Abstract
Objective
To characterize the serum metabolome of a primate model of in utero high fat exposure.
Study design
Serum from maternal and fetal (e130) macaques exposed to either a high fat or control diet were analyzed by gas chromatography-mass spectrometry (GC-MS). Multivariate data analysis (MVDA) was performed to reduce the generated data set. Candidate metabolites were further analyzed for significance using ANOVA and comparative t-tests.
Results
Approximately 1300 chromatographic features were detected. Through MVDA this number was reduced to 60 possible metabolites. Using comparative t-tests 22 metabolites had statistical significance (p<0.05) over the whole of the study. By virtue of maternal HF diet alone, fetal phenotypic differences are accompanied by altered metabolite concentrations of 7 metabolites (p<0.05).
Conclusions
In utero high fat diet exposure is associated with an altered fetal epigenome and parlays a characteristic modification in the fetal metabolite profile.
Keywords: Obesity, epigenetics, pregnancy, nutrition, fetal programming, metabolomics
INTRODUCTION
The current global health epidemic of obesity is considered a major contributor to the increasing prevalence of Type II diabetes, as well as atherosclerotic, cardiovascular, and hypertensive morbidity and mortality.1–6 In association with this rise in adult obesity throughout the developed and developing world, there is occurring a disproportionate earlier onset of obesity among children (infants to adolescents).7–9 Given such anticipation (e.g., tendency of disease to appear at an earlier age of onset and with increasing severity in successive generations), the increased prevalence of childhood obesity cannot be attributed to either environment or genetics alone.
According to Barker’s fetal origins of adult disease hypothesis, perturbations in the gestational milieu influence the development of diseases later in life.10,11 This supposition gains support from epidemiologic studies10–12 as well as animal models of nutritional constraint13–15 and uteroplacental insufficiency-induced intrauterine growth restriction (IUGR).16–18 These outcomes have been suggested to occur through the static reprogramming of gene expression via alterations in chromatin structure (epigenetic regulation). We and others have previously demonstrated that uteroplacental insufficiency and IUGR in the rat results in covalent modifications of chromatin structure16–18 or changes in DNA methylation patterns13–15 which leads to persistent changes in postnatal gene expression. In our initial studies establishing our non-human primate model of maternal high fat diet exposure in utero,19 we demonstrated that a maternal high fat/caloric dense diet altered fetal hepatic chromatin structure and lead to alterations in fetal and postnatal gene expression.20 These observations supported our hypothesis that a high fat maternal diet leading to obesity modifies the gestational milieu and profoundly influences the postnatal and fetal phenotype in association with fetal histone [H3] covalent modifications. These results lead us to question whether these fetal epigenetic modifications translate into meaningful alterations in the fetal and neonatal metabolome.
The metabolome is broadly defined as the complete set of the small molecules that contribute to metabolism.21,22 Metabolomics has emerged over the past near decade as the third major path of functional genomics, alongside its related “omics” of mRNA profiling (transcriptomics) and proteomics. It is a comparative analysis describing metabolic responses of living systems to pathophysiologic stimuli, genetic, or epigenetic modification.22 Comprehensive analyses of the metabolome in a biologic system occurs by the combined use of analytical methods and multivariate statistical analyses: metabolic fingerprints are acquired with an analytical platform (i.e., gas or liquid chromatography with mass spectroscopy), processed with multivariate analyses to allow for data visualization and discrimination (i.e., principal component analysis (PCA) and partial least squares discriminate analysis (PLS-DA)). The mass spectrum is used to determine the identity of discriminate metabolites by automated comparison to known metabolites in a databases.22 The synergistic integration of the metabolome with altered gene expression profiling allows for a comprehensive overview of living systems biology.
As metabolomics is unique from other "omics" in its capacity to efficiently decipher systems biology as a reflection of its close proximity to phenotype, we sought to employ its use in order to establish the maternal and fetal metabolic profile under conditions of a high fat maternal diet. With respect to the maternal phenotypic profile, we have previously described19,20 that over time (e.g., 3 to 5 years) two distinct maternal phenotypes become apparent among our dams. These dams can be considered as sensitive (HFD-S) or resistant (HFD-R) to our high fat diet. HFD-S dams display a unique metabolic phenotype in both the non-pregnant and pregnant state.19 By the 4th year on the high fat diet, the HFD-S group demonstrates a >35% increase in body weight, >3-fold increase in insulin AUC (area under the curve) in response to a glucose tolerance test, >5-fold increase in leptin levels (leptin/body weight ratio) and a near doubling in body fat.19 However, fasting glucose levels do not significantly differ in either the HFD-S or HFD-R groups despite a significant alterations (control, 2.53 +/− 0.50; HFD-R, 2.81 +/−0.70; HFD-S, 7.87 +/−1.50; P < 0.05 for HFD-S versus control) in the homeostasis model assessment of insulin resistance (or HOMAIR, which serves as a general measure of insulin resistance in reference to basal glucose and insulin). By comparison, HFD-R animals retained similar body weight and metabolic phenotype as the controls throughout the entire 4 years.
With respect to the fetus, we have similarly characterized the effect of in utero exposure to a high fat maternal diet.19,20 We have observed that while the maternal phenotype is relatively mild, regardless of maternal HFD-R or HFD-S status the fetus develops non-alcoholic fatty liver and displays an altered fetal hepatic epigenomic profile. Since the offspring are delivered in the early latter trimester of gestation (term being 167 days), appreciable differences in overall fetal weight are avoided.19,20 Of interest, we have shown that reversing the maternal diet in year 5 from high fat to control (“high fat diet reversal”) bears appreciable differences on the fetal phenotype with partial reversion trending towards to control values.19 However, it is unknown whether these phenotypic and epigonomic characteristics translate into observable alterations in the maternal and fetal metabolome. Given that we have observed significant alterations in the maternal and fetal phenotype and fetal epigenome and transcriptome following in utero exposure to a high fat maternal diet, we aimed to characterize the full spectrum of metabolite differences in our model system.
