Abstract
Background and Aims
Although many methods exist for quantifying the number of pollen grains in a sample, there are few standard methods that are user-friendly, inexpensive and reliable. The present contribution describes a new method of counting pollen using readily available, free image processing and analysis software.
Methods
Pollen was collected from anthers of two species, Carduus acanthoides and C. nutans (Asteraceae), then illuminated on slides and digitally photographed through a stereomicroscope. Using ImageJ (NIH), these digital images were processed to remove noise and sharpen individual pollen grains, then analysed to obtain a reliable total count of the number of grains present in the image. A macro was developed to analyse multiple images together. To assess the accuracy and consistency of pollen counting by ImageJ analysis, counts were compared with those made by the human eye.
Key Results and Conclusions
Image analysis produced pollen counts in 60 s or less per image, considerably faster than counting with the human eye (5–68 min). In addition, counts produced with the ImageJ procedure were similar to those obtained by eye. Because count parameters are adjustable, this image analysis protocol may be used for many other plant species. Thus, the method provides a quick, inexpensive and reliable solution to counting pollen from digital images, not only reducing the chance of error but also substantially lowering labour requirements.
Keywords: Carduus acanthoides, Carduus nutans, ImageJ, image processing, pollen grain count
INTRODUCTION
The quantification of pollen has been an important process in the field of plant reproductive biology. For example, the amount of pollen produced by a plant is a critical component of male reproductive function (Kearns and Inouye, 1993), and the fitness of different plant morphs (e.g. Harder and Barrett, 1993). Counting pollen can also be an important part of plant–pollinator studies, such as quantifying the amount of pollen that is removed and carried by pollinators (e.g. Thomson and Goodell, 2001; Adler and Irwin, 2006).
Numerous methods have been utilized to count pollen. These fall into three major categories: counting with the human eye, electronic or laser-based particle counters, and image processing algorithms. Counting visually does not require specialized tools other than microscopes but is very time-consuming. One can also count sub-samples and then estimate the total amount of pollen (Kannely, 2005). Haemocytometers, specialized slides with a grid, are also used to count sub-samples (e.g. Kearns and Inouye, 1993). However, pollen grains may settle disproportionately on grids, especially if the grains are large (which is often the case for animal-dispersed pollen), which may then produce erroneous estimates (Kannely, 2005). Photographing pollen grains followed by manual counting of the grains on the images is another simple method. However, these related methods are very labour intensive and prone to human error.
Electronic or laser-based counters physically detect pollen grains in order to count them. Electronic particle counters, including Elzone® and Coulter® counters, utilize electric currents in saline solutions to determine the presence of particles. When a particle passes through the detector, the change in current triggers a count. A particle counter counts every particle; unfortunately, this may include debris and clumped pollen (Kearns and Inouye, 1993). The clumping of pollen is particularly a problem for animal-dispersed pollen, which possesses a sticky coating (pollenkitt), resulting in a tendency to underestimate the true count unless the sample is sonicated or a surfactant is added. More recently, a laser-based counter, which detects pollen grains passing through a laser, has been developed to collect and count airborne pollen grains (Kawashima et al., 2007). Originally intended to aid in the study of pollinosis and the pollen of transgenic crops (Kawashima et al., 2007), this method may also be suitable for analysing pollen that has been sampled from the exterior of pollinators or directly from the anthers themselves (i.e. non-airborne pollen sources). Unfortunately, these particle counters can be expensive, which limits their operation to frequent users.
Image processing automates pollen counting from various forms of pollen grain images. This method requires use of computer software to scan images for objects and then count each separate object as a unit (e.g. Bechar et al., 1997; Aronne et al., 2001). A similar strategy is used with images of pollen that are collected and then allowed to fluoresce under actinic light; in this process, only the fluoresced pollen grains are counted (Fonseca et al., 2002). In addition, a computationally intensive image processing method has been developed not only to count pollen, but also to identify types of pollen based on surface texture (France et al., 2000). Like particle counters, most currently available image processing packages are also expensive (e.g. Bechar et al., 1997).
