Abstract
Background
Excessive proliferation of pulmonary artery smooth muscle cells (PASMCs) plays an important role in the development of idiopathic pulmonary arterial hypertension (IPAH), whereas a rise in cytosolic Ca2+ concentration triggers PASMC contraction and stimulates PASMC proliferation. Recently, we demonstrated that upregulation of the TRPC6 channel contributes to proliferation of PASMCs isolated from IPAH patients. This study sought to identify single-nucleotide polymorphisms (SNPs) in the TRPC6 gene promoter that are associated with IPAH and have functional significance in regulating TRPC6 activity in PASMCs.
Methods and Results
Genomic DNA was isolated from blood samples of 237 normal subjects and 268 IPAH patients. Three biallelic SNPs, −361 (A/T), −254(C/G), and −218 (C/T), were identified in the 2000-bp sequence upstream of the transcriptional start site of TRPC6. Although the allele frequencies of the −361 and −218 SNPs were not different between the groups, the allele frequency of the −254(C→G) SNP in IPAH patients (12%) was significantly higher than in normal subjects (6%; P<0.01). Genotype data showed that the percentage of −254G/G homozygotes in IPAH patients was 2.85 times that of normal subjects. Moreover, the −254(C→G) SNP creates a binding sequence for nuclear factor-κB. Functional analyses revealed that the −254(C→G) SNP enhanced nuclear factor-κB–mediated promoter activity and stimulated TRPC6 expression in PASMCs. Inhibition of nuclear factor-κB activity attenuated TRPC6 expression and decreased agonist-activated Ca2+ influx in PASMCs of IPAH patients harboring the −254G allele.
Conclusions
These results suggest that the −254(C→G) SNP may predispose individuals to an increased risk of IPAH by linking abnormal TRPC6 transcription to nuclear factor-κB, an inflammatory transcription factor.
Keywords: calcium; hypertension; pulmonary; ion channels; muscle, smooth; NF-kappa B
Pulmonary arterial hypertension (PAH) is a fatal and progressive disease characterized by elevated pulmonary vascular resistance resulting from severe pulmonary vascular remodeling.1-3 Approximately 6% of PAH patients have a family history of the condition and are referred to as having familial PAH; the rest are considered to have idiopathic PAH (IPAH). Although the cause of PAH remains unclear, elevated levels of mitogenic, angiogenic, and proinflammatory factors such as platelet-derived growth factor, endothelin-1, interleukin-1β/−6, soluble CD40 ligand, angiopoietin-1, and serotonin have been reported to correlate with the onset of IPAH.2-4 Other factors associated with IPAH include the downregulation and dysfunction of voltage-gated K+ channels5 and upregulation of the serotonin receptors and transporter.6 Moreover, mutations in the bone morphogenetic protein receptor-type II gene (BMPR2) have been demonstrated to associate with the development of familial PAH and IPAH.7,8 However, because BMPR2 mutations are present in only 15% to 20% of IPAH patients and the likelihood that clinical pulmonary hypertension will develop is only 10% to 20% in known carriers of BMPR2 mutations,9 additional genetic and environmental factors other than BMPR2 mutations may also contribute to the development of IPAH.
Regardless of the initial pathogenic trigger, the elevated pulmonary vascular resistance and pulmonary arterial pressure in IPAH patients are caused mainly by sustained pulmonary vasoconstriction, concentric vascular remodeling, obliteration of small arteries and arterioles, in situ thrombosis, and formation of the plexiform lesion.1-3 Neointimal and medial hypertrophy in small and medium-sized pulmonary arteries is a key aspect of pulmonary vascular remodeling in IPAH patients and is attributed to excessive pulmonary artery smooth muscle cell (PASMC) proliferation.1,2
Ca2+ operates as an important second messenger in cellular mechanisms leading to gene expression, cell proliferation, and contraction. A rise in cytosolic free Ca2+ concentration ([Ca2+]cyt) in PASMCs is a major trigger for pulmonary vasoconstriction and an important stimulus for PASMC proliferation and migration.10 Conventional Ca2+ channel blockers (ie, nifedipine and diltiazem), which inhibit voltage-dependent Ca2+ channels in PASMCs, have been used to treat 15% to 20% of IPAH patients in clinical studies,11 suggesting that increased [Ca2+]cyt may be an important link in cellular pathways that lead to IPAH. Elevation of [Ca2+]cyt in PASMCs results from Ca2+ release from intracellular stores and Ca2+ influx through plasmalemmal Ca2+ channels.12 In addition to voltage-dependent Ca2+ channels, it has been demonstrated that canonical transient receptor potential (TRPC) channels are responsible for Ca2+ entry in PASMCs.12-14
TRPC6 is an important isoform of TRPC channels expressed in the lungs and pulmonary artery.12-15 We previously observed that TRPC6 mRNA and protein expression in lung tissues and PASMCs isolated from IPAH patients was substantially elevated compared with normal subjects and control patients with cardiopulmonary diseases.16 TRPC6 upregulation is also a critical initial step in the elevation of [Ca2+]cyt required for mitogen-mediated PASMC proliferation and a critical contributor to the elevated [Ca2+]cyt in IPAH PASMCs.13 Downregulation of TRPC6 expression with siRNA significantly attenuates DNA synthesis and proliferation of PASMCs isolated from normotensive and IPAH patients.13,16 Together, these observations imply that upregulated TRPC6 gene transcription may promote the development of IPAH.17
To test this hypothesis, we sequenced the 5′-regulatory region of TRPC6 from 268 IPAH patients and identified a C to G (C→G) single-nucleotide polymorphism (SNP) at nucleotide −254 of the TRPC6 gene that is associated with IPAH. Moreover, the −254C→G change creates a canonical nuclear factor-κB (NF-κB) binding site (GGGGGTCTCC) in the promoter region of TRPC6 and significantly affects TRPC6 gene transcription and TRPC6 channel function in PASMCs from IPAH patients who carry the −254G allele.
Methods
Subjects
A total of 237 normal subjects and 268 IPAH patients (including 124 patients enrolled at the University of California, San Diego Medical Center, 60 patients at the Vanderbilt University Medical Center, and 84 patients in the Giessen Lung Center in Germany) who participated in the study. All control subjects (all white) and IPAH patients were white (including 2 Hispanics) and were unrelated. The control subjects and patients were very closely matched racially and ethnically. We did not include data from blacks in this report because of the small sample number. The basic demographics of age, gender, and race in normal subjects and IPAH patients and the hemodynamics of all patients from each of the 3 centers are shown in Table I of the online-only Data Supplement. No significant difference was found (P=0.68682) between normal subjects (87.7±9.35 [SD] mm Hg) and IPAH patients (88.3±14.76 mm Hg) from whom we collected blood/DNA samples for this study. The diagnosis of IPAH was based on the criteria used in the National Institutes of Health Registry on Primary Pulmonary Hypertension. Informed consent was obtained from all subjects, and the study was approved by the Institutional Review Board at the University of California, San Diego.