Leveraging these collective observations, we set out to first study the serum metabolome of HFD-R and HFD-S dams and to test if these observed phenotypes were transferred in utero to the developing fetus. We then characterized the fetal metabolome from dams fed control, high fat diet, or high fat diet reversal. We hypothesized that this approach would not only decipher the fetal and maternal serum metabolome under maternal high fat diet conditions leading to maternal obesity, but would potentially identify candidate biomarker(s) associated with the development of obesity.
MATERIALS AND METHODS
Non-human primate model of maternal high fat diet exposure
Our model is described in detail elsewhere.19,20 Briefly, all animal procedures were in accordance with the guidelines of Institutional Animal Care and Use Committee of the Oregon National Primate Research Center (ONPRC). Age and weight matched young (5–6 years of age at the start of the studies) adult female Japanese macaques were socially housed in indoor/outdoor enclosures in groups of 5–9 with 1–2 males per group. Animals were separated into two groups: The control group was fed standard monkey chow that provides 14% of calories from fat (Monkey Diet #5052, Lab Diet, Richmond, IN, USA) and the high fat diet group was fed a diet that supplied 32% of calories from fat (Custom Diet 5A1F, Test Diet) and included calorically dense treats.19,20 Both diets are sufficient in vitamin, mineral and protein content for normal growth. Prior to this study all animals were maintained on standard monkey chow in large outdoor enclosures and were naive to any experimental protocols. During the fifth year, a cohort of HF diet fed animals were switched back to the standard monkey chow specifically during the breeding/pregnancy season (“diet reversal”).
Dams were allowed to breed naturally. Both groups consisted of primiparous and multiparous gravidae and all pregnancies were singleton. Pregnancies were terminated by cesarean delivery, performed by ONPRC veterinarians, at gestational day 130 by sonographic fetal biometry. For the experiments involving postnatal (e.g., “juvenile”) offspring, the pregnancies were allowed to progress to term and vaginal birth occurred and offspring were subsequently maintained on the maternal diet after weaning.
Immediately after cesarean delivery or at the time of sacrifice, the fetuses/juvenile animals where delivered to necropsy where they were weighed and measured. The fetuses were deeply anesthetized with sodium pentobarbital (>30 mg/kg, iv) and then exsanguinated for attainment of serum and necropsy tissue. For metabolomic profiling, we studied serum samples from 47 dams and 40 fetuses collected in heparinized capillary tubes from the fetal aorta and centrifuged at 2,400 r.p.m. All samples were flash frozen and stored at −80°C in aliquots such that freeze-thawing was minimized.
Metabolite profiling
Protein removal from serum samples was performed using the method of Jiye et. al23 with the addition of 20 uniformly labeled stable isotope amino acids of known concentration as an internal standard.
To ensure metabolite volatility each sample underwent a two step derivatization process. The dried extracts were suspended in 50 µL of pyridine containing 20 mg/ml O-methylhydroxylamine hydrochloride, sonicated for five minutes then incubated at 60°C for 30 minutes. This was followed by the addition of 50 µl MSTFA +1% TMCS (Pierce, Rockford IL) and incubated at 37°C for thirty minutes. A retention index standard mixture consisting of fatty acid methyl esters was added and each sample immediately analyzed.
All samples were analyzed in a random order by gas chromatography-mass spectrometry. Using an autosampler 1 µL of each sample was injected into an Agilent 6890 gas chromatograph (Agilent, Santa Clara CA). The injector was set to a 2:1 split ratio and the temperature was held at 220°C. A 30 m Rtx-5MS with Integra guard column (Restek, Bellefonte, PA) was employed for separation. Helium was used as the carrier gas at a 1 mL/min flow rate. The transfer line into the mass spectrometer was held at 250°C. The temperature program was as follows, an initial 2 minute hold at 70°C, followed by an 8°C/minute ramp to 250°C then a second 20°C/minute ramp to 330°C which was held for 2 minutes. A MicroMass GCT Premier (Waters, Beverly, MA) was used for mass spectrometric analysis. Normal electron impact ionization conditions were used. Data was recorded using MassLynx (Waters, Beverly MA).
Model development and metabolite identification
Chromatographic peak detection and area analysis for each chromatogram was performed using MarkerLynx (Waters). To find possible variation in each sample group the data was transferred to SIMCA-P ver.12.0 (Umetrics, Kinnelon, NJ) where principle component analysis (PCA) and partial least squares-discriminate analysis (PLS-DA) was performed. This produced a possible list of metabolites that had changed due to diet or sensitivity to high fat diet. Each metabolite from this list was investigated further by extracting the area under the chromatographic curve and analyzing it using ANOVA and comparative t-tests. Metabolites were identified either using an in-house data base or the NIST MS Search 2.0. Unknown (un) metabolites were identified using their retention index (ri) and characteristic mass to charge ratio (m/z) in the following format un(ri_m/z).
Statistical analysis
Partial least squares-discriminate analysis (PLS-DA) was performed using SIMCA-P (Umetrics). This multivariate statistical analysis technique is similar to principle component analysis with the addition of a second data matrix identifying specific classes, thus it is a supervised method when compared to PCA which is unsupervised. ANOVA and comparative t-tests were performed for each metabolite using the JMP 7.0 statistics package (SAS). Excel (Microsoft, Redmond, WA) was used for spread sheet development and simple statistics.
RESULTS
Analytical analysis and chromatographic data deconvolution produced over 1300 chromatographic features. Through multivariate statistical analysis this number was reduced to 60 possible metabolites that contributed to PLS-DA model development. Integration of the chromatographic area of these individual metabolite peaks followed by statistical analysis produced 22 metabolites that had statistical significance (p<0.05) over the whole of the study.