The purpose of the present study was therefore to develop a fast, user-friendly, low-cost image processing method of counting pollen of known identity. An automated process is necessary to reduce the time and labour required for such analysis. ImageJ, a Java-based image analysis software provided by the US National Institutes of Health (http://rsbweb.nih.gov/ij/; Rasband, 1997–2009), was used. ImageJ allows processing and analysis of objects within an image, and may be used on any computer capable of running Java platforms. The software is free and can run on Mac OS X, Linux x86 or Microsoft Windows, making it an ideal product to use in most laboratories.
Pollen from two species from the Asteraceae, Carduus acanthoides (spiny plumeless thistle) and C. nutans (musk or nodding thistle), was used to test and optimize this novel technique. Pollen size in both these species (35–40 µm in diameter) falls within the range of the majority of angiosperm pollen grains (5–250 µm in diameter, Muller, 1979). In addition, these two species produce copious quantities of pollen, making manual counting of pollen grains impractical. For example, a capitulum (flower head) of C. acanthoides can produce at least 1 mg d−1 of pollen for several days (L. Russo, Pennsylvania State University, USA, pers. comm.).
METHODS AND MATERIALS
Study species description
Carduus acanthoides L. and C. nutans L. are short-lived, monocarpic perennials, native to Eurasia and invasive worldwide. In their invaded range, they are considered to be weeds of major economic importance (Skinner et al., 2000). In central Pennsylvania, where the present study was conducted, these two species inhabit disturbed areas such as roadsides and pastures. C. acanthoides produces numerous, clustered, small capitula whereas C. nutans produces few, solitary, large capitula (Desrochers et al., 1988). Both species are pollinated by generalist pollinators such as bees (Hymenoptera), butterflies (Lepidoptera) and flies (Diptera) (Giurfa and Núñez, 1992). Pollen counting may offer an important and tractable tool for understanding the reproductive success of these invasive species.
Pollen collection
Capitula from C. acanthoides and C. nutans were collected from an experimental population on the Pennsylvania State University's Russell E. Larson Research Farm at Rock Springs at an early stage of development in which florets were not in anthesis. One capitulum was sampled from 17 C. nutans and 15 C. acanthoides individuals for a total of 32 capitula. Each stem was brought into the laboratory, inserted into an Aquapic® (Syndicate Sales, Inc., Kokomo, IN, USA), which is a tube with a rubber cap that can hold a single stem in water, and isolated in a column (11·0 cm in diameter, 30·5 cm in height) which allowed pollen collection over time and protection from disturbances (such as air circulation) that might result in pollen loss. Pollen was collected twice daily from each capitulum for the duration of its life span. Both passive and active pollen collection was carried out in each individual capitulum, to avoid loss of any pollen. Passive collection involved collecting any grains that dropped from the florets to the bottom of the isolation chambers. Active collection involved removing dehisced pollen directly from the capitulum by tapping and using probes to loosen pollen grains from florets. For the present purposes, the end of a capitulum's life span, and associated pollen collection, occurred when the capitulum no longer appeared to produce pollen (approx. 5 d). A schematic of these pollen collection methods is shown in Supplementary Data Fig. S1 (available online).
Processing pollen grains for image preparation and capture
Once collected, pollen grains were placed on a flat glass slide (7·5 × 12·7 ×0·24 cm), with each slide containing only one capitulum's pollen from one collection time point. Because in initial exploratory analyses the accuracy of the software analysis was found to increase as the number of particles in an image decreases, pollen grains on a slide were separated into smaller sub-samples to allow approx. 5000 grains per sub-sample. A drop of dilute detergent solution (one drop of Palmolive® liquid dishwashing detergent in 250 mL of water) was added to each sub-sample to disperse the grains into a single dissipated layer (Kearns and Inouye, 1993). Slides were designed to allow optimal light conditions for digital image capture under a microscope. Pale yellow masking tape (1·8 cm width) was applied to the edges of the slides, overlapping the top of the slide on all sides. Snake lights (Leica KL 1500 LCD, Wetzlar, Germany) were placed on the tape and adjusted until the pollen had a slight glow and there were no shadows in the background beneath the pollen grains (Fig. 1). A schematic of these sample preparation methods is shown in Fig. S1.