Identification of SNP in the TRPC6 Gene Promoter Region
Genomic DNA was extracted from the blood samples of normal subjects and patients with a Wizard genomic DNA purification kit. Six paired amplification polymerase chain reaction (PCR) primers (online-only Data Supplement Table II) were designed to amplify 6 overlapping DNA segments spanning 2000 bp upstream and 110 bp downstream of the transcriptional start site of human TRPC6. The purified PCR products were sequenced, analyzed by Chromas software, and compared with known SNPs deposited in the NCBI SNP databank (see supplementary Materials).
Cell Preparation and Culture
PASMCs from IPAH patients and non–pulmonary hypertensive (NPH) patients were isolated from lung tissues of transplant patients, and PASMCs from normal subjects were purchased from Lonza (Walkersville, Md). PASMCs were cultured in 5% CO2 in air at 37°C in smooth muscle cell growth medium (Lonza) and used at the fourth to sixth passage.5,12 For tumor necrosis factor-α (TNF-α) stimulation experiments, the cells were growth arrested by culturing in smooth muscle cell basal medium (Lonza) for 24 hours before treatment.
Preparation of Cytoplasmic and Nuclear Extracts
Cytosolic and nuclear extracts from cultured PASMCs were collected using a modified protocol18 (see supplementary Materials).
Electrophoretic Mobility Shift Assay and Supershift Assay
Double-stranded oligonucleotide sequences from nucleotide −261 to −238 of TRPC6 containing the −254C wild-type (5′-ATCCTCGCGGGGTCTCCTCGGCCT-3′) or −254G mutated site (5′-ATCCTCGGGGGGTCTCCTCGGCCT-3′) were synthesized and labeled by the biotin 3′ end-labeling kit (Pierce Biotechnology Inc, Rockford, Ill). Each binding reaction (24°C for 40 minutes) contained 10 mmol/L Tris (pH 7.5), 50 mmol/L KCl, 1 mmol/L EDTA, 10 mmol/L dithiothreitol, 2.5% glycerol, 50 ng/mL poly(dI-dC), 5% albumin bovine, 4 μg nuclear extract, and 40 to 50 fmol biotin end-labeled target DNA. Competition tests were used to verify whether the observed shifted bands were specific. The nuclear extract was preincubated with 200-fold excess unlabeled −254C or −254G probe before electrophoretic mobility shift assay. For supershift assays, polyclonal antibodies against p50 or p65 were added to the binding reaction before addition of the biotin end-labeled probe. The DNA-protein complexes were electrophoresed by 6% DNA retardation gels in 0.5× TBE running buffer and electrotransferred to nitrocellulose membranes. The biotin-labeled oligonucleotides in the membrane were detected with streptavidin–horseradish peroxidase conjugate and a chemiluminescent substrate (Pierce).
TRPC6 Putative Promoter Region Cloning and Promoter Activity
The human TRPC6 putative promoter regions were amplified using genomic DNA isolated from PASMCs. The DNA fragments (−1682 to 110 bp) containing −254C or −254G were amplified by PCR. The PCR products were cloned into PCR 2.1 TOPO cloning vector. A transient expression system, pBlue-TOPO TA expression kit, was used to test the cloned TRPC6 putative promoter activities. DNA fragments were fused to the promoterless β-glucuronidase (LacZ) reporter gene vector. COS-7 cells were transiently transfected with promoter vectors and, 24 hours after transfection, replated onto Petri dishes coated with poly-l-lysine. The promoter activities were detected and quantified by measuring the absorbance at 420 nm. Transfection efficiencies were normalized to green fluorescence protein (GFP) expression from a cotransfected pRL-CMV-GFP vector. The relative promoter activity is expressed as fold induction relative to the basal level of promoterless empty pBlue-TOPO vector. For LacZ staining, the cells were fixed with 0.05% glutaraldehyde and stained in X-Gal solution containing 40 mmol/L HEPES (pH 7.4), 5 mmol/L K3[Fe(CN)6], 5 mmol/L K4[Fe(CN)6], 2 mmol/L MgCl2, 15 mmol/L NaCl, and 1 mg/mL X-Gal.
Generation of Recombinant Adenovirus Carrying Human TRPC6-Specific siRNA and Adenoviral Infection
Recombinant adenovirus carrying siRNA targeting human TRPC6 was generated with an Invitrogen expression kit (Invitrogen, Carlsbad, Calif). For adenovirus infection experiments, PASMCs were infected with the appropriate virus in smooth muscle cell basal medium containing 0.2% FBS for 4 hours and used for experiments 24 to 48 hours after adenoviral infection (see supplementary Materials).
Human TRPC6 cDNA Cloning and Transient Transfection
Human TRPC6 cDNA was purchased from Open Biosystems (Huntsville, Ala). It was tagged with a hemagglutinin HA epitope sequence at the C terminal by introducing an in-frame HA epitope coding sequence before the stop codon and subcloning the corresponding cDNA into a mammalian expression pCDNA3 vector (Invitrogen) and pCMS-EGFP vector (BD Bioscience, San Jose, Calif), respectively. Resulting clones were confirmed by DNA sequencing and were designated pcDNA3-hTRPC6HA and pCMS-EGFP-hTRPC6HA. For transient transfection, electroporation-mediated transfection by nucleofector (Amaxa Biosystems, Walkersville, Md) was used, following the PASMC-specific protocol.
Statistical Analysis
Values are expressed as mean±SEM. Statistical differences were assessed with unpaired Student t test or 1-way ANOVA with post hoc analysis. Differences were considered significant at values of P<0.05. We used χ2 analysis to compare the allele frequencies and genotype frequencies in normal subjects and patients. The odds ratio was estimated by the logistic regression model, assuming 95% as the CI, using the Woolf approximation. StatsDirect software (StatsDirect, Ltd, Cheshire, UK) was used for this analysis.
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
Three SNPs Were Identified in the Putative Promoter Region of TRPC6
The human TRPC6 gene contains 13 exons (Figure 1A). Using specifically designed primers, we sequenced the 5′-untranslated region (−2000 bp upstream of the start codon, ATG) of TRPC6 (Figure 1B) from 237 normal subjects and 268 IPAH patients. DNA sequence alignment with the human genomic TRPC6 sequence (AP003080) revealed 3 variants, −361(A→T), −254(C→G), and −218(C→T) (Figure 1B and 1C). All 3 SNPs have been reported and are listed in the NCBI SNP database: −254(C→G) SNP matches to rs3824934, −361(A→T) to rs41302375, and −218(C→T) to rs56134796. The −361(A→T) and −218(C→T) SNPs are in complete linkage disequilibrium.