Maternal Metabolic Profiles
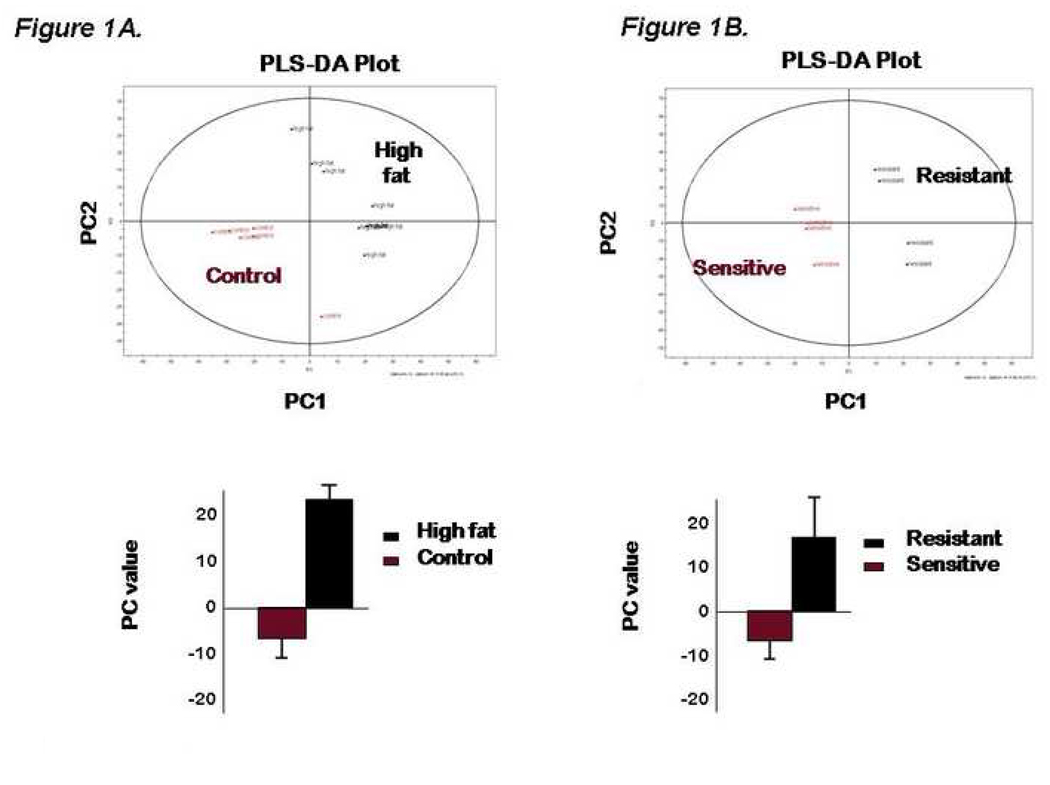
Our first study aim was to characterize the metabolomic footprint from dams fed a 13% fat by calories (control) diet and a 35% fat by calories (high fat; HF) diet. Chromatographic data from six control and eight HF-diet animals was processed to give the initial principal components analysis (PCA) plot (not shown). To thereafter quantitate and identify metabolite changes a partial least squares-discriminate analysis (PLS-DA) was performed (Figure 1A). The loadings plot from this model was used to find chromatographic features which differ between the control and HF animals. We found ten metabolites that differed significantly (p <0.05) between control and HF animals (Table I).
Figure 1.
(A–B). PLS-DA scatter plots of serum from dams maintained for two to three years on either the high fat (HFD) or control (con) diet (Figure 1A, upper panel) with mean differences as quantitated by bar graphs (lower panel). In accordance with their phenotype as previously described,19 PLS-DA analysis of serum metabolites from high fat diet animals was stratified into either high fat resistant (HFD-R) or sensitive (HFD-S) (Figure 1B). The loadings plot from these graphs was used to effectively search for metabolites that contribute to differences between groups based on mass spectrometry. Those metabolites achieving significance are as summarized in Table I.
TABLE I. Metabolites from maternal serum that differ significantly between high fat and control diet fed animals. The high fat diet fed dams are further stratified by sensitivity to the diet (e.g., develop obesity) and resistance to the diet (e.g., do not develop obesity despite multiple years of feeding). Control n=6; HFD=8.
Analysis of maternal metabolites contributing to the PLS-DA model in Figure 1. Significantly discrete serum metabolites by mass spectroscopy are as listed and correspond to the mass spectra data as presented in Figure 1.
| HFD versus Control | HFD-S versus Control | HFD-R versus control | ||||
|---|---|---|---|---|---|---|
| Metabolite | P value | Fold change (high fat/control) |
P value | Fold change (HFD-S/control) |
P value | Fold change (HFD-R/control) |
| cholesterol | 0.0093 | 1.5 | 0.0038 | 1.6 | 0.043 | 1.4 |
| α-tocopherol | 0.023 | 1.7 | 0.00059 | 2.2 | 0.36 | 1.2 |
| γ-tocopherol | 0.039 | 2.2 | 0.047 | 2.8 | 0.45 | 1.6 |
| glycine | 0.037 | 1.3 | 0.187 | 1.4 | 0.16 | 1.3 |
| aminomalonic acid | 0.024 | 1.5 | 0.041 | 1.6 | 0.27 | 1.4 |
| citric acid | 0.015 | 1.3 | 0.0089 | 1.3 | 0.16 | 1.3 |
| un(2252_203) | 0.016 | 1.7 | 0.015 | 2.1 | 0.26 | 1.3 |
| un(2281_131) | 0.047 | 1.5 | 0.023 | 1.9 | 0.41 | 1.2 |
| un(2298_144) | 0.038 | 1.5 | 0.015 | 1.9 | 0.41 | 1.1 |
| un(2682_116) | 0.0028 | 2.2 | 0.017 | 2.1 | 0.051 | 2.3 |
| un(823_200) | 0.065 | 1.4 | 0.018 | 1.5 | 0.24 | 1.2 |
| un(1346_104) | 0.086 | 1.8 | 0.012 | 2.4 | 0.48 | 1.3 |
PLS-DA, partial least squares-discriminate analysis, un(ri_m/z), unknown(retention index_mass/charge ratio)
As anticipated by virtue of their progression to obesity on the maternal HF diet, cholesterol levels were elevated 1.5 fold higher in high fat fed dams relative to those on control diet, while no significant differences could be detected in the levels of palmitic, stearic, lauric, oleic and elaidic free fatty acids; higher lipid species are not detectable using our GC-MS methods. We similarly observed elevated level of two primary constituent of the vitamin E complex: α-tocopherol and γ-tocopherol in the HFD dams. This is consistent with obesity physiology in response to a high fat diet as these lipid soluble compounds act as antioxidants, protecting the cell membrane by reacting with lipid radicals24 or triggering intracellular signaling cascades.25 Glycine, aminomalonic acid and four unknowns, un(2252_203), un(2281_131), un(2298_144 and un(2682_116) were also elevated in the HFD dams, while citric acid was decreased.