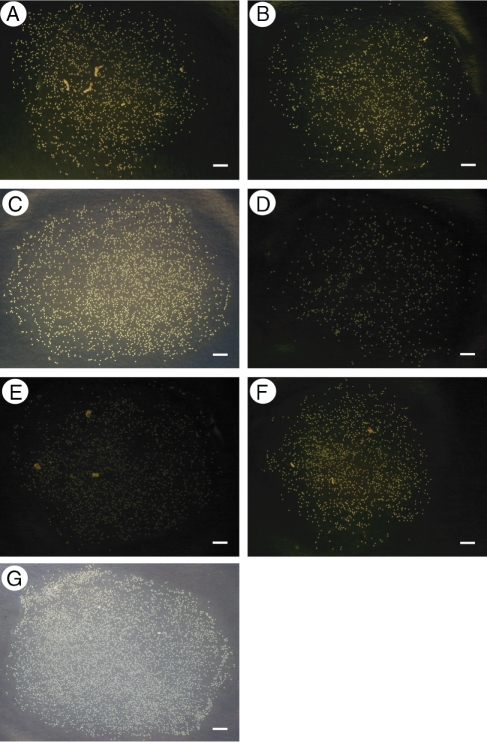
Fig. 1.
Images used in the comparison of single and multiple (macro) image analyses to visual counting. These images were chosen to illustrate the potential errors one may encounter with the ImageJ methods. All methods produced similar counts for each image except E, which was incorrectly counted by multiple image analysis. Scale bars = 100 pixels.
Imaging methods
A Nikon Coolpix 950 (Tokyo, Japan), with a C-mount adapter (model MA 151/40/50, Meiji Techno, San Jose, CA, USA), was used to capture digital images of each subsample of pollen through a Leica MZ 12·5 stereomicroscope (magnification range 5·1–25·6 × ). The microscopic magnification varied to accommodate the range of pollen grains in the sub-sample, as some sub-samples had a few hundred grains whereas others had a few thousand grains. The data were saved in JPEG format on a Compact Flash (CF) card and then transferred to a computer running Windows XP with 1·6-GHz single core processor.
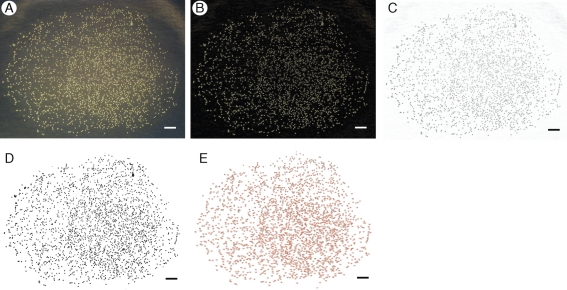
ImageJ was used for processing and analysis of the captured images (Rasband, 1997–2009), as summarized in Fig. 2. Each image was modified in order to minimize noise and sharpen the contrast between the pollen grains and the background. First, the original background was removed from each image, creating a new dark monochromatic background while keeping the pollen grains pale in colour (Fig. 2B). The image was then inverted to create a light background with the pollen as dark objects, because dark objects are recognized by the software as particles to be counted (Fig. 2C). Next, the image was converted to an 8-bit format, creating a bi-chromatic image. Using threshold adjustments, the contrast was modified under the red-scale setting in order to delineate each object more clearly (Fig. 2D). The software's watershed function was used to create breaks between any clusters of pollen that may have been scanned as a single image.
Fig. 2.
Appearance of images at each stage of the processing and analysis with ImageJ. Major steps are: (A) unprocessed image: JPEG format, image is coloured; (B) background subtraction: dark background, light-coloured objects; (C) 8-bit adjustment: inverted image, white background with black objects; (D) threshold adjustment: set contrast to define objects under the red-scale setting; and (E) analysis: Count particles with size 10 pixels2 to 100 pixels2 and circularity 0·5–1·0 (Carduus-specific settings). Scale bar = 100 pixels.