Figure 1.
Identification of 3 SNPs in the 5′-regulatory region of the human TRPC6 gene. A, Schematic diagram of the TRPC6 gene and locations of the amplified PCR segments. B, The location of 3 SNPs, −361(A→T), −254(C→G), and −218(C→T), in the 5′-regulatory region of TRPC6. C, Representative sequence chromatographs for the 3 SNPs in IPAH patients. Arrow denotes position of the SNP. UTR indicates untranslated region.
Allele and Genotype Frequencies of the −254C→GSNP in TRPC6 Are Significantly Higher in IPAH Patients Than in Normal Subjects
To examine whether the 3 SNPs of TRPC6 are associated with IPAH, we analyzed their allele frequencies and genotype frequencies in IPAH patients and normal control subjects. As shown in Table 1, the allele frequency of the −254G SNP was significantly higher in IPAH patients (12%) than in normal subjects (6%; P<0.01), whereas the allele frequencies of the −361(A→T) and −218(C→T) SNPs were comparable (P=0.83) between IPAH patients (25% and 25%) and normal subjects (25% and 25%). Both the homozygous and heterozygous variants of the −254(C→G) SNP were identified in IPAH patients (Figure 1C). Genotype analysis showed that 25 of 237 normal subjects (10.5%) were heterozygotes and 3 of 237 of normal subjects (1.3%) were homozygotes for the −254(C→G) SNP. In contrast, 43 of 268 of the IPAH patients (16.0%) were heterozygotes (−254C/G) and 10 of 268 patients (3.7%) were homozygotes (−254G/G) (P<0.02 between normal subjects and IPAH patients for both heterozygotes and homozygotes; Table 2). The −254C/G heterozygote and −254G/G homozygote frequencies in IPAH patients were 1.52 (16.0% versus 10.5%) and 2.85 (3.7% versus 1.3%) times, respectively, those of normal subjects (P<0.02). A full description of the allele frequencies for each patient cohort is provided in online-only Data Supplement Table III.
Table 1.
Allele Frequency of the −254(C→G) SNP Is Significantly Higher in IPAH Patients Than in Normal Subjects
| Position (nt.) | SNP | Allele | Normal Subjects (n=237), n (%) | IPAH (n=268), n (%) | Odds Ratio (95% CI) | P |
|---|---|---|---|---|---|---|
| −361 | A to T | A | 354 (74.7) | 402 (75.0) | ||
| T | 120 (25.3) | 134 (25.0) | 0.98 (0.74−1.31) | 0.91 | ||
| −254 | C to G | C | 443 (93.7) | 473 (88.2) | ||
| G | 30 (6.3) | 63 (11.8) | 1.97 (1.25−3.10) | 0.0029 |
P values, odds ratios, and 95% CIs are calculated by χ2 analysis for 2×2 contingency tables. The −361(A→T) and −218(C→T) SNPs are in complete linkage disequilibrium. Because values are identical, only the −361A→T SNP is shown for the sake of clarity.
Table 2.
The Homozygous −254G/G Genotype Has a Significantly Higher Frequency in IPAH Patients Than in Normal Subjects
| SNP | Genotype | Normal (n=237), n (%) | IPAH (n=268), n (%) | Odds Ratio (95% CI) | P |
|---|---|---|---|---|---|
| −361(A→T) | −361 A/A | 132 (55.7) | 152 (56.7) | ||
| −361 A/T | 90 (38.0) | 98 (36.6) | 0.96* (0.67−1.36) | 0.82* | |
| −361 T/T | 15 (6.3) | 18 (6.7) | |||
| −254(C→G) | −254 C/C | 209 (88.2) | 215 (80.2) | ||
| −254 C/G | 25 (10.5) | 43 (16.0) | 1.84† (1.12−3.02) | 0.015† | |
| −254 G/G | 3 (1.3) | 10 (3.7) |
P values, odds ratios, and 95% CIs are calculated by χ2 analysis for comparison of the genotype frequencies between IPAH patients and normal subjects.
Odds ratio and P value for (−361 A/T and −361 T/T) vs −361A/A and for (−218 C/T and −218 T/T) vs −218C/C.
Odds ratio and P value for (−254 C/G and −254 G/G) vs −254 C/C. The −361(A→T) and −218(C→T) SNPs are in complete linkage disequilibrium. Because their allele frequencies, odds ratio, and P values are identical, only the −361A→T SNP is shown for the sake of clarity.
The −254(C→G) SNP Generates an NF-κB Binding Site at the Promoter Region of TRPC6
Interestingly, the C-to-G conversion at nucleotide −254 results in the sequence alteration of “CGGGGTCTCC” (nucleotide −254 to −245) to “GGGGGTCTCC,” which matches the sequence of “GGGRNNYYCC” (where R=A or G; Y=C or T; N=any nucleotide), a consensus sequence for the transcription factor NF-κB. Therefore, the −254(C→G) SNP adds a putative NF-κB binding site into the 5′-untranslated region of TRPC6.
We performed an electrophoretic mobility shift assay to determine whether the −254(C→G) SNP confers functional NF-κB protein binding to the promoter region of TRPC6. Biotin-labeled oligoduplex probes with nucleotides encompassing sequences between nucleotide −238 and −261 containing −254C (−254C probe) or −254G (−254G probe) were incubated with nuclear extracts isolated from normal PASMCs. To evaluate the physiological or functional consequences, PASMCs were stimulated with TNF-α before the electrophoretic mobility shift assay was performed. As shown in Figure 2A, TNF-α (10 ng/mL) stimulated translocation of the p50 and p65 of the NF-κB components into the nucleus of PASMCs, indicating that TNF-α is an effective stimulator for nuclear translocation of NF-κB.
Figure 2.
Nuclear NF-κB proteins of PASMCs specifically bind to the −254(C→G) SNP–generated NF-κB binding site. A, NF-κB activation demonstrated by translocation of p50 and p65 from the cytoplasm to the nucleus. PASMCs from normal subjects were stimulated with (+) or without (−) TNF-α (10 ng/mL). B, Electrophoretic mobility shift assay and competition analysis. TNF-α–stimulated nuclear extracts from PASMCs were incubated with biotin-labeled double-stranded oligonucleotides corresponding to the 24-bp sequences containing the wild-type −254C or mutant −254G probes. Nuclear extract/oligonucleotide complexes (C1 and C2) are indicated by arrows. C, Antibody-mediated supershift assay. NF-κB p50 and p65 antibodies identified complexes C1 as p50/p65 and C2 as p50/p50 NF-κB–containing complexes.