Based on the previously described phenotypic differences, we sought to compare the maternal sensitive (i.e., obese and insulin resistant; n=4) and resistant (i.e., non-obese and insulin sensitive; n=4) metabolomic profiles (Figure 1B). Using a combination of PLS-DA and standard statistical analysis from the previously identified metabolites HFD-S animals contributed the bulk of change between HFD and control animals. Eleven metabolites were significantly altered as shown in Table I.. Nine of these metabolites are shared between the HFD versus control data set and two additional unknowns were also found. The fat soluble metabolites cholesterol, α-tocopherol and γ-tocopherol are increased in the HFD-S versus the control animals. Significant metabolite alterations as aminomalonic acid, citric acid and six unknowns; un(2252_203), un(2281_131), un(2298_144), un(2682_116), un(823_200), and un(1346_104). Only cholesterol (Table IC) was found to be significant in the HFD-R animals versus the control.
Fetal Metabolic Profiles
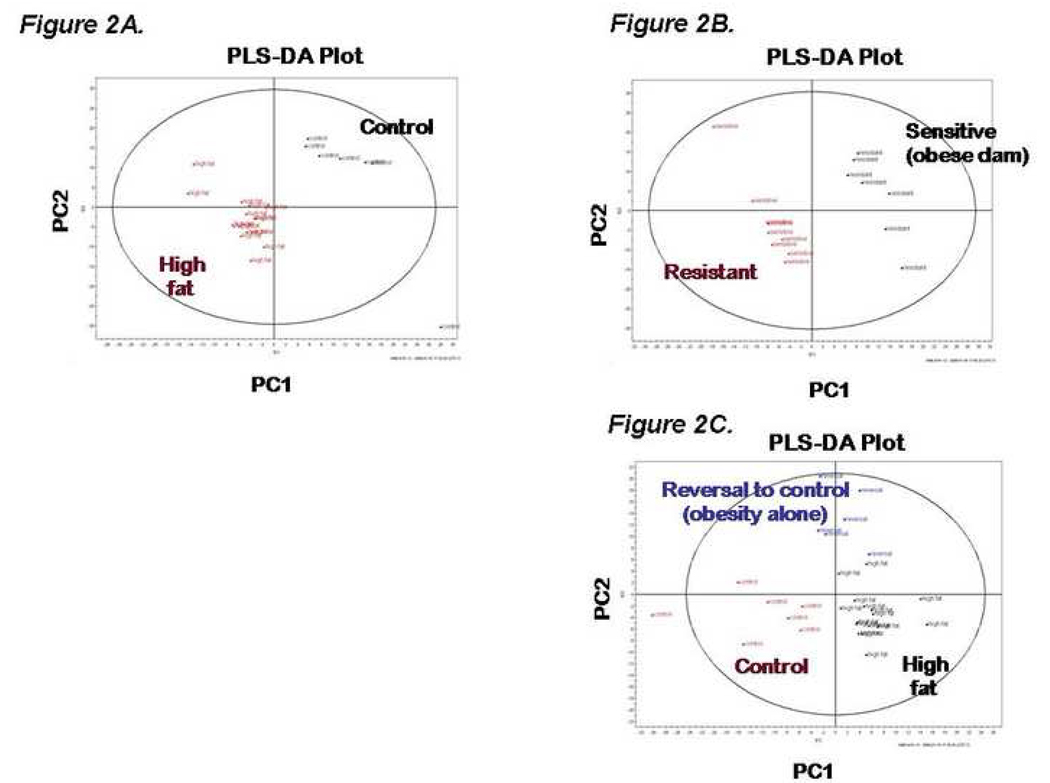
Our second objective was to characterize the fetal serum metabolome in order to determine its association to both maternal diet and phenotype. A PLS-DA scatter plot was generated comparing fetal serum from high fat diet fed dams (n=16) versus those on control diet (n 6; Figure 2A). Eight metabolites were found to have changed in control versus HFD exposed offspring (Table II), with cholesterol and α-tocopherol being found in both maternal and fetal serum but with inverse concentration profiles (i.e., lower fetal cholesterol under maternal high fat diet conditions). Of note, our findings have been similarly observed with attained maternal and fetal serum cholesterol values as measured in conventional clinical assays in a limited cohort (n=4 control versus n=5 HF diet; maternal mean serum cholesterol 81±5 vs 118±17 mg/dL, p=0.002; fetal 73±10 vs 63±7, p=0.143; Dr. Dan Marks, unpublished observations). Also interesting was the 8 fold decrease of ascorbic acid in HFD fetal serum when compared to control (p=0.0069). It has been proposed that ascorbic acid acts as part of the antioxidant network in which vitamin E participates.24
Figure 2.
