive DNA sequences were identified and designated Dmo with the corresponding plate coordinates.
Sequence analysis of these eight clones revealed six sequences ranging in size from 87 bp (DmoB22, EMBL accession no. FM212245) to 150 bp (DmoB8, DmoG14, DmoH14, DmoK14 and DmoO19) consisting of variable numbers of the Arabidopsis-like 5′-TTTAGGG-3′ telomeric sequences. The strong conservation without any degeneracy of the telomeric repeat motifs indicates their origin from the physical ends of the D. moschatum chromosomes.
The clone DmoO11 (EMBL accession no. FM212246) was identified as a microsatellite array of 30 copies of the dinucleotide GA. The clone DmoF14 (EMBL accession no. FM212247) has a length of 420 bp and showed no significant homologies with other sequences in the EMBL/GenBank database.
Genomic organization of repetitive sequences in Dendrobium species
Southern hybridization was performed to investigate the genomic organization and abundance of the repeats isolated from the D. moschatum c0t-1 library. DmoB22 as a representative clone containing conserved telomeric repeats was hybridized to genomic DNA of D. moschatum. The hybrization signals extended over a wide range of fragments detectable as a smear over all lanes up to 20 kb (data not shown). Conserved bands originating from proximal degenerated repeats were not detected.
For analysis of the genomic organization of the DmoO11 and DmoF14 repeats, Southern analyses were performed with genomic DNA of D. moschatum, D. aphyllum and D. aggregatum.
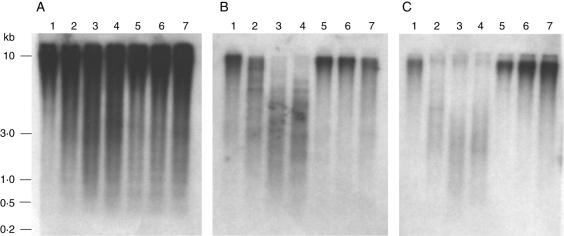
In D. moschatum, the (GA)n microsatellite DmoO11 showed hybridization signals of strong intensity over the whole range of separation, ranging from 3 to 10 kb (Fig. 1A). In contrast, the intensity of the hybridization in D. aphyllum and D. aggregatum was significantly weaker (e.g. HinfI and RsaI), indicating a lower abundance of the (GA)n microsatellite in these two species compared with D. moschatum (Fig. 1B and C). Moreover, D. aphyllum genomic DNA restricted with HinfI and RsaI (Fig. 1B, lanes 3 and 4) revealed hybridization signals only in the range 1·0 kb to approx. 5·0 kb and reduced signals with larger genomic fragments. A similar but weaker hybridization with smaller fragments was also observed in D. aggregatum (Fig. 1C, lanes 3 and 4).
Fig. 1.
Comparative Southern hybridization of the (GA)30 microsatellite DmoO11 with genomic DNA of D. moschatum (A), D. aphyllum (B) and D. aggregatum (C), digested with BamHI (lane 1), HaeII (lane 2), HinfI (lane 3), RsaI (lane 4), PstI (lane 5), HpaII (lane 6) and MspI (lane 7). Molecular size markers are given on the left.
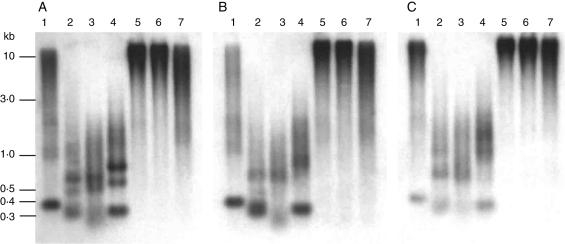
Hybridization of DmoF14 to genomic DNA of D. moschatum, D. aphyllum and D. aggregatum, each digested with BamHI, HaeIII, HinfI and RsaI, revealed strong signals detectable as conserved bands of different size and a smear over the whole lane, suggesting a dispersed organization of the DmoF14 repeat family (Fig. 2A–C). Of note, a prominent BamHI fragment of approximately 375 bp corresponding to the distance between two BamHI sites in DmoF14 was observed in D. moschatum and D. aphyllum (Fig. 2A, B, lane 1). A similar but slightly weaker pattern was observed in D. aggregatum (Fig. 2C, lane 1).
Fig. 2.
Comparative Southern hybridization of the dispersed repeat DmoF14 with genomic DNA of D. moschatum (A), D. aphyllum (B) and D. aggregatum (C), digested with BamHI (lane 1), HaeII (lane 2), HinfI (lane 3), RsaI (lane 4), PstI (lane 5), HpaII (lane 6) and MspI (lane 7). Molecular size markers are given on the left.
In D. moschatum the RsaI restriction revealed three strong fragments between 400 and 750 bp (Fig. 2A, lane 4). The 400-bp fragment was conserved in D. aphyllum whereas it was weaker in D. aggregatum (Fig. 2B, lane 4).
Distinct HaeIII and HinfI fragments ranging from approx. 400 to 700 bp were localized in all three Dendrobium species (Fig. 2A–C, lanes 2 and 3). The methylation-sensitive enzymes HpaII and MspI showed strong hybridization in a high-molecular-weight range (Fig. 2A–C, lanes 6 and 7). Similarly, the large PstI fragments indicated strong methylation of the DmoF14 repeat.
