Abstract
Background and Aims
Pre-harvest sprouting susceptibility in grain sorghum (Sorghum bicolor) is related to low seed dormancy and reduced embryo sensitivity to inhibition of germination by abscisic acid (ABA). Intra-specific variability for pre-harvest sprouting might involve differential regulation of ABA signalling genes.
Methods
Sorghum genes encoding homologues for ABA signalling components from other species (ABI5, ABI4, VP1, ABI1 and PKABA1) were studied at the transcriptional and protein level (ABI5) during grain imbibition for two sorghum lines with contrasting sprouting phenotypes and in response to hormones.
Key Results
Transcript levels of these genes and protein levels of ABI5 were higher in imbibed immature caryopses of the more dormant line. Dormancy loss was related to lower transcript levels of these genes and lower ABI5 protein levels in both genotypes. Exogenous ABA inhibited germination of isolated embryos but failed to prevent ABI5 rapid decrease supporting a role for the seed coat in regulating ABI5 levels.
Conclusions
Several genes involved in ABA signalling are regulated differently in imbibed caryopses from two sorghum lines with contrasting pre-harvest sprouting response before – but not after – physiological maturity. A role for ABI5 in the expression of dormancy during grain development is discussed.
Keywords: Sorghum bicolor, pre-harvest sprouting, abscisic acid, hormone signalling, ABI5, seed dormancy
INTRODUCTION
Seed dormancy is an internal block to germination (Finch-Savage and Leubner-Metzger, 2006). Domestication of crop species has favoured the presence of genotypes with low dormancy level at harvest, as it allows immediate malting or a rapid emergence and a uniform stand in the field. But dormancy loss before harvest can allow the crop to suffer pre-harvest sprouting (PHS) with a negative impact on grain quality and yield. In many species, dormancy is regulated hormonally (physiological dormancy) and involves the products of many different genes in interaction with the environment (Finch-Savage and Leubner-Metzger, 2006; Finkelstein et al., 2008), resulting in a very complex trait which is difficult to include in breeding programmes.
Similar to other cereal crops, grain sorghum (Sorghum bicolor) has a broad intra-specific variability for PHS: inbred line ‘IS9530’ can remain dormant until harvest maturity and is resistant to PHS, while inbred line ‘RedlandB2’ loses dormancy precociously and is highly susceptible to PHS (Steinbach et al., 1995). Previous research with both lines has shown that dormancy is established during early stages of seed development, and is regulated mainly by the antagonistic effects of abscisic acid (ABA; enhances dormancy and inhibits germination) and gibberellins (GAs; reduce dormancy and promote germination) (Steinbach et al., 1995, 1997). Dormancy in sorghum caryopses is imposed by tissues surrounding the embryo (endosperm, testa and pericarp), and there is no evidence of a mechanical restraint to radicle expansion exerted by these tissues like the endosperm layer in arabidopsis (Bethke et al., 2007). Isolated sorghum embryos from ‘IS9530’ and ‘RedlandB2’ caryopses germinate fully within 36 h when incubated in distilled water, and this has been related to a rapid release of ABA into the incubation medium (Gualano et al., 2007). Steinbach et al. (1995) observed that isolated embryos from ‘IS9530’ are far more sensitive to inhibition of germination by exogenous ABA than those from ‘RedlandB2’. Embryonic ABA levels in imbibed caryopses decreased similarly in both genotypes, despite their contrasting germination response (Benech-Arnold et al., 2003). Plants from both genotypes have normal responses to drought and environmental stress in the field, and their seeds develop and mature similarly; hence, differences in sensitivity to ABA are restricted only to seed germination.
As knowledge on the ABA signalling network is being widened it is being found to be increasingly complex, and involves multiple levels of control: over 50 genes have been identified as playing a role in ABA signalling in seeds (Nambara and Marion-Poll, 2003). Among these genes, ABI3/VP1, ABI4 and ABI5 encode transcription factors that mediate seed-specific and/or ABA-inducible gene expression (Nakamura et al., 2001; Finkelstein et al., 2002, 2008). Arabidopsis mutants abi3, abi5 and abi4 exhibit ABA-resistant germination. Seed dormancy is only affected in the abi3 mutant (Finkelstein et al., 2002) and maize vp1 mutants (orthologue of ABI3) are viviparous. Generation of aberrant transcripts due to natural missplicing of VP1 in wheat is related to PHS in this species (McKibbin et al., 2002). Although abi4 and abi5 mutations do not affect seed dormancy release in arabidopsis, ABI4 and ABI5 play crucial roles during germination: ABI4 mediates ABA inhibition of germination and lipid breakdown in the endosperm (Penfield et al., 2006), and ABI5 acts as the final common repressor of germination in response to changes in ABA and GA levels in arabidopsis (Piskurewicz et al., 2008). Transcription of ABI3, ABI4 and ABI5 genes is promoted by ABA in imbibed arabidopsis seeds (Penfield et al., 2006). ABA also regulates ABI5 activity by changing its phosphorylation status: ABI5 and TaABF (a wheat orthologue) proteins are phosphorylated and thus activated by SnRK2 kinases from arabidopsis (López-Molina et al., 2001; Fuji et al., 2007; Piskurewicz et al., 2008) and ABA-induced wheat PKABA1 (Johnson et al., 2002). ABI5 protein degradation in the proteasome is promoted by AFP, a negative regulator of ABA signalling in arabidopsis seeds (López-Molina et al., 2003).
Negative regulation of ABA signalling also involves protein phosphatases type-2C (PP2C) like ABI1 and ABI2 in arabidopsis and FsPP2C1 in Fagus sylvatica (Merlot et al., 2001; González-García et al., 2003). Protein–protein interaction analysis with ABI1 and ABI3, ABI4 and ABI5, failed to reveal binding to ABI1 (Nakamura et al., 2001) although other targets have been described (Himmelbach et al., 2002; Moes et al., 2008).