Two DNA-bound NF-κB complexes were identified with the −254G probe (designated C1 and C2), whereas NF-κB did not bind to the −254C probe (Figure 2B, left). Complex formation (as indicated by C1 and C2) was inhibited by the addition of excess unlabeled −254G probe but not by unlabeled −254C probe because the unlabeled −254G probe competitively bound to the same DNA binding site as NF-κB, whereas the unlabeled −254C probe failed to compete for binding to the NF-κB binding site. These results indicate that NF-κB specifically binds to the −254G probe (Figure 2B, right). Supershifted complexes were detected by the addition of the anti-p65 or anti-p50 antibody to the binding reaction, indicating that the NF-κB C1 and C2 complexes contain a p50/p65 heterodimer and a p50/p50 homodimer, respectively (Figure 2C).
The −254(C→G) SNP Affects NF-κB–Mediated TRPC6 Promoter Activity
Gene constructs were generated to define whether the −254(C→G) SNP introduces an active NF-κB–regulated promoter region in the TRPC6 gene. Three different-length constructs of the 5′ upstream region of TRPC6 containing −254C or −254G were cloned and inserted into a promoterless pBlue-TOPO vector upstream of a β-glucuronidase (LacZ) reporter gene. The resulting constructs were designated pBlue(−335/+110), pBlue(−456/+110), and pBlue(−1682/+110), respectively.
In COS-7 cells transiently transfected with these vectors, TNF-α (20 ng/mL for 24 hours), by causing nuclear translocation of NF-κB (p65 and p50), significantly enhanced promoter activity for the constructs [pBlue(−335/+110), P<0.001; pBlue(−456/+110), P<0.001; pBlue(−1682/+110), P=0.027] with the −254G mutation but negligibly affected promoter activity for the constructs with the −254C wild-type sequence (Figure 3A) [P=0.437 (−335/+110), P=0.889 (−456/+110), P=0.231 (−1682/+110)]. Compared with cells transfected with promoterless pBlue-TOPO vector, the basal promoter activity (or the relative induction of β-galactosidase activity in cells not treated with TNF-α) significantly increased when the 5′ upstream region was extended from nucleotide −335 to −456 and −1682 in the wild-type −254C template (Figure 3A). X-Gal staining confirmed these results by exhibiting an intense staining corresponding to β-galactosidase in the endoplasmic reticulum with the −254G pBlue(−335/+110) construct but not with the −254C construct (Figure 3B). Together, these data indicate that the −254(C→G) SNP modulates TRPC6 gene promoter activity and enhances TNF-α– and NF-κB–mediated TRPC6 transcription.
Figure 3.
The basal and TNF-α–mediated promoter activities are enhanced in PASMCs transfected with the reporter constructs containing −254G allele. A, Schematic diagram (left) of the reporter constructs used for the promoter assay and the relative promoter activity in control and TNF-α–treated cells (right). The relative β-galactosidase activity (right) represents the mean±SE (plotted on a log scale) of 3 independent experiments performed in quadruplicate. *P<0.05, ***P<0.001 vs control cells. B, COS-7 cells transfected with pBlue(−335/+110) constructs that contain −254C or −254G allele. X-Gal staining was used to detect the β-galactosidase–stained cells. Empty (or promoterless) pBlue TOPO vector was used as control.
Upregulated TRPC6 Expression Contributes to Regulating [Ca2+]cyt in PASMCs of IPAH Patients
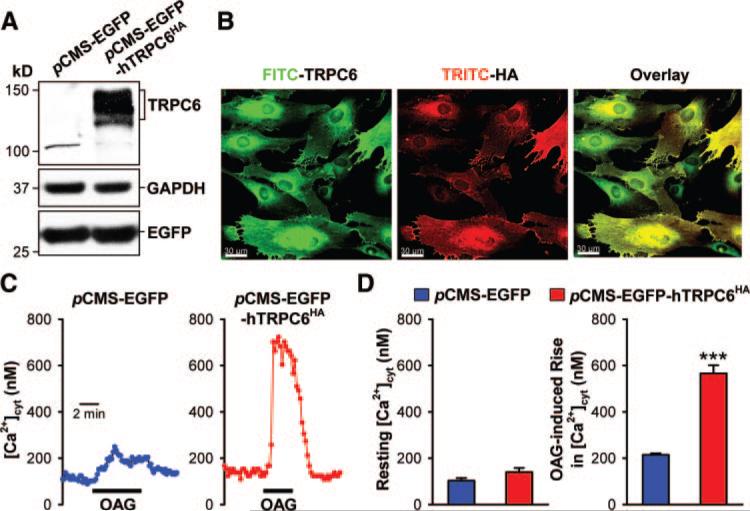
We previously reported that TRPC6 expression was significantly increased in IPAH PASMCs compared with normal PASMCs. To examine whether upregulated TRPC6 mRNA and protein expression in IPAH PASMCs (with the −254G allele) (Figure 4A) contributes to regulating [Ca2+]cyt, we measured and compared the resting [Ca2+]cyt and agonist-mediated Ca2+ influx in normal PASMCs and IPAH PASMCs. The resting [Ca2+]cyt and the increase in [Ca2+]cyt induced by 1-oleoyl-2-acetyl-sn-glycerol (OAG; 100 μmol/L), a membrane-permeable diacylglycerol analog known to activate TRPC6 channels, were both significantly enhanced in IPAH PASMCs compared with normal PASMCs (P<0.001 for resting and OAG-induced [Ca2+]cyt]; Figure 4B and 4C). The OAG-induced increase in whole-cell cation currents in IPAH PASMCs was also significantly enhanced compared with normal PASMCs (Figure 4D). Inhibition of TRPC6 expression in IPAH PASMCs using siRNA markedly attenuated the OAG-mediated increase in [Ca2+]cyt (P<0.001) and reduced the resting [Ca2+]cyt (P=0.003; Figure 5). In normal PASMCs (with the −254C/C genotype), overexpression of the human TRPC6 gene significantly enhanced the OAG-induced increase in [Ca2+]cyt (P<0.001; Figure 6) but not resting [Ca2+]cyt (P=0.075). These results indicate that upregulated TRPC6 expression is functionally involved in regulating resting [Ca2+]cyt and agonist-mediated Ca2+ entry in PASMCs from IPAH patients harboring the −254G allele.