The final step was to utilize the ‘analyse particle’ function to produce pollen counts, but it was first necessary to obtain values of particle size and circularity that were appropriate for the two species (Fig. 2E). This step is necessary to exclude objects that were too small or large, or of the wrong shape, to be pollen. To narrow the default range setting of 0 pixels2 to infinity pixels2 and 0·0–1·0 circularity [1·0 being a perfect circle, which is calculated by the software using 4π(area/perimeter2)], the function was first run on sample images without adjusting for size or circularity. The ranges were narrowed gradually until the counts produced changed drastically. Values from the step prior to the drastic count change were chosen as values appropriate to our two study species. In other words, as the ranges were narrowed, objects that were too big or too small, or too dissimilar in shape from the majority of the objects in the image (the pollen), were gradually discarded (not counted). A drastic decrease in counts corresponded to when the pollen grains themselves were excluded from the analysis, and therefore the range chosen just previous to this drastic decrease represented values that fit the size and shape of the pollen grains. In 8-bit image format, each individual pollen grain for the two study species had an average size ranging from 10 pixels2 to 100 pixels2, with a circularity of 0·5–1·0. These values were consistent with the average particle size calculated as part of the ‘analyse particles’ output.
With the exception of the size and circularity optimization process, which is necessarily species specific, the entire image improvement procedure and pollen count were automated by writing a macro (Supplementary Data, available online) designed to carry out each step on multiple images. This reduces the total amount of time required to process and analyse each image.
In total, 2568 images were processed. In order to determine the accuracy and consistency of the ImageJ counts, the counts produced by visual counting (with the unaided human eye) were compared with single and multiple (macro) image processing and analysis using the seven images depicted in Fig. 1. These seven images (from our collection of 2568 images) were selected to represent a range of image quality, such as the presence of artefacts (contaminants), poor image contrast and high numbers of pollen grains, to demonstrate the flexibility of the methods as well as potential sources of error. For each of these images, three separate users (including both novice and expert ImageJ users) conducted the counts with the different methods. Visual counting used a laser-printed negative (i.e. inverted) version of the unprocessed image. The single and multiple image analysis started with the same unprocessed images. Pearson's correlation coefficients were calculated to test for the relationship between pollen counts made visually and those obtained from ImageJ processing. In addition, differences in counts among the methods, within each image, were tested for via analysis of variance. All statistics were conducted using the statistical package R (R Development Core Team, 2009).
RESULTS
From the 2568 images, and using multiple image processing, a single C. nutans capitulum was found to produce 249 491·1 ± 22 533·04 (mean ± s.e., n = 17) over 5 d and a single C. acanthoides capitulum 125 744·1 ± 14 006·47 (n = 15) over 4 d.
Both single and multiple image processing were substantially faster than visual analysis. For the seven images in Fig. 1, visual counting of samples took on average 22·6 ± 7·0 min (mean ± s.e., range, 5–68 min) per image. The macro analysed each image in less than 3 s, an improvement of at least two orders of magnitude. The processing of single images using the software without the macro ranged between 30 and 60 s, an order of magnitude quicker than manual counting. Thus, visual analysis of our 2568 images would have taken 967·3 h (40·3 d), in contrast to the fastest method of multiple image analysis, which took approximately 2 h in total.
The counts produced by both ImageJ procedures were comparable to the counts obtained by visual analysis. Single image processing counts and visual analysis counts were highly correlated (Pearson's r = 0·9997, P < 0·0001, n = 7 for each count method). Similarly, pollen counts obtained by multiple image processing were highly correlated to the counts made by visual analysis (Pearson's r = 0·9540, P = 0·0009, n = 7 for each count method). For two of the seven images selected to exemplify the types of images encountered, the counts produced by the ImageJ and visual analysis methods were significantly different (Table 1; D: P = 0·0416, E: P < 0·0001, see Fig. 1D, E). For Fig. 1D, single image processing produced a much lower count than multiple image analysis and visual analysis (Table 1). For Fig. 1E, multiple image analysis did not count the number of pollen grains correctly (Table 1). However, in the complete set of 2568 images, multiple image analysis failed to detect pollen grains correctly only three times. Avoiding these errors is possible, and they are discussed below.
Table 1.