Different from sorghum, seed dormancy in arabidopsis is not related to an enhanced sensitivity to ABA, but to changes in ABA metabolism instead (Ali-Rachedi et al., 2004), and inhibition of germination in imbibed dormant seeds correlates with higher expression of ABA synthesis genes as well as down-regulation of ABA inactivation genes (Cadman et al., 2006; Millar et al., 2006). Known ABA signalling components do not have an obvious role in arabidopsis seed dormancy, although they might play a significant role during the expression of dormancy in other species like sorghum. Previous work (Carrari et al., 2001) showed a higher expression of sorghum VP1 during imbibition of dormant caryopses, indicating that differential regulation of this gene might be involved in intra-specific variability for sprouting response. It is proposed to extend this analysis to other ABA signalling components by studying their expression in sorghum grains from two genotypes with contrasting sprouting responses and harvested at different developmental stages. Also addressed is the possibility that there is a differential regulation of ABI5 at the protein level under a variety of dormancy situations.
In this paper, the following questions are specifically addressed: (a) Are ‘ABA signalling genes’ and ABI5 protein levels regulated differently in imbibed sorghum seeds from ‘IS9530’ and ‘RedlandB2’ that exhibit contrasting dormancy levels, or as a result of dormancy release during development? (b) Do ABI5 protein levels change in response to manipulation of the ABA/GA balance (that affects the germination response) in dormant grains and in isolated embryos (after removal of the seed coats)?
MATERIALS AND METHODS
Plant material
Two sorghum [Sorghum bicolor (L.) Moench] inbred lines, ‘IS9530’ and ‘RedlandB2’, were sown in the experimental field of the Facultad de Agronomía de la Universidad de Buenos Aires (Bs. As., Argentina), during the summers of 2004, 2005 and 2007. Three plots 24 m2 each were sown for each genotype in a completely randomized design. Anthesis date (anthers exposed in the middle third of the panicle) was recorded for each plant. For each experiment, seeds from the middle third of the panicle were harvested from ten plants with the same anthesis date and within a same field plot, and pooled together. Two field plots were used as biological repetitions for each genotype. Experiments began within the same day the seeds were harvested. To obtain samples of embryos for RNA, protein and hormone analysis, enough Petri dishes (9 cm diameter, plus 6 mL of distilled water and two discs of filter paper) containing 50 seeds each were incubated in chambers with constant temperature at 20 or 25 °C. Although the germination rate of sorghum grains can be higher at 25 °C compared with 20 °C, differences in dormancy between genotypes are equally expressed at any of both temperatures, and significant interactions between the genes studied and temperature were not expected within this temperature range. Incubation chambers had no artificial light, but seeds incubated in transparent plastic Petri dishes were exposed to ambient light repeatedly during experimental manipulation.
Sampling for RNA and protein extraction was done as follows. After different incubation periods, embryos were dissected from the surrounding tissues (endosperm, testa and pericarp) and stored in liquid N2. A single sample consisting of approx. 150 embryos was stored for each of two biological repetitions and for each incubation time. Portions of each of these samples were processed for RNA and protein extraction.
Incubation experiments were done with caryopses sampled at four different developmental stages: 30 (immature, before physiological maturity, PM), 37 (PM) and 42 and 47 d after pollination (DAP; after PM). Whole 37-DAP caryopses from ‘IS9530’ were also incubated in 50 µm ABA and 150 µm GA3+4 solutions (100 µm GA3 and 50 µm GA4; all hormones from Sigma). Isolated embryos from 30-DAP caryopses were incubated in either 4 mL of distilled water (control) or 50 µm ABA for different periods at 25 °C (50 embryos per Petri dish). Each experiment (except isolated embryos in ABA and whole grains with addition of ABA and GA) was repeated in at least two different years with similar results.
Germination response
Simultaneously with each experiment described above, seeds were also incubated (three replicates of 50 seeds on two layers of filter paper plus 6 mL of distilled water) under the same experimental conditions. Germination (radicle protrusion) was scored daily. Germinated embryos were not included in the samples for RNA. For long-term comparisons of germination response, a germination index (GI) was used as described in Steinbach et al. (1995), and which is calculated as follows:
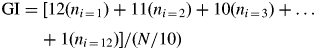 |
where ni = number of seeds germinated on day i of incubation, and N = number of incubated seeds. GI values range from 120 (all seeds germinated on the first day) to 0 (no germination after 12 d).
RNA extraction and real-time quantitative PCR (RT-QPCR)
Between 30 and 45 embryos (70–100 mg) per sample were ground to powder with mortar and pestle in liquid N2, and added to 600 µL of the RA1 extraction buffer included in the Nucleospin RNA plant mini-kit (Macherey-Nagel, Germany) for total RNA extraction. PVP-40 (Sigma) was added to the RA1 buffer to a final concentration of 1 % (wv). After 5-min centrifugation, clear supernatant was used in the extraction protocol as described in the kit's manual. RNA quality was assessed in 1 % agarose gel stained with EtBr. After reverse transcription (Promega reactives and dT15 primer) cDNA was diluted 1:5 in water, and used for quantitative PCR, in an ABI7500 equipment (Applied Biosystems) with ABI SYBR-green master mix. PCR standard conditions for ABI7500 were used. Each sample consisted of 150–200 seeds pooled together from plants in a single field plot, and incubated for a certain period of time (two biological repetitions – field plots – were done for each genotype). For each sample, two replicates for each PCR reaction (cDNA from the same sample as template) were pipetted in two wells within the same 96-well PCR plate, and the average was calculated for the values obtained in both reactions (which were accepted as valid when the difference in Ct values was less than one cycle). The ‘quantitative method’ as described in the ABI7500 user's manual was used, with standard curves for each amplicon. After a first PCR trial the size and unicity of the product was checked in an agarose gel, and the excised amplicon was purified (Nucleospin Extract kit, Macherey-Nagel) and used to create a set of serial dilutions (1:10 each step) covering a wide range of Ct values. Of these, five dilutions with Ct values ranging from 17 to 33 approx. were used to obtain the standard curve. An arbitrary number of 10000, 1000, 100, 10 and 1 molecules was assigned to the dilutions which had Ct values that ranged from 16 to 32 or 33. The relative number of copies obtained for each cDNA species was normalized against SbActin transcript values for each sample as an internal reference. SbActin levels were similar to those of SbUbiquitin (data not shown), and did not change significantly during incubation of whole grains or in response to hormone treatments in imbibed caryopses (see Fig. S3C in Supplementary data available online). SbActin expression was significantly inhibited by ABA in isolated embryos of both genotypes tested, but not SbUbiquitin, so the latter was used for normalization of gene expression in this experiment (Fig. S3A and B in Supplementary data).