(A–C). PLS-DA scatter plot of fetal serum from dams fed a high fat diet versus fetal serum from dams fed a control diet (Figure2A). PLS-DA scatter plot of fetal serum from dams fed a high fat and who were either HFD-S or HFD-R (see text, Figure 2B). PLS-DA scatter plot of fetal serum whose dam where fed a high fat, diet reversal or control diet (Figure 2C). The loadings plot from these graphs was used to identify metabolites that contribute to differences between groups based on mass spectrometry. Those metabolites achieving significance are as summarized in Table II and Table III.
TABLE II. Analysis of fetal metabolites contributing to the PLS-DA model in Figure 2. Control n=7; HFD n=15; Reversal n=6.
Analysis of fetal metabolites contributing to the PLS-DA model in Figure 2 stratified by in utero diet exposure and maternal phenotype. Significantly discrete serum metabolites by mass spectroscopy are as listed and correspond to the mass spectra data as presented in Figure 2.
| Maternal HFD vs Control | Reversal vs Control | Maternal HFD vs Reversal | |||||
|---|---|---|---|---|---|---|---|
| Metabolite | P value | Fold change (HFD/Con) |
P value | Fold change (DR/Con) |
P value | Fold change (HFD/DR) |
|
| 2-hydroxybutyrate | 0.003 | 0.59 | 0.00098 | 0.48 | 0.31 | 1.2 | |
| ascorbic acid | 0.0069 | 0.11 | 0.0078 | 0.38 | 0.66 | 0.29 | |
| α-tocopherol | 0.0031 | 0.53 | 0.029 | 0.71 | 0.15 | 0.75 | |
| cholesterol | 0.015 | 0.77 | 0.093 | 0.83 | 0.51 | 0.93 | |
| 3-hydroxybutyrate | 0.0055 | 2.3 | 0.0036 | 2.8 | 0.33 | 0.82 | |
| un(1462_100) | 0.014 | 0.41 | 0.0053 | 0.63 | 0.38 | 1.3 | |
| un(1574_304) | 0.071 | 0.71 | 0.012 | 0.59 | 0.16 | 1.2 | |
| un(1952_334) | 0.029 | 0.77 | 0.011 | 0.67 | 0.39 | 1.1 | |
PLS-DA, partial least squares-discriminate analysis, un(ri_m/z), unknown(retention index_mass/charge ratio); con, control; HF, high fat; DR, diet reversal
As an initial step in our effort to differentiate the effects of maternal obesity versus high fat diet exposure on the fetal metabolome, we characterized fetal serum profiles among a subgroup of dams whose high fat diet was reverted to control (reversal) in year 5 (e.g., control diet in an obese maternal environment). We used a PLS-DA scatter plot (Figure 2B) and the underlying loading profile to separate both maternal diet and obesity (i.e., horizontal plot groupings among HFD and reversal offspring distinct from control offspring). Metabolites from the loadings plot that contributed to this model were further analyzed and are detailed in Table II.. Fetal profiles could not be significantly distinguished by virtue of maternal diet (e.g., HFD relative to reversal) but could by virtue of maternal obesity (e.g., diet reversal relative to control) for all metabolites except cholesterol.
As a second step in differentiating the effects of maternal obesity versus high fat diet exposure on the fetal metabolome, we again stratified our analysis by virtue of maternal HFD sensitivity (Figure 2C). Six metabolites were found to be significantly altered in the HFD-S serum versus control, cholesterol, α-tocopherol, hypoxanthine, myo-inositol, un(698_103, and un(2298_144). When HFD-R is compared to control diet serum only α-tocopherol was found to be significantly altered. Three fetal metabolites were found to be significantly elevated when comparing HFD-S to HFD-R: hypoxanthine, myo-inositol and un(2298_144).
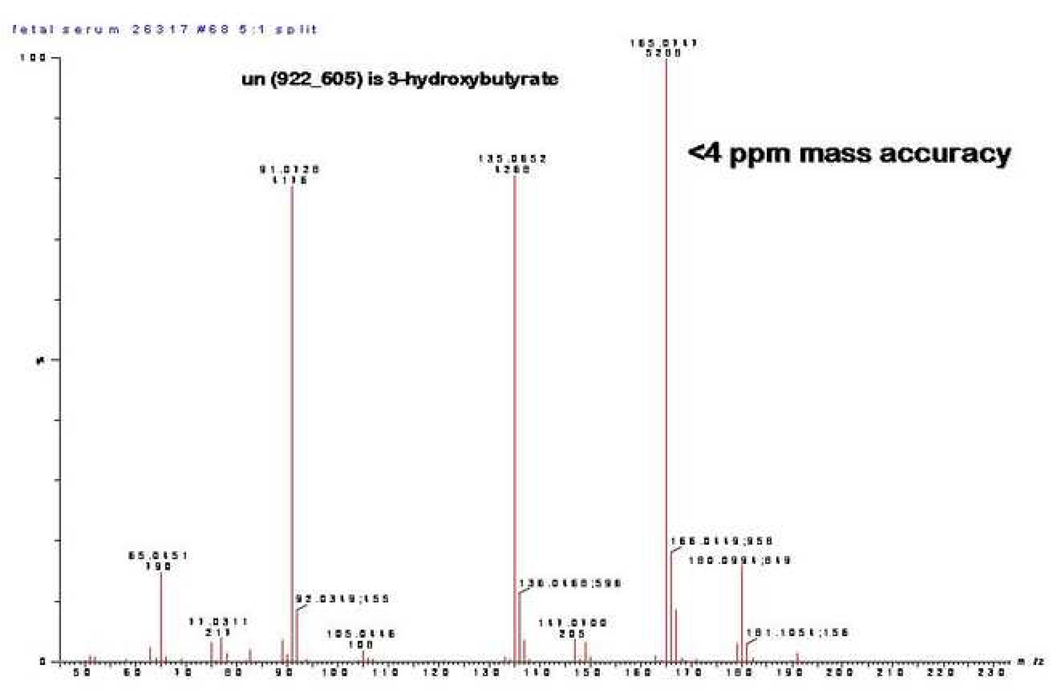
It bears mention that 3-hydroxybutyrate (initially identified as unknown 922_605 and later confirmed to be 3-hydroxybutyrate by mass spectrometry of purchased standard is unique among the significantly distinct fetal metabolites by virtue of its elevations in association with maternal obesity (Figure 3). Thus, fetal levels of 3-hydroxybutyrate can be distinguished with a maternal HFD and obesity (HFD versus control 2.3 fold, p=0.006) or obesity alone (reversal versus control 2.8 fold, p=0.004). In contrast, fetal levels are not distinguished in maternal HFD relative to reversal conditions (0.82 fold, p=0.33).