Divergence of the dispersed repeat DmoF14 in orchid species
As the Southern hybridization patterns of DmoF14 are highly conserved in the Dendrobium species analysed, other Dendrobium species and species from different orchid genera were included in comparative Southern hybridization experiments to elucidate whether DmoF14 is present in closely and distantly related species.
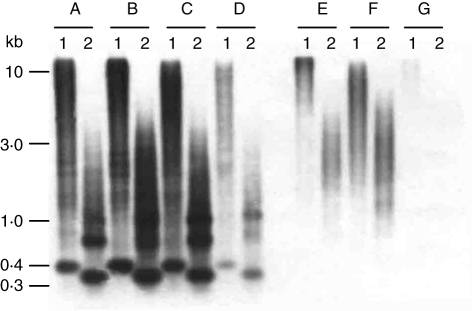
Therefore, genomic DNA of D. moschatum, D. anosmum, D. fimbriatum var. oculatum and D. palpebrae and species of different orchid genera (Nageliella angustifolia, Oncidium sphacelatum and Phalaenopsis lueddemanniana var. purpurea) was digested with BamHI and RsaI and hybridized with DmoF14. In both restriction enzymes, strong fragments of approx. 400 and 320 bp, respectively, were visible in all Dendrobium species (Fig. 3A–D, lanes 1 and 2). However, the strength of hybridization signals in D. palpebrae was considerably weaker (Fig. 3D).
Fig. 3.
Analysis of species distribution of DmoF14 by comparative Southern hybridization of D. moschatum (A), D. anosmum (B), D. fimbriatum var. oculatum (C), D. palpebrae (D), N. angustifolia (E), O. sphacelatum (F) and P. lueddemanniana var. purpurea (G). The DNA of each species was digested with BamHI (lane 1) and RsaI (lane 2). Molecular size markers are given on the left.
In contrast, a smear without conserved fragments was observed in Nageliella angustifolia and Oncidium sphacelatum (Fig. 3E, F) whereas in Phalaenopsis lueddemanniana var. purpurea, very faint hybridization was detected only after longer exposure (Fig. 3G).
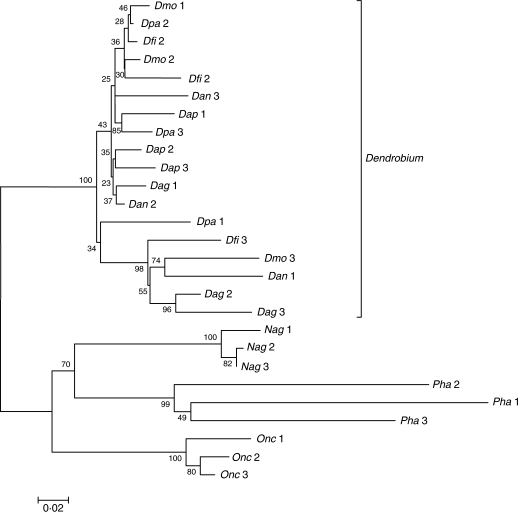
To investigate further the divergence of the DmoF14 repeat family in the orchid species studied and to complement the Southern hybridization results, repeat-specific primers were designed. PCR products were cloned and three independent clones from each species were sequenced (EMBL accession nos FM212248–FM212273). In order to display the structural relatedness of the species-specific DmoF14 sequences, a dendrogram was generated by a ClustalW alignment followed by a neighbour-joining analysis (Fig. 4). The tree showed two major clades supported by a 100 % bootstrap value. All Dendrobium DmoF14 repeats were grouped in one clade. The corresponding low bootstrap values indicate that many sequences analysed form a DmoF14 subfamily that is widespread in species of the genus Dendrobium. A second, more diverged subfamily consists of the Dendrobium repeats Dpa 1, Dfi 3, Dmo 3, Dan 1, Dag 2 and Dag 3.
Fig. 4.
Neighbour-joining analysis showing the sequence divergence of three DmoF14 repeats amplified from each of the six Dendrobium species and the species of other orchid genera. The number of bootstrap trials was 1000, and the resulting bootstrap values are indicated on the respective branches. The length of branches within the dendrogram represents the distance between the DmoF14 sequences, while the scale bar indicates the number of substitution events per site.
The species of the three other orchid genera were separately grouped in distinct genus-specific clades, as supported by the corresponding high bootstrap values. The overall dendrogram revealed that the DmoF14 sequence is considerably diverged in species outside the genus Dendrobium. The divergence among Dendrobium species ranged from 2·3 to 10 %. The highest divergence (32–34 %) was observed between Dendrobium species and P. lueddemanniana var. purpurea. Of particular note is a single 35–38-bp deletion in N. angustifolia, which did not strongly affect the position of the DmoF14 sequences of this species in the dendrogram.
Physical mapping of repetitive sequences along orchid chromosomes
FISH has been applied to physically map the identified repeats along chromosomes of Dendrobium species and to reveal aspects of chromosome evolution. As the chromosomes of orchid species are small and numerous and plant material is limited, cytogenetic analyses have to be performed with great care.