Data points on each graph are the mean of two biological repetitions (two field plots), and bars indicate standard error.
Protein extraction and ABI5 detection by western blot
Samples (80–100 mg of embryos) were ground to powder with mortar and pestle in liquid N2 and then added to 0·5 mL of extraction buffer (50 mm Tris, pH 7·5; 150 mm NaCl; 10 % glycerol; 10 mm EDTA, pH 8; 10 mm β-mercaptoethanol; 0·1 % sarcosyl; 1 mm PMSF). After a 5-min centrifugation, supernatant was transferred to a clean tube and stored at −80 °C. Total protein content in the samples was quantified by Bradford protocol with a spectrophotometer (UV-1700 Pharma Spec; Shimadzu) against a BSA standard curve. Western blots were performed according to Sambrook et al. (2001). On each lane, 50 µg of total protein were loaded plus 5 µL of cracking buffer (62·5 mm Tris, pH 6·8; 10 % glycerol; 2 % SDS; 0·72 m β-mercaptoethanol; 0·05 % BPB) and water to a final volume of 20 µL. After 0·1 % SDS–16·5 % PAGE, proteins were electroblotted onto a nitrocellulose membrane. Blue Ranger pre-stained marker (Pierce) was loaded as a reference for protein size. Membranes were reversibly stained with Ponceau S red to check equal loading and protein integrity. Serum containing polyclonal antibodies against ABI5 sorghum protein were obtained from rabbits (custom antibody service, Biosynthesis Inc., USA) immunized with the peptide MNFPGGSGRRQQQEPEHLPPMT. This sequence belongs to the N terminal of the predicted sorghum ABI5 protein, and is not present in other sorghum bZIP homologues (Fig. S6 in Supplementary data available online) and no other proteins in the sorghum genome were found that contained a similar sequence. Membranes were blocked overnight in TTBS–5 % low fat milk, and then incubated in TTBS–0·5 % milk plus crude serum at a final dilution of 1/800. Bound antibodies were detected using an anti-rabbit immunoglobulin-HRP (Bio-Rad) at a final dilution of 1/5000. The protein blot was detected with the ECL kit from Amersham Biosciences and CL-X Posure Film, clear blue (Pierce).
ABA measurements
Samples of 25–35 embryos were lyophilized, powdered, weighed and stored at –20 °C until being assayed for ABA content by radioimmunoassay as described in Steinbach et al. (1995). This method uses the monoclonal antibody AFRC MAC 252 (Quarrie et al., 1988) and tritiated-ABA (Amersham-Pharmacia). Each sample was assessed twice. The results presented are the mean value of two biological replicates ± standard error.
In silico search for sorghum sequences encoding homologues of ABA signalling components
Identification of sorghum sequences with high identity to the reference genes was done by searching the PlantGDB Sorghum bicolor database. The identification codes for the selected sorghum sequences are shown in Supplemental data available online (see Table S1) as well as the alignments between the predicted sorghum and reference proteins (Figs S5–S10). As a result of the in silico search, one sequence (locus Sb04g141930, chromosome 4) was identified encoding a protein with highest identity with ABI5 from maize (88·5 %) and rice (77·2 %), and with a weaker identity to ABI5 from barley and arabidopsis (54·7 and 37·5 %, respectively). Three homologues were also identified (Figs S5 and S6 in Supplementary data). Primers for QPCR were designed to discriminate among them. Only one sequence with high identity to ZmABI4 (Niu et al., 2002) was found. A unique sequence was also identified with high identity to both phosphatases AtABI1 and AtABI2, while two sequences with high identity to wheat PKABA1 were found and designated SbPKABA1.1 and SbPKABA1.2. Sorghum VP1 was cloned and sequenced by Carrari et al. (2001). RAB28 protein accumulates during late embryogenesis in both arabidopsis and maize, but only in maize is RAB28 transcription induced in vegetative tissues in response to ABA and water stress (Borrell et al., 2002). In arabidopsis seeds, functionality of this nuclear protein is associated to enhanced germination under low osmotic potential and salts. Although a role in seed dormancy has not been proposed, SbRAB28 expression was evaluated as a molecular marker of ABA signalling activity. Primers for QPCR used in this work are shown in Table 1, and were designed with Primer Express software (Applied Biosystems).
Table 1.
Sequences of primers used in this work for gene expression determination by real-time PCR after reverse transcription
| Gene name* | Forward | Reverse |
|---|---|---|
| SbABI5 | gcagcagacaatgctgtttcc | gcagcggaggcaccaa |
| SbPKABA1.1 | cacgctctacgtcatgctcg | tcttttggatcctcgaacgg |
| SbPKABA1.2 | tgagatcatgcagatcctggc | cgagaaccagctgctggaat |
| SbABI4 | gatcagcacgacgcttcttaatt | aaacttcaaaccgaccaagca |
| SbABI1 | cgaatgaagaggcatgcaaa | tgcgacaccattgttcttgtg |
| SbVP1 | acatcgaccagatcctcgac | ccagaacgggtttgtgttga |
| SbRAB28 | gagaggcacacccaaatctc | cctctggctcattgtcaagc |
| SbActin | gccgtcctctctctgtatgc | cgagcttctccttcatgt |
| SbBZIP16042 | ccgcggccgattcag | ccccaaagttgccgaagaa |
| SbBZIP70604 | caaggatttcggctccatga | cctccgcggtccagatg |
| SbBZIP48312 | cgcacgctcagccagaa | cggtgaagcccacgatct |
| SbUbiquitin | gctgtacctgcgtttgtctg | acacacgggacacaagacac |
* Sorghum genes were named after their homologues in reference species.