Figure 4.
Upregulated TRPC6 expression in PASMCs from IPAH patients increases the resting [Ca2+]cyt and enhances agonist-mediated Ca2+ influx. A, mRNA and protein expression of TRPC6, determined by RT-PCR (left), Western blot (middle), and immunocyto-chemistry (right), respectively, is significantly higher in IPAH PASMCs than in control PASMCs from NPH patients. B, Representative records of [Ca2+]cyt changes in response to OAG (100 μmol/L) in control (NPH) and IPAH PASMCs. C, Summarized data of the resting [Ca2+]cyt and the amplitude of the OAG-induced increase in [Ca2+]cyt in NPH and IPAH PASMCs. **P<0.01, ***P<0.001 vs NPH PASMCs. D, Representative currents recorded in NPH (left) and IPAH (middle) PASMCs before (Cont) and during (OAG) application with OAG (100 μmol/L). OAG-sensitive currents, generated by subtracting the currents recorded during OAG from the control currents, in NPH and IPAH PASMCs are shown in the right panel.
Figure 5.
Downregulation of TRPC6 with siRNA inhibits OAG-induced Ca2+ influx in PASMCs from IPAH patients. A, mRNA (left) and protein (right) expression levels of TRPC6, TRPC4, and GADPH are shown in IPAH PASMCs treated with scrambled siRNA, GADPH siRNA, or TRPC6 siRNA. B, Representative records of [Ca2+]cyt changes in response to OAG in IPAH PASMCs treated with scrambled siRNA (left) or TRPC6 siRNA (right). C, Summarized data of the resting [Ca2+]cyt (left) and OAG-induced [Ca2+]cyt increases (right) in IPAH PASMCs treated with scrambled or TRPC6 siRNA. **P<0.01, ***P<0.001 vs scrambled siRNA-treated control cells.
Figure 6.
Overexpression of TRPC6 in normal PASMCs (with −254C/C genotype) enhances the OAG-induced increase in [Ca2+]cyt. A, Western blot analysis of TRPC6 in cells transfected with pCMS-EGFP or pCMS-EGFP-hTRPC6HA. B, Exogenous human TRPC6HA expression was identified by immunofluorescence staining. Normal PASMCs were transfected with pcDNA3-hTRPC6HA. HA epitope tag was stained by TRITC-conjugated anti-HA monoclonal antibody. TRPC6 antibody, followed by FITC-conjugated secondary antibody, was used to detect TRPC6 expression. C, Representative records of [Ca2+]cyt changes in response to OAG in PASMCs transfected with pCMS-EGFP or pcMS-EGFP-hTRPC6HA. D, Summarized data of the resting [Ca2+]cyt and the amplitude of the OAG-induced [Ca2+]cyt increases in cells transfected with pCMS-EGFP or pCMS-EGFP-hTRPC6HA. ***P<0.001 vs pCMS-EGFP cells.
Inhibition of NF-κB Activity by IκBα Attenuates TRPC6 Expression and Function in PASMCs From IPAH Patients With the −254G Allele
IκBα is an inhibitory subunit of the NF-κB complex that binds to p50/p65 and keeps the complex in the cytoplasm. IκBα superrepressor is a nondegradable mutant dominant-negative variant of IκBα that inhibits NF-κB activation.19 Overexpression of the IκBα superrepressor with an adenoviral vector markedly inhibited translocation of p50 and p65 into the nucleus in PASMCs from IPAH patients (with the −254G allele) and significantly inhibited TNF-α–mediated upregulation of TRPC6 (Figure 7A). Western blot analysis indicated 2 bands corresponding to TRPC6 protein, 1 at ≈110 kDa and the other at 130 kDa. The ≈110-kDa band reflects nascent TRPC6 early in the secretory pathway, whereas the 130-kDa band reflects complex-glycosylated TRPC6 that appears to maintain a relatively long half-life.20 In IPAH PASMCs (−254G allele) treated with TNF-α, inhibition of NF-κB by overexpression of the IκBα superrepressor predominantly downregulated the nascent core-glycosylated isoform (the ≈110-kDa band) of TRPC6 (Figure 7A, right), implying that the effect is due to a reduction in TRPC6 gene transcription. Together, these data support the contention that NF-κB plays an important role in regulating TRPC6 gene transcription and expression in IPAH patients harboring the −254(C→G) SNP. This finding is supported by similar assays performed in PASMCs from −254C/C IPAH. Unlike the −254C/G individuals, TRPC6 protein expression was not significantly altered in −254C/C PASMCs treated with TNF-α (see online-only Data Supplement Figure I).
Figure 7.
Overexpression of the inhibitory subunit IκBα attenuates NF-κB–mediated TRPC6 expression and inhibits agonist-mediated Ca2+ influx in IPAH-PASMC. A, Overexpression of IκBα (Ad-IκB) inhibits translocation of NF-κB p50 and p65 into the nucleus (left) and diminishes TNF-α–stimulated TRPC6 expression (right). GAPDH, histone (H2A), and p84 antibodies were used as controls for nuclear and cytoplasmic proteins. Experiments were reproduced 3 times. B, Summarized results showing that TNF-α increases the resting [Ca2+]cyt (left) and enhanced OAG-induced [Ca2+]cyt rise (right) in IPAH PASMCs (−254C/G), whereas IκBα (Ad-IκB) abolishes the TNF-α–mediated enhancement of the resting [Ca2+]cyt and OAG-mediated Ca2+ influx. C, Summarized results showing that TNF-α negligibly affects the resting [Ca2+]cyt (left) and OAG-induced [Ca2+]cyt increase (right) in normal PASMCs (−254C/C).
To determine whether the enhanced basal TRPC6 expression and TNF-α–mediated TRPC6 upregulation in the −254C/G variant have functional significance, we compared the levels of resting [Ca2+]cyt and OAG-induced Ca2+ influx in unstimulated and TNF-α–stimulated −254C/G PASMCs from IPAH patients to the levels in PASMCs from −254C/C normal subjects. As shown in Figure 7B and 7C, the resting [Ca2+]cyt and the OAG-induced [Ca2+]cyt increase in unstimulated −254C/G IPAH PASMCs were both significantly greater than in −254C/C normal PASMCs. TNF-α treatment significantly increased the resting [Ca2+]cyt and enhanced the OAG-induced [Ca2+]cyt (P<0.001) increase in PASMCs from −254C/G IPAH patients (Figure 7B) but not in normal PASMCs from −254C/C subjects (P=0.424; Figure 7C). Furthermore, overexpression of IκBα abolished the TNF-α–mediated increase in the resting [Ca2+]cyt and enhancement of the OAG-mediated Ca2+ influx (P<0.001) in PASMCs from −254C/G IPAH patients (Figure 7B). These results provide strong evidence that the −254(C→G) SNP in the TRPC6 gene has functional significance in regulating basal [Ca2+]cyt and agonist-mediated Ca2+ entry in PASMCs.