Comparison of counts produced by three forms of counting methods: multiple (macro) image processing using ImageJ, single image processing using ImageJ and counting by eye
| Pollen count (±s.e.) |
||||
|---|---|---|---|---|
| Image | Multiple (macro) image processing using ImageJ | Single image processing using ImageJ | Visual analysis by human eye | P-value |
| A | 1362 (±0) | 1380 (±55) | 1439 (±50) | 0·5599 |
| B | 1231 (±0) | 1194 (±19) | 1225 (±4) | 0·1955 |
| C | 2567 (±0) | 2531 (±24) | 2571 (±4) | 0·2320 |
| D | 679 (±0) | 645 (±9) | 671 (±6) | 0·0416 |
| E | 6 (±0) | 1671 (±44) | 1741 (±16) | <0·0001 |
| F | 1410 (±0) | 1383 (±20) | 1425 (±14) | 0·2295 |
| G | 6511 (±0) | 6122 (±(191) | 6601 (±27) | 0·0851 |
The last column shows the P value from ANOVA, testing for significant differences for each image (Fig. 1A–G) among the counts produced by each method. The macro analysis of all seven images was completed in 20 s (approx. 3 s per image) while single analysis of each image ranged from 30 to 60 s. Human visual counting took between 5 and 68 minutes per image.
The level of reliability of both single and multiple image analysis can be seen when compared with visual analysis (Table 1). The macro procedure consistently provided the same pollen count each time the same image was tested, whereas considerable variation in count was found with single image analysis (Table 1).
DISCUSSION
The method described here for counting pollen grains from digital images is not only user-friendly, but also efficient, accurate and consistent. Both novice and expert ImageJ users were able to process and analyse images easily and quickly, with and without the macro. Although the multiple image analysis can very occasionally fail to detect pollen grains correctly, multiple image analysis counts were more consistent than single image analysis counts. Taking this trade-off into consideration will thus be important when deciding upon which of these two analysis methods to employ for a particular research question.
Understanding the source of counting error by the software is crucial to minimizing error occurrence, such as the erroneous count by multiple image processing of Fig. 1E (Table 1). At first sight this appears to be due to a lack of contrast between the background and objects. However, Fig. 1D has a similarly poor contrast, but was counted correctly. In fact, the error arose because Fig. 1E contained artefacts that were brighter and larger than the pollen grains present. The macro removed the pollen grains along with the background, and counted the artefacts as the objects of interest. Visual inspection of the image showed the presence of many pollen grains, however. So, upon encountering this very low count (Table 1), we were easily able to determine that an error had occurred. It is important to note that the presence of artefacts did not automatically lead to an erroneous count. In Fig. 1A a correct pollen count was found even in the presence of artefacts because the contrast between the pollen and the background was higher than the contrast between the artefact and the background. Images that have low counts should be visually inspected, and then processed with the single image analysis method, as adjusting software settings is usually much simpler than removing artefacts from samples.
Over-estimation is another potential source of image analysis error. However, this scenario is relatively unlikely. Owing to the adjustment of the range of particle size (in the form of pixel size, as described in the methods), the counted objects are limited to the sizes specified. Circularity of objects is also used to reduce the possibility of over-estimation. Any object that is not in the shape range of the pollen grains will not be counted; this is more likely to lead to underestimates of pollen counts if the detergent or other surfactant treatment fails to separate clumped particles, and these are consequently ignored. Thus, to prevent over-estimation, selecting the correct range and circularity values for the species of interest is of crucial importance.
Single image analysis produced more variation between counts of the same image than multiple image analysis, and a much lower count for Fig. 1D (Table 1). We attribute this variation to the individual decisions made by each user as they conducted each image adjustment step. For example, some users may tend to remove more noise than others, and in the case of the low-contrast image in Fig. 1D, this probably included some of the dimmer pollen grains. However, in our experience, a user can become adept at single image analysis with sufficient training, which then minimizes the variation in counts produced by the single image method. Although single analysis of images is based on personal skill and decision-making, the macro requires little expertise, ensures greater consistency, and is very time and labour efficient.
Compared with other image analysis software packages, ImageJ is free and provides the tools necessary to manage, process and analyse images of pollen grains with ease. Although other image analysis programs can be similarly effective, they are often not freely available and require intimate knowledge of specialized software. For example, similar image software described by Bechar et al. (1997) costs nearly US $1500. The methods proposed by Bechar et al. (1997) should be comparable with those described here; however, without purchasing this expensive software package, we were not able to make any direct comparisons. We suggest that users of the Bechar et al. (1997) method confirm concordance with our methods before making any changes in protocol.