To compare the chromosomal distribution pattern of the three isolated Dendrobium repeats, FISH was performed on metaphase chromosomes of D. moschatum (2n = 2x = 40), D. aphyllum (2n = 2x = 38) and D. aggregatum (2n = 2x = 38).
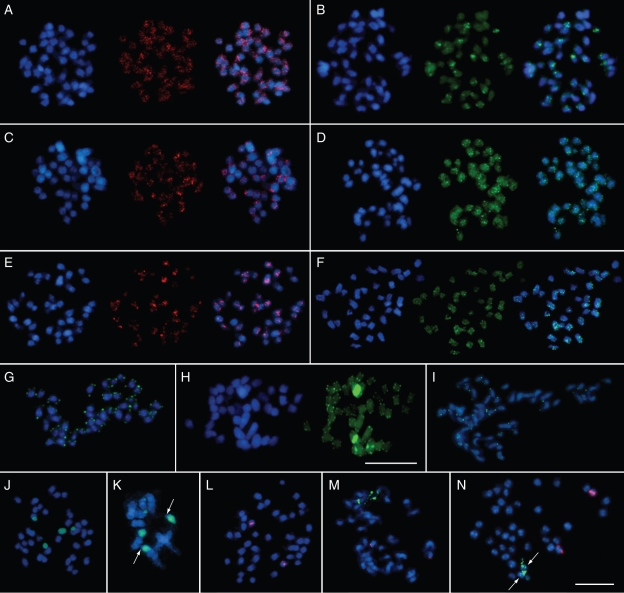
The repetitive DNA probe DmoF14 identified dispersed fluorescent signals of different intensity on all chromosomes of the Dendrobium species (Fig. 5A, C, E). In D. moschatum and D. aphyllum, DmoF14 is widely scattered over most chromosome arms detectable as interstitial signals (Fig. 5A, C). Furthermore, 12–14 subtelomeric signals were observed in D. aphyllum. In D. aggregatum, almost half of the signals included pericentromeric regions (Fig. 5E).
Fig. 5.
Fluorescence in situ hybridization to metaphase chromosomes of three Dendrobium species. In panels A–F, the DAPI-stained DNA (blue fluorescence) shows the morphology of the chromosomes. The images on the left show the dispersed distribution of the DmoF14 fragment on the chromosomes of D. moschatum (A), D. aphyllum (C) and D. aggregatum (E), in red fluorescence (middle and overlay image in each panel). The images on the right show the dispersed localization of the microsatellite DmoO11 on the chromosomes of D. moschatum (B), D. aphyllum (D) and D. aggregatum (F), respectively, as green fluorescence (middle and overlay image in each panel). (G–I) Chromosomal localization of the telomeric sequence DmoB22 (green fluorescence) on chromosomes of D. moschatum, D. aphyllum and D. aggregatum, respectively. The internal positions of telomeric sequence motifs on two pairs of D. aphyllum chromosomes are visible as bright yellow–green signals (H, right). (J–N) rDNA FISH analysis on D. moschatum (J, K and L), D. aphyllum (M) and D. aggregatum (N) metaphases. Red signals indicate 5S rDNA repeats and green signals represent 18S–5·8S–25S rRNA arrays. Terminal regions lacking 18S–5·8S–25S rRNA genes on one pair of D. moschatum chromosomes are indicated by arrows in a partial metaphase (K, close-up). The minor 18S–5·8S–25S rRNA site on two D. aggregatum chromosomes is shown by an arrow in N. The scale bar in N corresponds to 10 µm and is valid for all images except H containing an individual 10 µm scale bar.
The microsatellite probe DmoO11 consisting of GA dinucleotides was located on all chromosomes in D. aphyllum and D. aggregatum and on most chromosomes in D. moschatum (Fig. 5B, D, F). The majority of strong signals were found in interstitial regions of D. moschatum chromosomes (Fig. 5B). In D. aphyllum DmoO11 was dispersed over all chromosome arms with stronger hybridization in some interstitial sites (Fig. 5D). In D. aggregatum most of the hybridization of DmoO11 was confined to 2–3 interstitial sites per chromosome (Fig. 5F).
Six of eight clones of the c0t-1 plasmid library contained telomeric repeats. As a representative clone, DmoB22 has been used as a FISH probe. Metaphase chromosomes of D. moschatum, D. aphyllum and D aggregatum showed an identical hybridization pattern, revealing the presence of telomeric sequences at all chromosome termini (Fig. 5G–I). Of note, in D. aphyllum very strong hybridization signals were detected in an internal region of two chromosomes, while two strong signals were present at an intercalary position of another chromosome pair (Fig. 5H).
Physical mapping of ribosomal genes
FISH was applied to localize the tandemly arranged 18S–5·8S–25S rRNA and 5S rRNA genes on Dendrobium chromosomes, using heterologous probes. The clone pTa71 from Triticum aestivum (Gerlach and Bedbrook, 1979) was used to detect 18S–5·8S–25S rDNA, while the probe pXV1 from Beta vulgaris (Schmidt et al., 1994) enabled the localization of 5S rRNA.