Figure 3.
Identification of “un(922_165)” as 3-hydroxybutyrate. We found unknown compound of 922_165 to be of particular interest. Subsequent analysis by mass spectrometry against known metabolic libraries identified it as 3-hydroxybutyrate (>97% certainty).
COMMENT
Maternal obesity is thought to increase a child’s risk of juvenile obesity and metabolic diseases; however the mechanism(s) whereby excess maternal nutrition impacts fetal development remains poorly understood. According to the developmental origins of adult disease hypothesis, perturbations in the gestational milieu influence the development of diseases later in life through the static reprogramming of genes. Phenotypic characterization in our primate model strongly implicate in utero exposure to excess maternal lipids during early and mid fetal development as an independent risk factor for the development of non-alcoholic fatty liver disease (NAFLD) and metabolic diseases during postnatal life as evidenced by alterations in the epigenome and transcriptome.19,20 Whether these epigenomic alterations would parlay into meaningful alterations in the fetal functional genome (e.g., the fetal metabolome) had not yet been addressed.
To begin to address this issue, we used a nonhuman primate model to determine the effect of chronic maternal consumption of a high fat/high calorie diet on the development of metabolic systems in the fetal offspring. We have previously demonstrated that only a portion of the adult female monkeys chronically consuming a high fat diet become obese and insulin resistant.19 However, irrespective of maternal obesity and insulin resistance, all fetal offspring of dams fed a high fat diet demonstrated clinically and histologically evident NAFLD, including hepatic inflammation, oxidative stress/damage, triglyceride accumulation and premature gluconeogenic gene activation.19 In this study we have extended our phenotypic and fetal epigenomic characterization with metabolomic analysis of maternal and fetal serum. Altered serum concentrations of cholesterol, members of the vitamin E tocopherol complex, glycine, aminomalonic acid, citric acid and three unknowns was found between HFD versus control maternal serum. Associated increases in maternal cholesterol are most likely attributable to the high fat diet itself as elevations were observed among both HFD-R and HFD-S dams (Figure 1, Table I). By contrast, α and γ-tocopherol were not associated with HFD diet fed dams with the resistant phenotype, suggestive of a reduced ability to metabolize fats in the HFD-S animals that results in the observed phenotype. Vitamin E lipid soluble compounds also act as antioxidants, protecting the cell membrane by reacting with lipid radicals24 or triggering intracellular signaling cascades.25 The inability of HFD-S animals to metabolize this compound could be suggestive of the inability of these animals to cope with oxidative stress.
In order to question the effects of maternal obesity and high fat diet exposure in utero, we characterized the effects of diet, maternal sensitivity to high fat diet, and diet reversal with persistent obesity on the fetal metabolic serum profile. When we extended our metabolic profiling of fetal serum to include offspring from animals reverted to a control diet after 4 years (DR cohort; Table II) we observed that associated fetal profiles could not be significantly distinguished by virtue of maternal diet (e.g, HFD relative to DR), but could be by virtue of maternal obesity (e.g., DR relative to control) for all metabolites except cholesterol (Table II).
The observed decrease in HFD animals of three metabolites that are involved in oxidative stress (2-hydroxybutyrate, ascorbic acid and α-tocopherol) with concomitant increase in the gluconeogenic metabolite 3-hydroxybutyrate deserves further comment. The HFD serum metabolomic phenotype of lowered antioxidant levels and the increased metabolite 3-hydroxybutyrate persists in the fetal metabolome even after diet reversal occurs. This is of particular interest, as other investigators have demonstrated in obese children that as insulin and insulin resistance increases, plasma levels of non-esterified fatty acids and 3-hydroxybutrate decrease.26 However, in biopsy-proven NAFLD adult subjects with confirmed peripheral insulin resistance, plasma 3-hydroxybutrate levels increase in concert with hypertriglyceridemia independent of BMI.27 Based on these observations and others, it has been proposed that NAFLD may result from increased delivery of circulating lipids to the liver secondary to insulin functioning as an antilipolytic agent in adipose tissue. Within the liver, non-esterified fatty acids are then oxidized and re-esterified and circulate as triglyceride lipoproteins. Since this drives a shift in energy metabolism toward fatty acid β-oxidation and ketogenesis, the liver retains excessive lipids and steatosis (NAFLD) ensues. These findings are consistent with our findings in the fetal metabolomic profile, as we similarly observe NAFLD with elevated plasma 3-hydroxybutyrate both by virtue of maternal diet and phenotype (Table II). Taken together, we speculate that our model serves as the first evidence that in primate fetal life maternal obesity and in utero high fat diet exposure associate with elevations in 3-hydroxybutyrate which may serve as a marker for a correlation between fetal lipid oxidation, hypertriglyceridemia, and NAFLD. If this were to hold true in human studies, this serum marker could be used to screen for NAFLD and disposition towards obesity.