RESULTS
Both dormancy depth and embryo sensitivity to ABA change during seed development
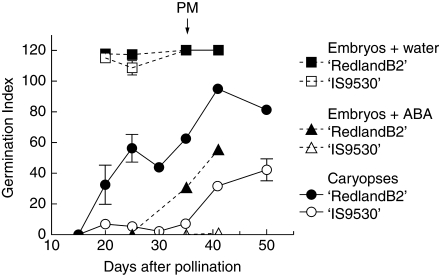
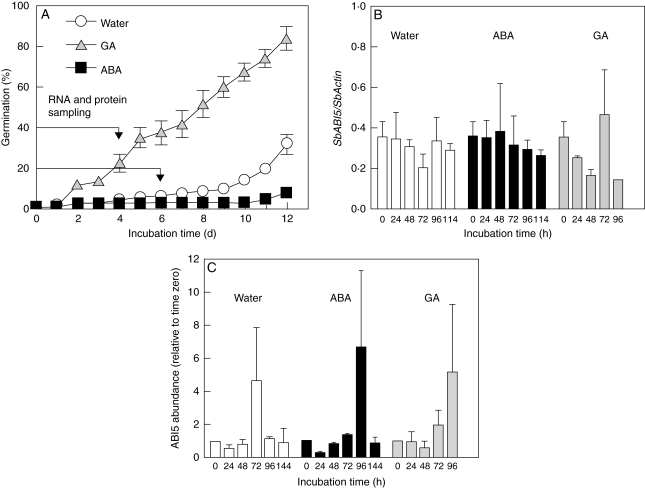
Dormancy depth was followed for both ‘IS9530’ and ‘RedlandB2’ through a germination index (GI; Fig. 1). Embryonic sensitivity to ABA was evaluated at 25, 35 and 42 DAP through incubation of isolated embryos in a 50 µm ABA solution. Dormancy release (detected as an increase in the GI of caryopses) began earlier in ‘RedlandB2’ as compared with ‘IS9530’, and this correlated also with an early loss of sensitivity to ABA in ‘RedlandB2’ (PM; Fig. 1). Isolated embryos incubated in distilled water germinated fully within 24 h since 20 DAP and similarly for both genotypes (Fig. 1).
Fig. 1.
Germination index for sorghum lines ‘IS9530’ and ‘RedlandB2’. Caryopses were harvested on different days after pollination (DAP), and incubated for 12 d in water at 25 °C (circles). Embryos (axis plus scutellum; Fig. S4 in Supplementary data, available online) were dissected and incubated at 25 °C in distilled water (squares) or 50 µm ABA (triangles). Each data point is the mean value of two biological replicates (i.e. two field plots) assessed in triplicate. Bars indicate standard error with n = 2. Time of physiological maturity is indicated with an arrow.
Embryonic ABA content during imbibition does not correlate with germination response
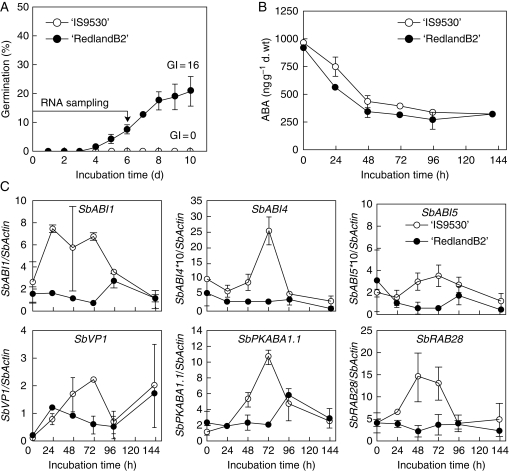
To discount a possible contribution of ABA metabolism to the different germination responses in the experiments (Fig. 2A and 3A), ABA levels within the embryo (axis plus scutellum) were quantified by radio-immuno assay in 30 and 47-DAP caryopses (before and after PM). In 30-DAP grains, embryonic ABA content decreased similarly for both genotypes (Fig. 2B) and was not related to the different responses to germination (Fig. 2A). Embryonic ABA content during incubation of 47-DAP grains (Fig. 3B) did not decrease, but remained unchanged in ‘RedlandB2’ and increased slightly in ‘IS9530’, from 300 ng g−1 d. wt up to a value of 400 ng g−1 d. wt at 64 h. After a 12-day incubation peiod, ABA content in non-germinated ‘IS9530’ dormant caryopses was close to 450 ng g−1 d. wt, suggesting that a steady state for ABA content (between 300 and 450 ng ABA g−1 d. wt) is reached under a wide range of situations regardless of the germination response.
Fig. 2.
(A) Germination response at 20 °C for immature caryopses (30 DAP) from two sorghum inbred lines: sprouting resistant ‘IS9530’ and sprouting-susceptible ‘RedlandB2’. (B) ABA content in the embryo during incubation of whole caryopses. ABA content was determined by radioimmunoassay. (C) Expression (determined by RT-QPCR; relative to SbActin) of sorghum genes encoding homologues for ABA signalling proteins: transcription factors SbABI4, SbABI5, SbVP1, protein-kinase SbPKABA1 and a PP2C-type protein kinase, SbABI1. SbRAB28 was included as an ABA-responsive gene (see Materials and Methods). Data in (A), (B) and (C) belong to the same experiment (year 2006). Each value is the mean of two biological replicates coming from two independent field plots.
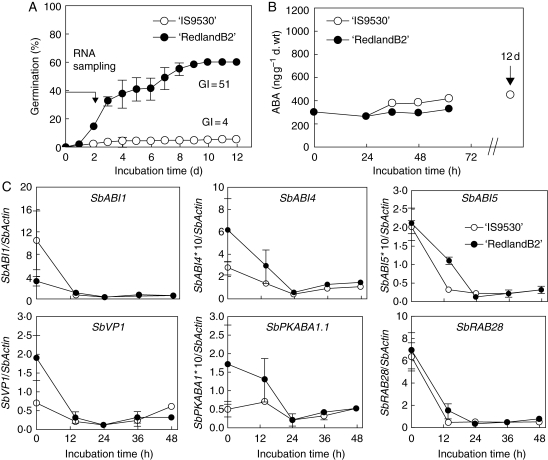
Fig. 3.
(A) Germination response for 47-DAP (after PM) caryopses incubated in distilled water at 20 °C, from two sorghum inbred lines: ‘IS9530’ (sprouting resistant) and ‘RedlandB2’ (sprouting susceptible). (B) ABA content in the embryo during incubation of whole caryopses. (C) Transcript levels (relative to SbActin mRNA levels) for the genes studied. Data in (A), (B) and (C) were all obtained from the same experiment. Each value is the mean of two biological replicates coming from two independent field plots.