In D. moschatum the 18S–5·8S–25S rRNA genes are located on three pairs of chromosomes (Fig. 5J, green fluorescence). Two major sites showing bright signals labelled almost completely a chromosome pair. Nevertheless, closer inspection of the pTa71 hybridization signals on these two chromosomes revealed the absence of pTa71 in telomeric/subtelomeric regions (Fig. 5K, arrows). Two other strong signals were restricted to one arm of another chromosome pair, while the minor pair of sites was located at the subtelomeric region of two other chromosomes (Fig. 5J). In contrast, D. aphyllum showed only two sites of 18S–5·8S–25S rRNA gene signals (Fig. 5M, green fluorescence). The signals were localized in the terminal position of one chromosome pair and showed not fully condensed ribosomal gene arrays (Fig. 5M). In D. aggregatum a major 18S–5·8S–25S rRNA gene site was also observed in the terminal position of one chromosome pair (Fig. 5N, green signals). Additionally, minor sites were detected on the same pair of chromosomes (Fig. 5N, arrows).
FISH for the detection of 5S rRNA genes revealed two signals in D. moschatum, D. aphyllum and D. aggregatum (Fig. 5L–N; red fluorescence). In each orchid species investigated, the chromosomal position of the 5S rRNA gene was different. Two sites were located in an intercalary region in D. moschatum (Fig. 5L). In D. aphyllum and D. aggregatum the two 5S rRNA gene signals were observed in the terminal region of two other chromosomes (Fig. 5M, N).
DISCUSSION
The genus Dendrobium comprises approx. 1200 species, but only a few of these species have been characterized regarding genome size (Jones et al., 1998; Leitch et al., 2009).
Molecular studies are available describing Dendrobium-specific rDNA sequences (Cheng et al., 2004; Tsai et al., 2004). However, thus far, there is no information regarding the abundance and chromosomal distribution of rDNA and other repetitive DNA families in Dendrobium. The present paper describes the isolation of repetitive sequences from Dendrobium moschatum, and comparative Southern analyses in other Dendrobium species and species from three different orchid genera.
Repetitive sequences can be isolated by several strategies. Restriction satellites are detectable as prominent bands after gel electrophoresis based on conserved recognition sites in repeating units. Other methods include ultracentrifugation with complexing dyes or shotgun cloning. Genome sequencing with next-generation technologies is becoming an increasing method for the detection of families of repetitive DNA, but these technologies have not yet been applied to Dendrobium species. In order to identify Dendrobium-specific satellite repeats, 13 restriction enzymes with recognition sites covering four to six nucleotides with AT- or GC-rich motifs were tested. However, no prominent fragments of the typical size of satellite DNA (150–180 bp or multimers thereof) were detected following gel electrophoresis.
A rapid and efficient method for the identification of repetitive DNA is the isolation of c0t-1 DNA, which is enriched for repeated sequences. This method has been used for the isolation of highly abundant repeats in many plant species (Yuan et al., 2003; Hribová et al., 2007; Anamthawat-Jonsson et al., 2009). Based on a small D. moschatum c0t-1 plasmid library, a telomere-like motif, a (GA)n microsatellite and a dispersed repeat with no homology to EMBL/GenBank entries were identified. The abundance of these repeats indicates the suitability of c0t-1 DNA for further isolation of highly repetitive orchid DNA sequences.
Microsatellites are useful markers to assess the organization of genomes and their diversity on the species and population level in many higher plant species (Bell and Ecker, 1994; Becker and Heun, 1995; Röder et al., 1995). It has been shown that different microsatellites are major components of the repeated DNA fraction of plant genomes (Schmidt et al., 1993; Schmidt and Heslop-Harrison, 1996). In orchids, several studies aiming at the isolation and application of microsatellites have been undertaken in order to investigate intra- and interspecific variation (Pellegrino et al., 2001; Helena et al., 2008). Yue et al. (2006) developed simple sequence repeat markers for Dendrobium species, but their study included only hybrids, mostly selected as ornamental plants. Here microsatellites were isolated from D. moschatum that consisted of a perfect array of GA dinucleotides repeats. Although polymorphic hybridization patterns were not detected, it is noteworthy that the abundance of GA dinucleotides varies considerably between D. moschatum, D. aggregatum and D. aphyllum. This suggests that it might be possible to differentiate Dendrobium species and accessions by DNA fingerprinting, although the level of intra- or interspecific polymorphism has to be tested and more microsatellite motifs should be included. If Southern hybridization is used, the level of resolution depends on the combination of microsatellite motif and restriction enzyme, often resulting in contrasting hybridization patterns in different plant species (Schmidt and Heslop-Harrison, 1996; Schmidt et al., 2000).
With FISH, a genome-wide (GA)n distribution has been observed, particularly in D. aphyllum. The position of microsatellites included interstitial and pericentromeric chromosomal regions. The relatively uniform coverage of the chromosomes is complemented by a local clustering in some regions as observed in D. moschatum. The distribution pattern is dispersed and similar to that reported in Beta vulgaris (Schmidt and Heslop-Harrison, 1996). In contrast, Hudakova et al. (2001) reported (GA)n sequences in centromeric regions of barley chromosomes, suggesting that the chromosomal position of microsatellites is variable.