Discussion
We report 4 primary findings in this study: (1) A “gain-in-function” −254(C→G) SNP in TRPC6 is associated with IPAH; (2) the −254(C→G) SNP generates an active NF-κB binding site that regulates TRPC6 transcription; (3) nuclear translocation of NF-κB upregulates TRPC6 expression and enhances agonist-induced Ca2+ influx in IPAH PASMCs with the −254G allele; and (4) inhibition of nuclear translocation of NF-κB attenuates TRPC6 expression and function in PASMCs from IPAH patients harboring a −254G allele.
Pulmonary arterial neointimal and medial hypertrophy is a major pathological feature in IPAH.1-3 One of the important mechanisms in pulmonary vascular wall thickening is increased PASMC proliferation and migration resulting from a rise in [Ca2+]cyt.12-16 As a critical signaling element, intracellular Ca2+ is responsible for activating multiple signal transduction cascades that trigger PASMC contraction, activate gene expression, and stimulate PASMC proliferation.12-16,21 Upregulated TRPC6 plays an important role in the development of pulmonary vascular remodeling in IPAH patients.16
TRPC6 is a canonical transient receptor potential channel isoform believed to be involved in forming receptor-operated Ca2+ channels.17,22,23 Studies on TRPC6 knockout mice revealed that TRPC6 plays a central role in receptor-operated control of vascular smooth muscle tone.24 In cardiac myocytes, overexpressed TRPC6 plays an important role in forming a Ca2+-dependent calcineurin-NFAT-TRPC6 loop that leads to pathological cardiac hypertrophy and remodeling.25 In PASMCs, TRPC6 is involved in forming functional Ca2+ channels that are activated by vasoconstrictor and mitogenic factors.13,14,26 Downregulation of TRPC6 inhibits PASMC proliferation.13,16
Compared with normal subjects and normotensive patients, mRNA and protein expression levels of TRPC6 in lung tissue and PASMCs were significantly higher in IPAH patients, suggesting an abnormal TRPC6 gene transcription in these patients.16 In the present study, we found that the allele frequency of the −254(C→G) SNP in TRPC6 was significantly higher in IPAH patients than in normal subjects. In contrast, the allele frequencies of the 2 neighboring SNPs, −361(A→T) and −218(C→T), were comparable between normal subjects and IPAH patients. Moreover, 3.7% of IPAH patients possess a homozygous −254G/G genotype, whereas 1.3% of the normal subjects have the homozygous −254G/G genotype. These data imply that the −254(C→G) SNP may be responsible for the abnormal TRPC6 expression and function in a subpopulation of IPAH patients.
The C→G conversion in IPAH patients harboring the −254(C→G) SNP introduces a known NF-κB binding site (GGGGGTCTCC)27 in the 5′-regulatory region of TRPC6 and confers NF-κB–mediated transcriptional activation of TRPC6. The −254G-generated NF-κB binding sequence in IPAH PASMCs showed measurable binding affinity for the p50/p50 homodimer and p50/p65 heterodimer of the NF-κB complex. Furthermore, TNF-α–mediated activation of NF-κB significantly upregulated TRPC6 expression, increased the resting [Ca2+]cyt, and enhanced the agonist-induced Ca2+ influx in PASMCs from IPAH patients with the −254(C→G) SNP. Inhibition of NF-κB with a mutant dominant-negative IκBα markedly attenuated TNF-α–induced enhancement of TRPC6 expression, resting [Ca2+]cyt, and OAG-induced [Ca2+]cyt rise in IPAH PASMCs with the −254G allele. These results support a functional role for the −254(C→G) SNP in NF-κB–induced TRPC6 gene transcription and TRPC channel activity.
In addition, our observations showed that the −254(C→G) SNP significantly increased the basal promoter activity of TRPC6 that is not related to NF-κB. The increase in basal promoter activity occurred when the pBlue(−335/+110) construct was used and was maintained when the pBlue(−456/+110) and pBlue(−1682/+110) constructs were used. These results suggest that the −254(C→G) SNP not only enhances NF-κB–mediated TRPC6 transcription but also may enhance basal transcription of TRPC6 in PASMCs. Because many binding sites are present in the promoter region of TRPC6, it is possible that the −254G-generated NF-κB binding site may facilitate interactions of various transcription factors (eg, NFAT, AP-1) to increase TRPC6 transcription.25
The pathophysiological significance of insertion of an NF-κB binding site into the TRPC6 gene may underlie the potential linkage of immune or inflammatory responses to the upregulation of TRPC6 channels in the pulmonary vasculature. The NF-κB transcription factor family plays an essential role in immune and inflammatory responses and maintains an antiapoptotic function in normal and malignant cells.28,29 Given the fact that viral and bacterial infection, along with the inflammatory response resulting from the infection, is related to the development of the plexiform lesion and vascular remodeling in IPAH patients,30,31 the −254(C→G) SNP in TRPC6 may serve as an important genetic variation that links inflammatory response to the occurrence of IPAH and predisposes the −254G allele carriers to an increased risk for inflammation-mediated pulmonary arteriopathy. Furthermore, a potential exists that the −254(C→G) SNP in TRPC6 may portend more importantly for patients with PAH secondary to connective tissue diseases (eg, scleroderma), HIV/AIDS, and schistosomiasis, who bear a significant inflammatory burden. Further study is needed to define whether the −254(C→G) SNP in TRPC6 (or other genetic variations in genes involved in the NF-κB pathway) may be an important factor in separating the scleroderma (or HIV/AIDS or schistosomiasis) patients who develop pulmonary hypertension from those who do not.
IPAH appears to have a heterogeneous origin involving multiple genetic, molecular, and cellular abnormalities.2,3,32 SNPs in genes encoding BMPR2,7-9 activin receptor-like kinase 1 (ALK1),33 and serotonin transporter (5-HTT)6 have been linked to familial PAH and IPAH. Our data also link the −254(C→G) SNP in TRPC6 to IPAH and indicate that the heterozygous −254C/G and homozygous −254G/G genotypes are associated with the occurrence of IPAH. However, mutations or SNPs in all these genes have been found in only a small portion of IPAH patients. It seems that the abnormality in each of the genes is important by itself, but none of them is sufficient to cause the disease. Therefore, the causal and pathogenic mechanisms of IPAH may involve abnormalities in multiple genes and gene products. It may be explained by the notion of multiple-hit theory proposed by many investigators.2,34,35 For instance, inheritance of mutations in TRPC6 and other genes (eg, BMPR2, 5-HTT, ALK1), followed by exposure to viral infection,31,36 inflammatory factors,4,37 and anorexic drugs or anorexigens,38 results in pathogenic changes in the pulmonary vasculature and the development of IPAH.