Sorghum homologues for ABA signalling genes are induced transiently in dormant-immature grains of ‘IS9530’, but not in less-dormant ‘RedlandB2’
Transcriptional activity of several genes encoding sorghum ABI3/VP1 and homologues for ABI4, ABI5 and PKABA1 (positive regulators of ABA signalling) was induced during incubation of dormant ‘IS9530’ immature caryopses (30 DAP, before physiological maturity, PM), but not in less-dormant ‘RedlandB2’ (Fig. 2). In ‘IS9530’, an 1·8- to 3·0-fold increase in transcript levels after 24 or 48 h of incubation was observed for SbABI5, SbABI4, SbABI1 and SbRAB28, and this induction was even stronger for SbPKABA1.1 and SbVP1 (11·9- and 22- fold increase, respectively) compared with time zero of incubation. In ‘RedlandB2’, an increase in transcript abundance within this time window was only observed for SbVP1 (4·5-fold), while no significant changes were observed for SbPKABA1.1, SbRAB28, SbABI5, SbABI4 and SbABI1. No consistent differences between genotypes were observed for other three homologue genes encoding bZIP transcription factors, and a second homologue for PKABA1, SbPKABA1.2, was induced similarly (about 8 times) in both genotypes after a 6-d incubation period (Fig. S1A in Supplementary data available online). A synchronized peak was observed after 72 h of incubation and several days before the first germinated caryopses were detected in ‘IS9530’, while expression levels remained low in less-dormant ‘RedlandB2’ grains. After 96 h of incubation and despite the contrasting germination responses, the expression levels for all the genes under study were similar in both genotypes.
Dormancy release between 30 and 47 DAP was related to changes in gene expression profiles during grain imbibition
Partial dormancy release in both genotypes between 30 and 47 DAP (Fig. 1) was accompanied by substantial changes in the transcriptional profiles of the studied genes after imbibition (Figs 2 and 3). The transient increase in mRNA levels observed during incubation of 30-DAP ‘IS9530’ caryopses was absent in more mature, 47-DAP caryopses. Similarly, the GI increase between 30 and 47 DAP (from 16 to 51) observed for ‘RedlandB2’ also correlated with significant changes in gene expression profiles (Figs 2 and 3, and Fig. S11 in Supplementary Data, available online); transcript levels of the studied genes were maintained (and increased for SbVP1) during the first 48 h of incubation in 30-DAP grains, but in less-dormant 47-DAP grains mRNA levels decreased significantly during the first 24 h and remained low afterwards. This decrease was about 10-fold for SbABI1, SbABI5 and SbVP1, and 23-fold for SbRAB28 after a 24-h incubation period and relative to time zero of imbibition.
Differences in dormancy between genotypes after physiological maturity are not explained by the expression profiles of the genes studied
During incubation of 47-DAP grains gene expression profiles were similar for both genotypes despite the contrasting germination responses (Fig. 3). These results indicate that inhibition of germination after physiological maturity does not involve differential regulation of the genes under study. One possibility is that differences in sensitivity to ABA that still exist at this stage (Fig. 1) involve post-translational regulation of some of these components, and this was further explored for the sorghum ABI5 homologue.
Polyclonal antibodies (raised against a 22-aa peptide corresponding to the N-terminal of sorghum ABI5; see Fig. S6 in Supplementary data, and Materials and Methods) were used to detect sorghum ABI5 protein. The size of the band observed was between 28 and 39 kDa (Fig. S2 in Supplementary Data). The predicted length for ABI5 protein was 342 aa (as deduced from sorghum ESTs (expressed sequence tags; Fig. S5 in Supplementary Data) with a molecular weight of 38 kDa, but the detected protein was closer to the 28-kDa mark indicating that the protein migrates with a smaller apparent size or that it may be smaller than predicted. In some cases (mostly in 30-DAP caryopses) a secondary band above 43 kDa was observed, with a lower but constant level not related to ABI5 pattern (Fig. S2A in Supplementary data). A similar non-specific signal was reported in arabidopsis by López-Molina et al. (2001).
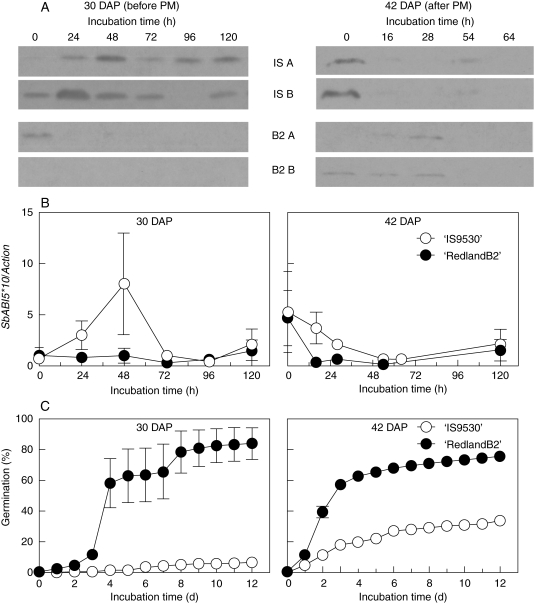
ABI5 protein levels are induced in dormant developing (30-DAP) grains, but not in maturing (42-DAP) grains
Embryonic ABI5 protein levels in 30-DAP (deeply dormant) ‘IS9530’ caryopses were induced after 24- and 48-h incubation periods, while in less-dormant ‘RedlandB2’, ABI5 decreased rapidly and was not detected after 24 h of incubation. Despite high variability for SbABI5 mRNA levels in ‘IS9530’ (Fig. 4B, left) between 24 and 48 h, ABI5 protein levels appear to be regulated at the transcriptional level until 72 h. Contrasting ABI5 levels between both genotypes observed after 72 h can be ascribed to different regulation at the protein level (Fig. 4A, B, left).
Fig. 4.
(A) Western blot showing ABI5 protein levels during incubation in water of grains of 30 DAP (before physiological maturity, PM) and 42 DAP (after PM) from ‘IS9530’ (dormant, PHS resistant) and ‘RedlandB2’ (less dormant, PHS susceptible) panicles. Caryopses were incubated at 25 °C and embryos were excised after different incubation times, and used for RNA and protein extraction. Biological replicates (‘A’ and ‘B’) for both genotypes are shown and belong to different field plots. (B) SbABI5 transcript levels relative to SbActin. (C) Germination response for ‘IS9530’ and ‘Redland B2’ grains used in the experiments above. Horizontal bars indicate the sampling period for RNA and proteins. Data in (A), (B) and (C) were all obtained from the same samples. Each data point in (B) and (C) is the mean of two biological repetitions (two field plots).