We similarly find the inverse correlation of cholesterol (elevated in obese dams but diminished in the fetus) to be of further interest. Consistent with an association of an altered fetal metabolome with maternal HFD intake as well as obese phenotype, we again observed significant differences among offspring of HFD-R versus HFD-S dams (Figure 2, Table III). We speculate that fetal hypocholesterolemia relative to maternal hypercholesterolemia may result from three possible (and non-mutually exclusive) mechanisms. First, it is possible that there is altered deposition in the fetus. Ergo circulating fetal cholesterol could be less but deposition could be increased. Others have previously observed that maternal hypercholesterolemia is associated with accelerated fetal plaque formation26 and we have observed fetal NAFLD, both supporting such a mechanism.19,20 Second, it is possible that maternal hypercholesterolemia alters placental lipid and cholesterol metabolism to decrease fetal de novo synthesis resulting in relatively decreased circulating fetal levels. Our observations pertaining to significantly decreased fetal cholesterol levels preferentially in HFD-S dams (Table III) supports this notion. Finally, it is possible that the maternal high fat diet and obesity differentially alter fetal de novo synthesis of both lipid and oxidative stress pathways. Such a “developmental plasticity” model gains support from others observations in animal models as well as human studies.24–28 Further consistent with such a “developmental plasticity” model, the observed absence of ascorbic acid in both high fat and diet reversal cohorts is indicative of persistent oxidative stress (Table II).
Table III. Fetal metabolites that differ between high fat sensitive and resistant.
Analysis of fetal metabolites contributing to the PLS-DA model in Figure 2 stratified by maternal phenotype. Significantly discrete serum metabolites by mass spectroscopy are as listed and correspond to the mass spectra data as presented in Figure 2.
| HFD-S vs. HFD-R | HFD-S vs. Con | HFD-R vs. Con. | ||||
|---|---|---|---|---|---|---|
| Metabolite | P value | Fold change (HFD-S/HFD-R) |
P value | Fold change (HFD-S/Con) |
P value | Fold change (HFD-S/Con) |
| cholesterol | 0.0099 | 0.73 | 0.16 | 0.81 | 0.45 | 0.89 |
| α-tocopherol | 0.0030 | 0.53 | 0.0038 | 0.53 | 0.98 | 1.0 |
| Hypoxanthine* | 0.025 | 1.6 | 0.63 | 1.1 | 0.016 | 1.4 |
| myo-inositol* | 0.040 | 1.9 | 0.99 | 1 | 0.042 | 1.9 |
| un(698_103) | 0.0025 | 0.79 | 0.15 | .86 | 0.44 | 1.1 |
| un(2298_144) | 0.022 | 1.9 | 0.98 | 0.99 | 0.029 | 1.9 |
=metabolites found through the second round of GC-MS, HF-S n=5, HF-R n=3, Con n=6 un(ri_m/z), unknown(retention index_mass/charge ratio); Con, control, HFD-S, high fat diet sensitive; HFD-R, high fat diet resistent.
The strengths of our study our multifold. We have utilized high throughput discovery “functional genomics” and characterized the metabolomic profile of maternal and fetal serum under conditions of a high fat maternal diet leading to obesity in non human primates. Our model system has advantages over other animal models on the developmental origins of human disease in so much as we utilize primates rather than phylogenetically dissimilar litter mate mammals. By stratification with respect to maternal diet sensitivity and diet reversal we are able to distinguish effects of diet from the obese maternal environment. Thus our insights are well characterized and representative of human development.
There are inherent limitations to primate research. First, the time and expense in establishing such a model system prohibits large numbers of animals and may limit statistically significant discovery. Second, as with humans, the animals behavioral and social hierarchy may confound our findings. Third, monitoring of metabolic parameters is restrictive relative to other model systems. Thus we cannot provide a longitudinal analysis as we cannot support a significant cohort of fetuses over time.
The maternal and fetal serum metabolomic profiling presented herein gives further insight into our complex biologic model. Moreover, since the fetal metabolome is proximal to the fetal phenotype our observations lend support to our prior findings on the effects of maternal high fat diet exposure on the fetal epigenome. Taken together, these data suggest that high fat diet exposure as well as maternal obese phenotype alters the fetal epigenome to result in functional transcriptome and metabolome variations. Further ongoing characterization of our unique model will provide insight and understanding of the role maternal diet and obesity play in fetal metabolism.
Acknowledgements
Support for this work came from the NIH Director New Innovator Pioneer Award DP21DP2OD001500-01 (KAT), and NIH grants 1R01DK080558 (RHL and KAT), DK60685-0351 (KLG), 1R01DK079194 (KLG), and RR00163 (KLG-ONPRC)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Strauss RS, Pollack HA. Epidemic increase in childhood overweight, 1986–1998. JAMA. 2001;286:2845–2848. doi: 10.1001/jama.286.22.2845. [DOI] [PubMed] [Google Scholar]
- 2.Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 3.Field A, et al. Impact of overweight on the risk of developing common chronic diseases during a 10 year period. Arch Intern Med. 2001;161:1581–1586. doi: 10.1001/archinte.161.13.1581. [DOI] [PubMed] [Google Scholar]
- 4.Finkelstein EA, Ruhm CJ, Kosa KM. Economic causes and consequences of obesity. Annu Rev Public Health. 2005;26:239–257. doi: 10.1146/annurev.publhealth.26.021304.144628. [DOI] [PubMed] [Google Scholar]
- 5.Rennie KL, Jebb SA. Prevalence of obesity in Great Britain. Obes Rev. 2005;6:11–20. doi: 10.1111/j.1467-789X.2005.00164.x. [DOI] [PubMed] [Google Scholar]