The dispersed repeat DmoF14 has been isolated from the c0t-1 DNA fraction of D. moschatum herein. Comparative studies of plant repetitive DNA sequences support the investigation of phylogenetic relationships between plant species (Kamm et al., 1995; Orgaard et al., 1995; Frello and Heslop-Harrison, 2000). Southern hybridization showed that DmoF14 is conserved and has a similar genomic organization in D. moschatum, D. aphyllum, D. aggregatum and other Dendrobium species such as D. anosmum, D. fimbriatum and D. palpebrae. Moreover, sequencing of Dmo14 repeats of species from different genera revealed genus-specific diversification, and the presence of DmoF14 in distantly related orchid genera suggests that this repeat is an ancient component of orchid genomes which has probably been present in the last common ancestor of the species investigated. The evolutionary origin of DmoF14 remains elusive. In the six Dendrobium species investigated, Southern hybridization revealed a strong BamHI fragment of approximately 350 bp which is consistent with conserved BamHI restriction sites at nucleotide positions 36 and 378 in DmoF14. Retrotransposons are ancient and highly amplified DNA sequences of plant genomes, and it is widely accepted that the majority of dispersed sequences in plant genomes are derived from retrotransposons, in particular from LTR (long terminal repeat) retrotransposons (Flavell, 1986). Therefore, it is possible that DmoF14 is part of or is derived from an LTR of an ancient or yet unknown orchid retroelement.
Telomeres consist in most mono- and dicotyledonous angiosperms, gymnosperms and bryophytes of the highly conserved Arabidopsis-type sequence repeats (TTTAGGG)n (Richards and Ausubel, 1988; Fuchs et al., 1995), while some genera of the Asparagales have a different type of telomeric sequence (Adams et al., 2001; Rotkova et al., 2004).
Here, an Arabidopsis-type telomeric sequence was observed on the chromosome termini of three Dendrobium species by FISH. Of note, the telomeric probe in D. aphyllum detected additional strong signals at pericentromeric and intercalary positions on two pairs of chromosomes (Fig. 5H). Several mechanisms can be postulated to explain this chromosomal rearrangement. In the phylogeny of D. aphyllum, Robertsonian fusions might have occurred, resulting in two pairs of submetacentric to metacentric chromosomes. The ancestral chromosomes could have been two acrocentrics (Schubert, 1992), but chromosomes with other position of the centromeres are also possible ancestors. However, as dicentric chromosomes are not stable, one centromere of the fused chromosomes must have been functionally inactivated. It is well known that Robertsonian fusions played an important role in altering karyotypes of animals and plants and therefore are a widespread mechanism in speciation (Meyne et al., 1990; Cox et al., 1993; Fuchs et al., 1995). However, internal telomeric repeats indicating a Robertsonian fusion have not been detected in D. aggregatum (having 2n = 38 chromosomes). Alternatively, the internal telomeric arrays can also result from DNA repair mechanisms, which has been observed in many plant species (Fitzgerald et al., 2001). The telomerase binds to short homologous stretches in single strand gaps and synthesizes short telomeric arrays. It is known that di- to trinucleotide homologies are sufficient to act as telomerase substrate. Subsequently, newly synthesized telomeric repeats could have been amplified by slippage replication. Another scenario might involve either para- or pericentric inversion as a result of a breakage in the telomeric block at the end of the chromosomes, with a part of the broken telomere array being transferred to an internal position. Similarly, an inversion of a chromosome arm or part thereof can transfer the telomeric block to an internal position requiring the healing of the newly formed chromosome end by telomerase activity, as described for wheat (Tsujimoto et al., 1999). Nevertheless, more Dendrobium species need to be analysed for the presence of internal telomeric sequences to further unravel the mechanisms leading to ectopic positions of telomere repeats on Dendrobium chromosomes.
In addition to other repetitive sequence classes, the localization of rRNA gene arrays on plant chromosomes by FISH has been widely used (Leitch and Heslop-Harrison, 1992; Schmidt et al., 1994), with both the 5S rRNA and the 18S–5·8S–25S rRNA genes providing useful markers for chromosome identification and karyotyping (Doudrick et al., 1995; Brown et al., 1999). In orchids just a few cytogenetic studies have been performed. Chromosome staining has been applied to show karyotype variation in Phalaenopsis and the related species Doritis pulchirrima (Kao et al., 2001). Schwarzacher et al. (1980), Schwarzacher and Schweizer (1982) and Koehler et al. (2008) used banding techniques for karyotype analyses in Austrian Cephalanthera species and species of the genus Christensonella, respectively. The genomic organization and relationships in Phalaenopsis species were investigated by genomic in situ hybridization (Lin et al., 2005). Telomeric sequences and rDNA genes were mapped on chromosomes of Cephalanthera species (Moscone et al., 2007). The chromosomal positions of 5S rRNA and the 18S–5·8S–25S rRNA genes have also been analysed in some wild orchids and hybrids from Italy, and in species of the genus Maxillaria (Demerico et al., 2001; Cabral et al., 2006).