At a later stage of seed development (42 DAP, after PM), a rapid decrease and overall lower ABI5 protein and transcript abundance was observed (Fig. 4A, B, right). Similar to previous results with mature grains (47 DAP; Fig. 3), SbABI5 mRNA and protein levels do not explain the contrasting germination response between ‘IS9530’ and ‘RedlandB2’.
As ABI5 protein stability and gene expression have been shown to be strongly up-regulated by ABA in other species, this possibility was tested in sorghum and also the effect of germination-promoting GAs.
Exogenous ABA and GA had opposite effects on germination of dormant grains but did not significantly affect ABI5 levels
Freshly harvested 37-DAP caryopses (moment of PM) of ‘IS9530’ were incubated in water, 50 µm ABA or 150 µm GA3+4 solutions. Germination was effectively promoted by GA3+4, and further inhibited by ABA (Fig. 5A). ABI5 protein levels (Fig. 5C) decreased slightly after 24 h both during incubation in water and ABA, peaked transiently at 72 h and 96 h (in water and ABA, respectively) and decreased afterwards to the original level. In contrast to what was expected, GA treatment did not affect negatively ABI5 protein levels, and a similar tendency to increase towards 96 h of incubation was observed (Fig. 5C). Incubation of grains in ABA or GA had no significant effect on SbABI5 transcript abundance (Fig. 5B) (nor did it affect SbABI4 and SbPKABA1.1; Fig. S1B in Supplementary Data).
Fig. 5.
Germination response (A), SbABI5 mRNA levels relative to SbActin (B) and ABI5 protein levels (C) in embryos dissected from dormant ‘IS9530’ caryopses (37 DAP) after different incubation periods at 25 °C in water, 50 µm ABA or 150 µm GA3 + 4 solutions. Each data point is the mean value for two biological replicates (seeds from two different field plots). Western blots were repeated up to five times and the obtained signals were quantified and expressed as relative to time zero of incubation. Bars represent s.e. for the two biological replicates.
ABI5 protein levels decreased rapidly and were not induced by ABA during incubation of isolated embryos
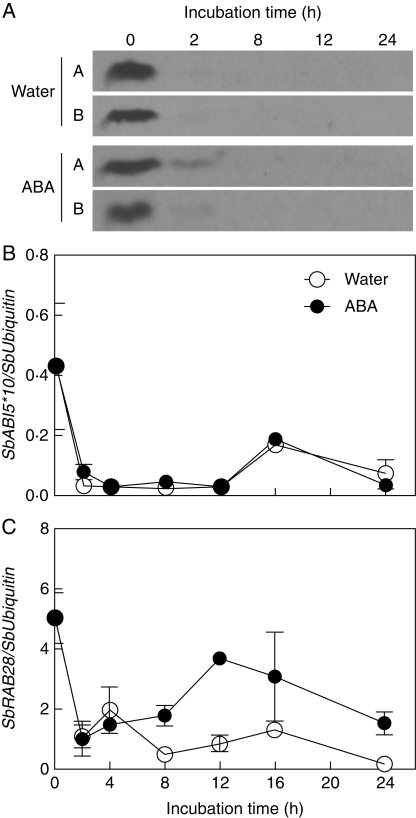
Regulation of ABI5 transcript and protein levels by ABA was also evaluated in isolated embryos. Excised embryos from ‘IS9530’ 30-DAP caryopses were incubated at 25 °C in water or in 50 µm ABA. ABA inhibited germination of isolated embryos which, otherwise, germinated rapidly in water (100 % germination between 24 and 36 h; Fig. S4 in Supplementary Data), thus providing extreme responses and also the possibility of evaluating the effect of the seed coats, if any, on the regulation of ABI5 levels by ABA.
ABI5 protein was detected in dry isolated embryos, but was undetectable after a 2-h incubation period in water (Fig. 6A). Incubation in 50-μm ABA only delayed complete disappearance of ABI5 for 2 h, and was undetectable thereafter. These results show that regulation of ABI5 levels by ABA does not occur in the isolated embryo, and suggest that some factor related to the seed coat is required for ABI5 stability. SbABI5 transcripts also decreased and maintained a similar level in both water and ABA during the 24-h incubation period (Fig. 6B). Induction of the SbRAB28 gene by ABA was observed, suggesting that ABA-mediated induction of this gene does not rely on ABI5, and other components of ABA signalling pathway are still active. Most important, these results show that ABA inhibition of germination of the isolated embryo does not rely entirely on ABI5 levels, and suggest that ABI5 is not essential for inhibition of germination but more likely acts as a modulating factor of ABA signalling during the expression of dormancy in the imbibed seed.
Fig. 6.
ABI5 protein blot (A) and SbABI5 and SbRAB28 transcript levels relative to SbUbiquitin (B), during incubation of isolated embryos that were dissected from 30-DAP ‘IS9530’ caryopses, and incubated in either distilled water or a 50 µm ABA solution at 25 °C. Replicates from a single seed lot are shown (seeds from ten plants pooled together). Vertical bars indicate the s.e. for two replicates (n = 2). Germination was completed between 24 and 36 h during incubation in water, and was inhibited by ABA (0 % after 12 d).
DISCUSSION
Transcription of ABA signalling genes and dormancy
Naturally occurring regulatory sites for ABA signalling in the context of seed dormancy were investigated in grain sorghum (S. bicolor). The possibility of a differential expression for several sorghum genes encoding putative ABA signalling components was addressed in two sorghum genotypes with contrasting PHS behaviour (‘IS9530’, resistant; ‘RedlandB2’, susceptible to PHS). Sorghum genes encoding homologues for ABI5 (and other bZIP homologues), ABI4, ABI1/2 and PKABA1 were identified after in silico search. Transcript abundance of these genes in the embryo was analysed by RT-QPCR during grain imbibition for both lines and at two developmental stages (before and after PM).