- 6.Popkin BM, Gordon-Larsen P. The nutrition transition: worldwide obesity dynamics and their determinants. Int J Obesity. 2004;28:S2–S9. doi: 10.1038/sj.ijo.0802804. [DOI] [PubMed] [Google Scholar]
- 7.Grove KL, Grayson BE, Glavas MM, Xiao XQ, Smith MS. Development of metabolic systems. Physiol Behav. 2005;86:646–660. doi: 10.1016/j.physbeh.2005.08.063. [DOI] [PubMed] [Google Scholar]
- 8.Bhargava SK, Sachdev HS, Fall CH, Osmond C, Lakshmy R, Barker DJ, Biswas SK, Ramji S, Prabhakaran D, Reddy KS. Relation of serial changes in childhood body-mass index to impaired glucose tolerance in young adulthood. NEJM. 2004;350:865–875. doi: 10.1056/NEJMoa035698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speiser PW, et al. (Obesity Consensus Working Group) Childhood obesity. J Clin Endocrinol Metab. 2005;90:1871–1887. doi: 10.1210/jc.2004-1389. [DOI] [PubMed] [Google Scholar]
- 10.Barker DJ. Fetal origins of coronary heart disease. BMJ. 1995;311:171–174. doi: 10.1136/bmj.311.6998.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eriksson JG, Forsen T, Tuomilheto J, Osmond C, Barker DJP. Early growth and coronary heart disease in later life: longitudinal study. BMJ. 2001;322:949–953. doi: 10.1136/bmj.322.7292.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klebanoff MA, Meirik O, Berendes HW. Second-generation consequences of small-for-dates birth. Pediatrics. 1989;84:243–247. [PubMed] [Google Scholar]
- 13.Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Philips ES, Beedle AS, Burdge GC, Hanson MA. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. PNAS. 2007;104:12796–12800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillicrop KA. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. B J Nutrit. 2007;97:435–439. doi: 10.1017/S0007114507352392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferace-1 expression is involved in impaired DNA methylation changes in histone modifications. B J Nutrit. 2007;97:1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacLennan NK, James SJ, Melnyk S, Piroozi A, Jernigan S, Hsu JL, Janke SM, Pham TD, Lane RH. Uteroplacental insufficiency alters DNA methylation, one-carbon metabolism, and histone acetylation in IUGR rats. Physiol Genomics. 2004;18:43–50. doi: 10.1152/physiolgenomics.00042.2004. 2004. [DOI] [PubMed] [Google Scholar]
- 17.Fu Q, McKnight RA, Yu X, Callaway CW, Lane RH. Growth retardation alters the epigenetic characteristics of hepatic dual specificity phosphatase 5. FASEB. 2006 doi: 10.1096/fj.06-6179fje. [PMID: 16940436] [DOI] [PubMed] [Google Scholar]
- 18.Fu Q, McKnight RA, Yu X, Wang L, Callaway CW, Lane RH. Uteroplacental insufficiency induces site-specific changes in histone H3 covalent modifications and affects DNA-histone H3 positioning day 0 IUGR rat liver. Physiol Genomics. 2004;20:108–116. doi: 10.1152/physiolgenomics.00175.2004. [DOI] [PubMed] [Google Scholar]
- 19.McCurdy CE, Bishop JM, Williams SM, Smith MS, Friedman JE, Grove KL. Chronic maternal high fat diet triggers fetal hepatic reprogramming and fatty liver in the nonhuman primate. J Clin Invest. 2008 doi: 10.1172/JCI32661. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, Lane RH. Developmental origins of disease and determinants of chromatin structure: Maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41(2):91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loftus N, Miseki K, Iida J, Gika HG, Theodoridis G, Wilson ID. Profiling and biomarker identification in serum from different Zucker rat strains via high mass accuracy multistage mass spectrometric analysis using liquid chromatography/mass spectrometry with a quadrupole ion trap-time of flight mass spectrometer. Rapid Commun Mass Spectrom. 2008;22:2547–2554. doi: 10.1002/rcm.3640. [DOI] [PubMed] [Google Scholar]
- 22.Werner E, Heiler J-F, Ducruiz C, Ezan E, Junot C, Tabet J-C. Mass spectrometry for the identification of the discriminating signals from metabolomics: Current status and future trends. J Chromat. 2008;871:143–163. doi: 10.1016/j.jchromb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Trygg AJ, Gullberg J, Johansson A, Jonsson P, Antti H, Marklund S, Moritz T. Extraction and GC/MS Analysis of the Human Blood Serum Metabolome. Anal. Cham. 2005;77:8086–8094. doi: 10.1021/ac051211v. [DOI] [PubMed] [Google Scholar]
- 24.Briegelieus-Flohe R, Traber MG. Vitamin E: function and metabolism. FASEB. 1999;13:1145–1155. [PubMed] [Google Scholar]
- 25.Azzi A, Aratri E, Boscoboinik D, Clement S, Ozer NK, Ricciarelli R, Spycher S. Molecular basis of alpha-tocopherol control of smooth muscle cell proliferation. Biofactors. 2008;7:3–14. doi: 10.1002/biof.5520070102. [DOI] [PubMed] [Google Scholar]
- 26.Bonet B, Viana M, Sánchez-Vera I, Quintanar A, Martínez J, Espino M. Adipose tissue and liver lipid metabolism in obese children: role of the body mass index and the presence of acanthosis nigricans. Diabet Med. 2007;24(11):1192–1198. doi: 10.1111/j.1464-5491.2007.02230.x. [DOI] [PubMed] [Google Scholar]
- 27.Bugianesi E, Gastaldelli A, Vanni E, Gambino R, Cassader M, Baldi S, Ponti V, Pagano G, Ferrannini E, Rizzetto M. Insulin resistance in non-diabetic patients with non-alcoholic fatty liver disease: sites and mechanisms. Diabetologia. 2005;48(4):634–642. doi: 10.1007/s00125-005-1682-x. [DOI] [PubMed] [Google Scholar]
- 28.Palinski W, Napoli C. The fetal origins of atherosclerosis: maternal hypercholesterolemia, and cholesterol-lowering or antioxidant treatment during pregnancy influence in utero programming and postnatal susceptibility to atherogenesis. FASEB. 2002;16:1348–1360. doi: 10.1096/fj.02-0226rev. [DOI] [PubMed] [Google Scholar]