Here a comparative analysis of the distribution of rRNA gene arrays on chromosomes of three Dendrobium species was performed. In D. moschatum, D. aphyllum and D. aggregatum, the number of 18S–5·8S–25S and 5S rDNA sites was clearly determined. Apart from one chromosome pair in D. moschatum, all rDNA sites are at terminal positions. This particular D. moschatum chromosome pair showed a very strong FISH signal, suggesting that most of the DNA consists of 18S–5·8S–25S rRNA genes, which is probably the result of rRNA gene array expansion. In synthetic Nicotiana hybrids it has been shown that rDNA arrays undergo rapid changes including expansion or reduction (Kovarik et al., 2008). However, each of the three Dendrobium species analysed here has a characteristic number and position of rDNA sites enabling the unequivocal identification of single chromosomes in a complement of the small and numerous orchid chromosomes.
Further analyses will need to prove the suitability of rDNA probes as well as probes from repetitive sequence families using the c0t-1 DNA library of D. moschatum as a resource for genome analysis in the genus. Nevertheless, more Dendrobium species should be included in comparative molecular–cytogenetic analyses to provide a valuable complementation to taxonomic data based on morphological characters.
ACKNOWLEDGEMENTS
We are grateful to the DAAD (German Academic Exchange Service) for providing scholarship to R.B. during her research work at Dresden University of Technology. R.B. also thanks the Ministry of Education, Peoples Republic of Bangladesh, for approving her study leave. We are very grateful to I. Walter, F. Zakrzewski and K. Koelling for excellent technical assistance. We thank B. Ditsch, Dresden Botanical Garden, for providing research materials.
LITERATURE CITED
- Adams SP, Hartman TP, Lim KY, et al. Loss and recovery of Arabidopsis-type telomere repeat sequences 5′-( TTTAGGG) (n)-3′ in the evolution of a major radiation of flowering plants. Proceedings. Biological Sciences/The Royal Society. 2001;268:1541–1546. doi: 10.1098/rspb.2001.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anamthawat-Jonsson K, Wenke T, Thorsson AT, Sveinsson S, Zakrzewski F, Schmidt T. Evolutionary diversification of satellite DNA sequences from Leymus (Poaceae: Triticeae) Genome. 2009;52:381–390. doi: 10.1139/g09-013. [DOI] [PubMed] [Google Scholar]
- Becker J, Heun M. Barley microsatellites: allele variation and mapping. Plant Molecular Biology. 1995;27:835–845. doi: 10.1007/BF00020238. [DOI] [PubMed] [Google Scholar]
- Begum R, Alam SS. Karyomorphological studies in two orchid species. Dhaka University Journal of Biological Science. 2004;13:99–101. [Google Scholar]
- Begum R, Alam SS. Karyotype analysis of seven orchid species from Bangladesh. Bangladesh Journal of Botany. 2005;34:31–36. [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Brown SE, Stephens JL, Lapitan NLV, Knudson DL. FISH landmarks for barley chromosomes (Hordeum vulgare L.) Genome. 1999;42:274–281. [PubMed] [Google Scholar]
- Cabral JS, Felix LP, Guerra M. Heterochromatin diversity and its co-localization with 5S and 45S rDNA sites in chromosomes of four Maxillaria species (Orchidaceae) Genetics and Molecular Biology. 2006;29:659–664. [Google Scholar]
- Cheng KT, Lo SF, Lee CY, Chen CC, Tsay HS. The rDNA sequence analysis of three Dendrobium species. Journal of Food and Drug Analysis. 2004;12:367–369. [Google Scholar]
- Clements MA. Molecular phylogenetic systematics in the Dendrobiinae (Orchidaceae), with emphasis on Dendrobium section Pedilonum. Telopea. 2003;10:247–296. [Google Scholar]
- Cox AV, Bennet ST, Parokonny AS, Kenton A, Callimassia MA, Bennett MD. Comparison of plant telomere locations using a PCR- generated synthetic probe. Annals of Botany. 1993;72:239–247. [Google Scholar]
- Demerico SD, Galasso I, Pignone D, Scrugli A. Localization of rDNA loci by fluorescent in situ hybridization in some wild orchids from Italy (Orchidaceae) Caryologia. 2001;54:31–36. [Google Scholar]
- Doudrick RL, Heslop-Harrison JS, Nelson CD, Schmidt T, Nance WL, Schwarzacher T. Karyotype of slash pine (Pinus elliottii var. elliottii) using patterns of fluorescence in situ hybridization and fluorochrome banding. The Journal of Heredity. 1995;86:289–296. [Google Scholar]
- Dressler RL. The orchids: natural history and classification. Cambridge, MA: Harvard University Press; 1981. [Google Scholar]
- Dressler RL. Phylogeny and classification of the orchid family. Portland, OR: Discorides Press; 1993. [Google Scholar]
- Fitzgerald MS, Shakirov EV, Hood EE, McKnight TD, Shippen DE. Different modes of de novo telomere formation by plant telomerases. Plant Journal. 2001;26:77–87. doi: 10.1046/j.1365-313x.2001.01010.x. [DOI] [PubMed] [Google Scholar]