Conclusions
We have identified a unique genetic variation, the −254(C→G) SNP in TRPC6, which may link the inflammatory response to upregulation of TRPC6, aberrant regulation of cytoplasmic Ca2+ in PASMCs, and ultimately alterations in the pulmonary vasculature. The enhanced transcriptional regulation of TRPC6 and augmented function of TRPC6 channels resulting from the −254(C→G) SNP may predispose individuals who have this mutation to an increased risk of developing IPAH.
CLINICAL PERSPECTIVE.
Genetic modifications of key genes have been associated with increased pulmonary vascular remodeling and incidence of disease in idiopathic pulmonary arterial hypertension (IPAH). We previously reported that upregulation of canonical transient receptor potential 6 (TRPC6) may be responsible for the abnormal pulmonary artery smooth muscle cell proliferation and pulmonary vascular medial hypertrophy in IPAH patients. This study identifies a gain-in-function single-nucleotide polymorphism (SNP) in the TRPC6 promoter −254(C→G) with a significantly higher allele frequency in IPAH patients from the USA and Germany. This −254(C→G) variation generates a functional binding site for nuclear factor-κB, which results in regulation of TRPC6 expression and function in modulating cytosolic Ca2+ levels in pulmonary artery smooth muscle cells. Examination of isolated pulmonary artery smooth muscle cells from IPAH patients further confirms that the presence of the −254(C→G) single-nucleotide polymorphism is linked to TRPC6 expression and function. These findings provide a strong putative link between the inflammatory response (or activated nuclear factor-κB), which has been suggested in part to underlie IPAH, and altered TRPC6 channel function, providing an alternative therapeutic strategy to treat IPAH patients and to prevent the occurrence of IPAH.
Supplementary Material
SUPPLEMENTAL MATERIAL
Determination of pulmonary hemodynamics. A flow directed, balloon tipped Swan-Ganz catheter was positioned into right ventricle and then pulmonary artery via the internal jugular vein. Hemodynamic measurements, such as mean PAP, were obtained by a pressure transducer (Namic/Boston Scientific, Natick, MA) connected to a Mac-Laboratory 7000 hemodynamic and electrocardiographic monitoring system (GE Medical System, Waukesha, WI). Cardiac output (CO) was measured by a thermodilution technique, and PVR was calculated according to the measured mean PAP, pulmonary artery wedge pressure (Pwedge) and CO by the monitoring system.
Supplementary Table 1. Demographic and hemodynamic characteristics of normal subjects and IPAH patients
The blood samples (for DNA extraction) from IPAH patients were obtained from IPAH patients underwent right heart catheterization. The normal subjects (non-smokers) had no history of cardiopulmonary disease, drug abuse, and HIV/AIDS. The blood samples from normal subjects were collected by phlebotomists, while the blood samples from IPAH patients were collected during right heart catheterization via the catheter.
Identification of SNP in TRPC6 gene promoter region. To identify SNPs in the 5′- promoter region of TRPC6, approximately a 2000-bp size of TRPC6 5'-regulatory region sequence and total TRPC6 gene sequence were obtained from the human chromosome 11 clone: RP11−223E3 (AP003080) in human genomic database. Six paired amplification PCR primers (Supplementary Table 2) were designed to amplify six overlapping DNA segments spanning 2000 bp upstream and 110 bp downstream of the transcriptional start site of human TRPC6.
PCR amplification of genomic DNA encoding the putative TRPC6 promoter region was performed by a GeneAmp PCR System (Perkin Elmer) using a Platinum PCR Supermix Kit (Invitrogen). The PCR products were detected by agarose gel electrophoresis, purified with a Gel extraction kit (Qiagen), and sequenced. The sequencing data were analyzed by Chromas software and compared with known SNPs deposited in the NCBI SNP databank. Vertebrate transcription factor binding sites within the 5’-regulatory region of human TRPC6 were identified by Dragon TF association miner (http://research.i2r.a-star.edu.sg/DRAGON/TFAM/v1.html).
Supplementary Table 2. PCR Primers used for amplifying six overlapping DNA segments of the human TRPC6 gene
Supplementary Table 3. Allele frequency of the TRPC6 promoter SNPs sorted by cohort
Preparation of cytoplasmic and nuclear extracts from PASMCs. Cytosolic and nuclear extracts from cultured PASMCs were collected using a modified protocol. Briefly, cells were washed with PBS and incubated in cytoplasmic extraction buffer [10 mM Tris-HCl (pH 7.9), 60 mM KCl, 1 mM EDTA, 0.4% NP-40, 1mM DTT and the protease inhibitor cocktail (Complete Mini, Roche Diagnostics)] for 10 min on ice, the samples were then scraped and collected. After centrifuged at 1000× g for 5 min, extracted soluble material representing the cytoplasmic fraction was isolated. The pellet was further extracted in the nuclear extraction buffer [50 mM Tris-HCl (pH 8.0), 410 mM NaCl, 1.5 mM MgCl2, 25% glycerol] for 10 min on ice to isolate the nuclear fraction. Extracted soluble material representing the nuclear fraction was isolated after centrifuged at 16000× g for 10 min at 4°C. The paired cytoplasmic and nuclear fractions from PASMCs were stored at −20°C before use.
RNA isolation and RT-PCR. Total RNA was isolated from PASMC using TRIzol reagent (Invitrogen). Two μg of RNA was used to synthesize first-strand cDNA using the Super-ScriptJ First-Strand Synthesis System (Invitrogen). PCR was performed by a GeneAmp PCR System (Perkin–Elmer) using the Platinum PCR Supermix (Invitrogen). The sense and antisense primers are specifically designed from the coding regions of TRPC genes (TRPC4 and TRPC6) and GAPDH (Supplementary Table 4).
Supplementary Table 4. Oligonucleotide sequences of primers used for RT-PCR*
Western blot analysis. The cytoplasmic extract (10 μg of protein), the nuclear extract (20 μg of protein) or total cell lysates (30−50 μg of protein) were electrophoresed on NuPAGE® 4−12% Bis-Tris gels (Invitrogen) and transferred to a nitrocellulose membrane. Polyclonal antibodies against p50, p65 (Upstate), IκBα (Cell Signaling) and TRPC6 (Chemicon); monoclonal antibodies against p84, histone H2, and GAPDH were used as primary antibodies. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG was used as secondary antibodies. The bound antibody was detected using an enhanced chemiluminescence detection system (Amersham).