In conclusion, the accessibility of required materials and the simplicity and negligible cost of ImageJ make our methods an ideal tool for counting pollen very cheaply, efficiently and reliably. We were able to process images, taken by a point and shoot camera with a microscope adapter, through a standard laboratory computer. The cost of the entire process is very low as most required items are generally available in any standard biology laboratory. This method is widely applicable, because ImageJ can be modified to count various types and sizes of pollen, and the appropriate adjustments to the desired size, circularity and object type can be easily established in the macro (Supplementary Data). Image processing using ImageJ not only reduces the chance of error but also ensures greater consistency between images and substantially lowers labour requirements necessary for reliable pollen counting.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Katriona Shea, who not only generously allowed us to use her laboratory equipment and personnel (especially Eelke Jongejans, Rui Zhang, Katie Myers Marchetto, Maria Stevens, Megan Lundgren, Mason Heberling, Maggie Wilkens, Zak DeWalt, Evan Stover, Chris DeMarco,and Richard Ashby) to conduct our research, but also provided us with comments that improved a previous version of this manuscript. We also thank Laura Russo, Andy Stephenson, Simon Hiscock and two anonymous reviewers for their input. This work was supported by the National Science Foundation (grant number DEB-0815373 to K. Shea, Biological Informatics Postdoctoral Fellowship to S.Y.) and Pennsylvania State University Department of Crop and Soil Sciences to S.Y.
LITERATURE CITED
- Adler LS, Irwin RE. Comparison of pollen transfer dynamics by multiple floral visitors: experiments with pollen and fluorescent dye. Annals of Botany. 2006;97:141–150. doi: 10.1093/aob/mcj012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronne G, Cavuoto D, Eduardo P. Classification and counting of fluorescent pollen using an image analysis system. Biotechnic & Histochemistry. 2001;76:35–40. doi: 10.1080/bih.76.1.35.40. [DOI] [PubMed] [Google Scholar]
- Bechar A, Gan-Mor S, Vaknin Y, Shmulevich I, Ronen B, Eisikowitch D. An image-analysis technique for accurate counting of pollen on stigmas. New Phytologist. 1997;137:639–643. [Google Scholar]
- Desrochers AM, Bain JF, Warwick SI. The biology of Canadian weeds. 89. Carduus nutans L and Carduus acanthoides L. Canadian Journal of Plant Science. 1988;68:1053–1068. [Google Scholar]
- Fonseca AE, Westgate ME, Doyle RT. Application of fluorescence microscopy and image analysis for quantifying dynamics of maize pollen shed. Crop Science. 2002;42:2201–2206. [Google Scholar]
- France I, Duller AWG, Duller GAT, Lamb HF. A new approach to automated pollen analysis. Quaternary Science Reviews. 2000;19:537–546. [Google Scholar]
- Giurfa M, Núñez JA. Foraging by honeybees on Carduus acanthoides – pattern and efficiency. Ecological Entomology. 1992;17:326–330. [Google Scholar]
- Harder LD, Barrett SCH. Pollen removal from tristylous Pontederia cordata: effects of anther position and pollinator specialization. Ecology. 1993;74:1059–1072. [Google Scholar]
- Kannely A. Preparation and quantification of entomophilous pollen using sonication and an area-counting technique. Madroño. 2005;52:267–269. [Google Scholar]
- Kawashima S, Clot B, Fujita T, Takahashi Y, Nakamura K. An algorithm and a device for counting airborne pollen automatically using laser optics. Atmospheric Environment. 2007;41:7987–7993. [Google Scholar]
- Kearns CA, Inouye DW. Techniques for pollination biologists. Niwot, CO: University Press of Colorado; 1993. [Google Scholar]
- Muller J. Form and function in angiosperm pollen. Annals of the Missouri Botanical Garden. 1979;66:593–632. [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Rasband WS. ImageJ. Bethesda, MD: National Institutes of Health; 1997–2009. [Google Scholar]
- Skinner K, Smith L, Rice P. Using noxious weed lists to prioritize targets for developing weed management strategies. Weed Science. 2000;48:640–644. [Google Scholar]
- Thomson JD, Goodell K. Pollen removal and deposition by honeybee and bumblebee visitors to apple and almond flowers. Journal of Applied Ecology. 2001;38:1032–1044. [Google Scholar]