- Flavell RB. Repetitive DNA and chromosome evolution in plants. Philosophical Transactions of the Royal Society of London B. 1986;312:227–242. doi: 10.1098/rstb.1986.0004. [DOI] [PubMed] [Google Scholar]
- Frello S, Heslop-Harrison JS. Chromosomal variation in Crocus vernus Hill (Iridaceae) investigated by in situ hybridisation of rDNA and a tandemly repeated sequence. Annals of Botany. 2000;86:317–322. [Google Scholar]
- Fuchs J, Brandes A, Schubert I. Telomere sequence localization and karyotype evolution in higher plants. Plant Systematics and Evolution. 1995;196:227–241. [Google Scholar]
- Ganal MW, Lapitan NLV, Tanksley SD. Macrostructure of tomato telomeres. Plant Cell. 1991;3:87–94. doi: 10.1105/tpc.3.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Research. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helena CC, Monteiro FA, Sousa ES, Fay MF, Chase MW, Pais MS. Conservation Genetics. 2008. Isolation and characterization of novel polymorphic nuclear microsatellite markers from Ophyris fusca (Orchidaceae) and cross-species amplification. online first (doi: 10.1007/s10592-008-9634-x) [Google Scholar]
- Hemleben V, Schmidt T, Torres-Ruiz RA, Zentgraf U. Molecular cell biology: role of repetitive DNA in nuclear architecture and chromosome structure. Progress in Botany. 2000;61:91–117. [Google Scholar]
- Heslop-Harrison JS. Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. The Plant Cell. 2000;12:617–636. doi: 10.1105/tpc.12.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T, Anamthawat-Jonsson K, Leitch AR, Shi M, Leitch IJ. In-situ hybridization with automated chromosome denaturation. Technique. 1991;3:109–116. [Google Scholar]
- Hossain ABME. A taxonomic report on the genus Dendrobium SW. (Orchidaceae) from Bangladesh. Bangladesh Journal of Plant Taxonomy. 2002;9:47–55. [Google Scholar]
- Hribová E, Dolezelová M, Town CD, Macas J, Dolezel J. Isolation and characterization of the highly repeated fraction of the banana genome. Cytogenetic and Genome Research. 2007;119:268–274. doi: 10.1159/000112073. [DOI] [PubMed] [Google Scholar]
- Hudakova S, Michalek W, Presting GG, et al. Sequence organization of barley centromeres. Nucleic Acids Research. 2001;29:5029–5035. doi: 10.1093/nar/29.24.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WE, Kuehnle AR, Arumuganathan K. Nuclear DNA content of 26 orchid (Orchidaceae) genera with emphasis on Dendrobium. Annals of Botany. 1998;82:189–194. [Google Scholar]
- Kamm A, Galasso I, Schmidt T, Heslop-Harrison JS. Analysis of a repetitive DNA family from Arabidopsis arenosa and relationships between Arabidopsis species. Plant Molecular Biology. 1995;27:853–862. doi: 10.1007/BF00037014. [DOI] [PubMed] [Google Scholar]
- Kao YY, Chang SB, Lin TY, et al. Differential accumulation of heterochromatin as a cause for karyotype variation in Phalaenopsis orchids. Annals of Botany. 2001;87:387–395. [Google Scholar]
- Koehler S, Cabral JS, Whitten WM, et al. Molecular phylogeny of the neotropical genus Christensonella (Orchidaceae, Maxillariinae): species delimitation and insights into chromosome evolution. Annals of Botany. 2008;102:491–507. doi: 10.1093/aob/mcn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A, Dadejova M, Lim YK, et al. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Annals of Botany. 2008;101:815–823. doi: 10.1093/aob/mcn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Heslop-Harrison JS. Physical mapping of the 18S–5·8S–26S rRNA genes in barley by in situ hybridization. Genome. 1992;35:1013–1018. [Google Scholar]
- Leitch IJ, Kahandawala I, Suda J, et al. Genome size diversity in orchids: consequences and evolution. Annaly of Botany. 2009;104 doi: 10.1093/aob/mcp003. in press. doi: 10.1093/aob/mcp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Chen YH, Chen WH, Chen CC, Kao YY. Genome organization and relationships of Phalaenopsis orchids inferred from genomic in situ hybridization. Botanical Bulletin of Academia Sinica. 2005;46:339–345. [Google Scholar]
- Meyne J, Baker RJ, Hobart HH, et al. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990;99:3–10. doi: 10.1007/BF01737283. [DOI] [PubMed] [Google Scholar]
- Moscone EA, Samuel R, Schwarzacher T, Schweizer D, Pedrosa-Harand A. Complex rearrangements are involved in Cephalanthera (Orchidaceae) chromosome evolution. Chromosome Research. 2007;15:931–943. doi: 10.1007/s10577-007-1174-6. [DOI] [PubMed] [Google Scholar]
- Orgaard M, Jacobsen N, Heslop-Harrison JS. The hybrid origin of two cultivers of Crocus (Iridaceae) analysed by molecular cytogenetics including genomic southern and in situ hybridization. Annals of Botany. 1995;76:253–262. [Google Scholar]