The present results support that maintenance of deep dormancy in ‘IS9530’ immature (before PM) caryopses is related to a transient and co-ordinated up-regulation of several genes involved in ABA signalling (SbABI3, SbABI4, SbABI5 and SbPKABA1) together with ABI5 protein levels (Figs 2 and 4), while low dormancy in immature ‘RedlandB2’ caryopses was related to the absence of an up-regulation for these genes. After PM a second phase of dormancy release took place in both genotypes (Fig. 1), and a lower dormancy level after 40 DAP was accompanied by a lower expression of the genes studied and ABI5 protein abundance (Figs 3 and 4). These results altogether support that up-regulation of these genes and ABI5 protein are involved in maintaining deep dormancy in ‘IS9530’ immature grains, and this mechanism is attenuated precociously (or is not functional at all) in immature ‘RedlandB2’ and later on in mature ‘IS9530’ caryopses as a result of partial loss of dormancy. In addition to changes in sensitivity to ABA during dormancy loss in developing sorghum grains, initial ABA content may have contributed to dormancy release which occurred between 30 and 47 DAP, as ABA content decreased to about one-third between these two developmental stages and in both lines (Figs 2B and 3B).
Although protein levels and activity of VP1, ABI4 and PKABA1 were not evaluated in this study, assuming that they roughly reflect transcriptional activity, it can be concluded that in mature (>40 DAP) but still partially dormant caryopses from both lines, differential expression of dormancy (associated with embryo sensitivity to ABA) depends on other components of the ABA signalling pathway different from those studied here. The possibility of different alleles in both lines was discarded after sequencing the genes studied (data not shown).
PP2C-type phosphatases like ABI1 work as negative regulators of ABA signalling (Merlot et al., 2001; González-García et al., 2003). We hypothesized that lower sensitivity to ABA in ‘RedlandB2’ as compared with ‘IS9530’ could be due to a higher expression of a sorghum gene encoding an ABI1 homologue. Contrary to this hypothesis, SbABI1 expression was higher in dormant ‘IS9530’ grains and its pattern resembled that of the SbRAB28 gene (Fig. 2C). Again, expression of SbABI1 mimicked the SbRAB28 in more mature, 47-DAP grains (Fig. 3C). Therefore, the different expression of SbABI1 in ‘IS9530’ and ‘RedlandB2’ immature grains seems to be the consequence – and not the cause – of the different ABA signalling activities operating in these genotypes.
Regulation of sorghum ABI5 by ABA, GA and the seed coat
Up-regulation of ABI5 mRNA and protein abundance in imbibed seeds in response to addition of ABA is well known in arabidopsis (López-Molina et al., 2001). More recently, down-regulation of ABI5 levels in response to GA was reported by Piskurewicz et al. (2008). Considering the ABA/GA balance as governing ABI5 levels and the germination response in arabidopsis, we hypothesized that sorghum ABI5 levels could be modulated by exogenous ABA and GA (Fig. 5). Both treatments had opposite effects on germination but, contrary to what was expected, evolution of ABI5 protein and transcript levels was not affected significantly (Fig. 5). Piskurewicz et al. (2008) demonstrated that high levels of ABI5 do not imply strong activity, and phosphorylation is necessary for ABI5 activation and inhibition of seed germination. In the present experiments with sorghum, migration of ABI5 protein in SDS–PAGE was not visibly affected by addition of ABA or GA to the incubation medium (Fig. S2D in Supplementary data available online). Nevertheless, differential phosphorylation of sorghum ABI5 protein cannot be discounted.
ABI5 transcript and protein levels were not significantly up-regulated by exogenous ABA during incubation of isolated embryos (Fig. 6), despite germination being completely inhibited. This result supports the fact that ABA-induced inhibition of germination does not rely on ABI5. In addition, the fast disappearance of ABI5 protein in isolated embryos as compared with whole grains suggests that ABI5 stability is strongly regulated by some factor provided or controlled by the seed coat which cannot be replaced by exogenous ABA. It is well known in arabidopsis that ABI5 is localized in the nucleus and its abundance can be strongly down-regulated by degradation in the proteasome (López-Molina et al., 2003). In the presence of ABA, ABI5 is phosphorylated and avoids degradation. Rapid disappearance of sorghum ABI5 observed in isolated embryos could be due to changes in its phosphorylation status that result in an enhanced targeting of ABI5 to the proteasome.
Role of sorghum ABI5 in the expression of dormancy, if any, is restricted to immature caryopses
Different from sorghum, regulation of barley ABI5 by ABA was observed in mature, non-dormant barley caryopses (Casaretto et al., 2005). Similar to its arabidopsis orthologue, HvABI5 appears to be related to post-germination tolerance to environmental stress. A more specific role for an ABI5 orthologue in seed dormancy has been reported by Johnson et al. (2002) in wheat: TaABF transcript abundance decreased during imbibition of non-dormant (after-ripened) seeds, but increased transiently in mature dormant grains. Johnson et al. (2002) suggested that TaABF may play a critical role in maintaining dormancy in mature wheat grains. In contrast to this observation in wheat, the present results support a possible role for sorghum ABI5 in ABA-induced inhibition of germination only in immature dormant grains, but not after PM.
Transcription factors included in this work (VP1, ABI4 and ABI5) are known to interact physically in transcriptional complexes (Himmelbach et al., 2003) and extensive cross-regulation among them exists in arabidopsis (Söderman et al., 2000) and is probably conserved in other species. Thus, it is not surprising that in sorghum these genes exhibit similar expression patterns and behave as a ‘signalling package’ under the control of a more hierarchical, still unidentified component regulating dormancy. Ongoing QTL (quantitative trait locus) analysis with RILs (recombinant isogenic lines) from a segregating population derived from sorghum lines ‘IS9530’ and ‘RedlandB2’ might provide new information on the mechanisms controlling dormancy in this species. In parallel, confirmation of a role in the expression of dormancy for the differentially expressed genes found in this work should arise after manipulating their expression with techniques such as RNAi, and observing the dormancy phenotype. Results presented so far provide us with firm candidate genes for future manipulation in sorghum. Most studies on seed dormancy have focused on mature, after-ripened seeds from species in which ABA metabolism is the key process involved in the maintenance of dormancy (reviewed by Finkelstein et al., 2008). In this context, the present work provides new evidence on the mechanisms involved in the expression of dormancy in a species in which this process is related to changes in ABA sensitivity instead of ABA metabolism, and also supports the existence of different mechanisms operating at different developmental stages.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We want to thank Ms Mirta N.Tinaro, Silvina Enciso (technicians), Renata Cantoro (doctoral student), Martín León and Damián Samarro (undergraduate students) for exceptional help and technical assistance with field and laboratory work, and Dr Rodolfo Sánchez for his comments on the manuscript. This work was funded by the Interamerican Bank for Development (BID) grant PICT 1728 OC/AR # 01565, and a Research Award for Projects for Regional Development from Banco Santander – Río S.A. group.