- Ostermeier M, Nixon AE, Shim JH, Benkovic SJ. Combinatorial protein engineering by incremental truncation. Proceedings of the National Academy of Sciences of the USA. 1999;96:3562–3567. doi: 10.1073/pnas.96.7.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino G, Cafasso D, Widmer A, Soliva M, Musacchio A, Cozzolino S. Isolation and characterization of microsatellite loci from the orchid Serapias vomeracea (Orchidaceae) and cross-priming to other Serapias species. Molecular Ecology Notes. 2001;1:279–280. [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Röder MS, Plaschke J, König SU, et al. Abundance, variability and chromosomal location of microsatellites in wheat. Molecular and General Genetics. 1995;246:327–333. doi: 10.1007/BF00288605. [DOI] [PubMed] [Google Scholar]
- Rotkova G, Sklenickova M, Durackova M, Skyorova E, Leitch AR, Fajkus J. An evolutionary change in telomere sequence motif within the plant section Asparagales had significance for telomere nucleoprotein complexes. Cytogenetic and Genome Research. 2004;107:132–138. doi: 10.1159/000079584. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmidt A, Doudrick RL, Heslop-Harrison JS, Schmidt T. The contribution of short repeats of low sequence complexity to larger conifer genomes. Theoretical and Applied Genetics. 2000;101:7–14. [Google Scholar]
- Schmidt T, Heslop-Harrison JS. The physical and genomic organization of microsatellites in sugar beet. Proceedings of the National Academy of Sciences of the USA. 1996;93:8761–8765. doi: 10.1073/pnas.93.16.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends in Plant Science. 1998;3:195–199. [Google Scholar]
- Schmidt T, Boblenz K, Metzlaff M, Kaemmer D, Weising K, Kahl G. DNA fingerprinting in sugar beet (Beta vulgaris) — identification of double-haploid breeding lines. Theoretical and Applied Genetics. 1993;85:653–657. doi: 10.1007/BF00225001. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Schwarzacher T, Heslop-Harrison JS. Physical mapping of rRNA genes by fluorescent in-situ hybridization and structural analysis of 5S rRNA genes and intergenic spacer sequences in sugar beet (Beta vulgaris) Theoretical and Applied Genetics. 1994;88:629–636. doi: 10.1007/BF01253964. [DOI] [PubMed] [Google Scholar]
- Schubert I. Telomeric polymorphism in Vicia faba. Biologisches Zentralblatt. 1992;111:164–168. [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS. Practical in situ hybridization. Oxford: BIOS Scientific Publishers Limited; 2000. [Google Scholar]
- Schwarzacher T, Schweizer D. Karyotype analysis and heterochromatin differentiation with Giemsa C-banding and fluorescent counterstaining in Cephalanthera (Orchidaceae) Plant Systematics and Evolution. 1982;141:91–113. [Google Scholar]
- Schwarzacher T, Ambros P, Schweizer D. Application of Giemsa banding to orchid karyotype analysis. Plant Systematics and Evolution. 1980;134:293–297. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4·0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Peng CI, Huang SC, Huang PL, Chou CH. Determination of the genetic relationship of Dendrobium species (Orchidaceae) in Taiwan based on the sequence of the internal transcribed spacer of ribosomal DNA. Scientia Horticulturae. 2004;101:315–325. [Google Scholar]
- Tsujimoto H, Usami N, Hasegawa K, Yamada T, Nagaki K, Sasakuma T. De novo synthesis of telomere sequences at the healed breakpoints of wheat deletion chromosomes. Molecular and General Genetics. 1999;262:851–856. doi: 10.1007/s004380051150. [DOI] [PubMed] [Google Scholar]
- Yuan Y, SanMiguel PJ, Bennetzen JL. High-Cot sequence analysis of the maize genome. Plant Journal. 2003;34:249–255. doi: 10.1046/j.1365-313x.2003.01716.x. [DOI] [PubMed] [Google Scholar]
- Yue GH, Lam-Chan LT, Hong Y. Development of simple sequence repeat (SSR) markers and their use in identification of Dendrobium varieties. Molecular Ecology Notes. 2006;6:832–834. [Google Scholar]
- Yukawa T, Ohba H, Cameron KM, Chase MW. Chloroplast DNA phylogeny of subtribe Dendrobiinae (Orchidaceae): insights from a combined analysis based on rbcL sequences and restriction site variation. Journal of Plant Research. 1996;109:169–176. [Google Scholar]
- Yukawa T, Kita K, Handa T. DNA phylogeny and morphological diversification of Australian Dendrobium (Orchidaceae) In: Wilson KL, Morrison DA., editors. Monocots: systematics and evolution. Melbourne: CSIRO Publishing; 2000. pp. 465–471. [Google Scholar]
- Zwick MS, Hanson RE, Islam-Faridi N, et al. A rapid procedure for the isolation of C0t-1 DNA from plants. Genome. 1997;40:138–142. doi: 10.1139/g97-020. [DOI] [PubMed] [Google Scholar]