LITERATURE CITED
- Ali-Rachedi S, Bouinot D, Wagner MH, et al. Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the CVi ecotype, the dormant model of Arabidopsis thaliana. Planta. 2004;219:479–488. doi: 10.1007/s00425-004-1251-4. [DOI] [PubMed] [Google Scholar]
- Benech-Arnold RL, Enciso S, Sánchez RA, Rodríguez MV. On the hormonal nature of the stimulatory effect of high incubation temperatures on germination of dormant sorghum caryopses. New Phytologist. 2003;160:371–377. doi: 10.1046/j.1469-8137.2003.00900.x. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Aoyama N, Chung YY, Still DW, Jones RL. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiology. 2007;143:1173–1188. doi: 10.1104/pp.106.093435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell A, Cutanda MC, Lumbreras V, et al. Arabidopsis thaliana AtRAB28: a nuclear targeted protein related to germination and toxic cation tolerance. Plant Molecular Biology. 2002;50:249–259. doi: 10.1023/a:1016084615014. [DOI] [PubMed] [Google Scholar]
- Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- Carrari F, Pérez-Flores L, Lijavetzky D, et al. Cloning and expression of a sorghum gene with homology to maize vp1. Its potential involvement in pre-harvest sprouting resistance. Plant Molecular Biology. 2001;45:631–640. doi: 10.1023/a:1010648420669. [DOI] [PubMed] [Google Scholar]
- Casaretto JA, Tuan-hua DH. Transcriptional regulation by abscisic acid in barley (Hordeum vulgare L.) seeds involves autoregulation of the transcription factor HvABI5. Plant Molecular Biology. 2005;57:21–34. doi: 10.1007/s11103-004-6520-x. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Tansley Review. Seed dormancy and the control of germination. New Phytologist. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. The Plant Cell. Supplement 2002. 2002. Abscisic acid signalling in seeds and seedlings; pp. S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annual Review of Plant Biology. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. The Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Rodrıguez D, Nicolas C, Rodríguez PL, Nicolas G, Lorenzo O. Negative regulation of abscisic acid signalling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiology. 2003;133:135–144. doi: 10.1104/pp.103.025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualano N, Carrari F, Rodríguez MV, et al. Reduced embryo sensitivity to ABA in sprouting susceptible sorghum (Sorghum bicolor) variety is associated with an altered ABA signalling. Seed Science Research. 2007;17:81–90. [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Höhener B, Grill E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO Journal. 2002;21:3029–3038. doi: 10.1093/emboj/cdf316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Current Opinion in Plant Biology. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiology. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Molina L, Mongrand S, Kinoshita N, Chua N-H. AFP is a novel negative regulator of ABA signalling that promotes ABI5 protein degradation. Genes and Development. 2003;17:410–418. doi: 10.1101/gad.1055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKibbin RS, Wilkinson MD, Bailey PC, Flintham JE, Andrew LM. Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proceedings of the National Academy of Sciences of the USA. 2002;99:10203–10208. doi: 10.1073/pnas.152318599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, le Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. The Plant Journal. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, et al. Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. The Plant Journal. 2006;45:942–954. doi: 10.1111/j.1365-313X.2006.02659.x. [DOI] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E. Nuclear localization of the mutant protein phosphatases abi1 is required for insensitivity towards ABA responses in Arabidopsis. The Plant Journal. 2008;54:806–819. doi: 10.1111/j.1365-313X.2008.03454.x. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR. Physical interactions between ABA response loci of Arabidopsis. The Plant Journal. 2001;26:627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. ABA action and interaction in seeds. Trends in Plant Science. 2003;8:213–217. doi: 10.1016/S1360-1385(03)00060-8. [DOI] [PubMed] [Google Scholar]
- Niu X, Helentjaris T, Bate J. Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. The Plant Cell. 2002;14:2565–2575. doi: 10.1105/tpc.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. The Plant Cell. 2006;18:1887–1899. doi: 10.1105/tpc.106.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. The gibberellic acid signalling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 Activity. The Plant Cell. 2008;20:2729–2745. doi: 10.1105/tpc.108.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie SA, Whithford PN, Appleford NEJ, et al. A monoclonal antibody to (S)-abscisic acid: its characterization and use in a radioimmunoassay for measuring abscisic acid in crude extracts of cereal and lupin leaves. Planta. 1988;163:330–339. doi: 10.1007/BF00401020. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. 2nd edition. [Google Scholar]
- Söderman E, Brocard I, Lynch T, Finkelstein R. Regulation and function of the Arabidopsis ABA-insensitive4 (ABI4) gene in seed and ABA response signaling networks. Plant Physiology. 2000;124:1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach HS, Benech-Arnold RL, Kristof G, Sánchez RA, Marcucci-Poltri S. Physiological basis of pre-harvest sprouting resistance in Sorghum bicolor (L.) Moench. ABA levels and sensitivity in developing embryos of sprouting-resistant and sprouting-susceptible varieties. Journal of Experimental Botany. 1995;46:701–709. [Google Scholar]
- Steinbach HS, Benech-Arnold RL, Sánchez RA. Hormonal regulation of dormancy in developing sorghum seeds. Plant Physiology. 1997;113:149–154. doi: 10.1104/pp.113.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Programmed cell death during endosperm development. Plant Molecular Biology. 2000;a 44:283–301. doi: 10.1023/a:1026588408152. [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Regulation of programmed cell death in maize endosperm by abscisic acid. Plant Molecular Biology. 2000;b 42:397–414. doi: 10.1023/a:1006333103342. [DOI] [PubMed] [Google Scholar]