Immunofluorescence staining. PASMCs on cover slips were fixed in 4% paraformaldehyde/PBS, permeabilized and exposed to rabbit anti-TRPC6 antibody (Chemicon). FITC-conjugated goat anti-rabbit IgG was used to detect TRPC6. HA epitope tag was stained by TRITC-conjugated anti-HA mouse antibody. The cells on cover slips were visualized using a confocal laser scanning microscope (LSM510 META, Zeiss).
Generation of recombinant adenovirus carrying human IκBαSR. The IκBα-super repressor (mutant IκBα, IκBSR) cDNA (S32A/S36A) was cloned into the pShuttle by digesting the pCMV-IκBαM with BamHI/HindIII and ligating the resulting fragments into the Xba1 site of the pShuttle vector. The pAdeno-X/IκBSR (Ad-IκB) was constructed by digesting pShuttle constructs with PI-SceI/I-CeuI and ligating the resulting fragments into the PI-SceI/I-CeuI sites of the pAdeno-X adenoviral vector. Recombinant adenoviral plasmids were packaged into infectious adenoviral particles by transfecting HEK-293 cells using LipofectAMINE PLUS reagent. The adenoviral particles were propagated in HEK-293 cells and purified on CsCl ultracentrifugation. Titers of the adenoviral stock were determined by standard plaque assay. Recombinant adenoviruses were screened for expression of the introduced gene by Western blot. pAdeno-X, which was the recombinant replication-incompetent adenovirus carrying no IκBSR cDNA insert, was used as a control adenovirus (Ad-null).
Generation of recombinant adenovirus carrying human TRPC6 specific siRNA. Recombinant adenovirus carrying siRNA targeting on human TRPC6 was generated using an Invitrogen Expression Kit. We also constructed DNA-based vector (pSilencer 2.0-U6-TRPC6) that expressed TRPC6 siRNA targeting on AAGGTCTTTATGCAATTGCTG (nucleotide 2214−2234 of TRPC6 mRNA, NM_004621). The siRNA expression DNA cassettes, including RNA Polymerase III promoter U6, a hairpin siRNA target sequence and terminator, were subcloned into pENTR1A entry vector (Invitrogen). A scrambled control vector and GAPDH specific siRNA vector were also constructed. After efficient recombination of the entry vector into the promoterless pAd/PL-DEST Gateway® vectors, followed by viral production and transduction. The adenoviral particles for TRPC6-siRNA, GAPDH-siRNA and scramble control were plaque-purified and amplified in 293A cells. The titer of the adenoviral stock was measured by spectrophotometer. For adenovirus infection experiments, cells were infected with the appropriate virus in SMBM containing 0.2% FBS for 4 hrs and used for experiments 24 to 48 hours after adenoviral infection.
Cytosolic [Ca2+] measurement. Cells were loaded with fura-2/AM (3 μM) in culture media for 30 min, then mounted in a perfusion chamber on the stage of a Zeiss Axiovert 35 microscope equipped with a 40× plan-Neofluar objective (NA 1.30 oil). Cells were then washed for 40 min by Ringer's solution to remove extracellular dye and allow intracellular esterases to cleave cytosolic fura 2-AM into active fura 2. Alternating 340 or 380 nm light was controlled by a wave length switcher (Lambda DG-4, Sutter Instruments) to elicit fura-2 fluorescence. A 400 nm dichroic mirror and 480 nm long-pass emission filter (Chroma) supplied light to a Photometrics CoolSNAP HQ CCD camera (Roper Scientific). Images were acquired with Metafluor software (Universal Imaging Corp.). [Ca2+]cyt was calculated from fura-2 fluorescent emission excited at 340 and 380 nm (F340/F380) using the ratio method based on the following equation: [Ca2+]cyt = Kd × (Sf2/Sb2) × (R − Rmin)/(Rmax − R), where Kd (225 nM) is the dissociation constant for Ca2+, Sf2 and Sb2 are emission fluorescence values at 380-nm excitation, R is the measured fluorescence ratio, and Rmin and Rmax are minimal and maximal ratios, respectively. The [Ca2+]cyt in each condition was determined as the average from 10 to 20 cells, and experiments were repeated at least four times.
Electrophysiological measurements. PASMCs were cultured on cover slips and mounted in a perfusable recording chamber on a Zeiss inverted microscope immediately before use. Membrane currents and capacitance were recorded with an EPC-9 amplifier coupled to ITC16 interface using whole-cell patch-clamp technique. Patch pipettes (2−2.5 MΩ) were pulled on a Sutter electrode puller using borosilicate glass capillary tubes, and fire polished on a Narishige microforge. Na+-containing Ringer solution was used as the extracellular (bath) solution and contained (in mM): NaCl 160, KCl 4, CaCl2 2, MgCl2 1, and HEPES 10 (pH 7.4). The pipette (intracellular) solution contained (in mM): cesium aspartate (CsAsp) 145, MgCl2 2, CaCl2 0.3, EGTA 10, and HPES 10 (pH 7.2 and pCa 8). Voltage ramps ranging from −100 mV to +100 mV were applied from a holding potential of 0 mV (which inactivates voltage-gated Na+ and Ca2+ channels). All experiments were performed at room temperature (24°C). The TRPC6 currents were activated by application of 100 μM OAG for 10−15 min.
The −254(G-to-C) SNP in normal subjects and IPAH patients in African Americans. In addition to Caucasian patients, the −254(C→G) SNP in the TRPC6 gene was also found in two African American IPAH patients; the frequency of the −254G allele in African American patients is also significantly higher than in African American controls. As shown in Supplementary Table 5, the allele frequency of the −254(C→G) SNP is 18.8% in African American IPAH patients (n=8; vs. 1.3% in African American normal controls, n=38). One of the two African American IPAH patients is a homozygote (−254G/G), and the other is a heterozygote (−254C/G). These data imply that the difference is not limited in Caucasians.
Supplement Table 5. The −254G allele frequency is significantly higher than in IPAH patients than in normal subjects in African Americans
Differential regulation of NF-κB expression in −254C/G and −254C/C IPAH patient PASMC. We have isolated PASMC from 4 IPAH patients, three of whom have a −254C/G genotype and one has the −254C/C genotype. Despite strenuous efforts, we have been unable to get tissue from a −254G/G allele patient prior to publication of the current study. Patient demographics and hemodynamics are reported in Supplement Table 6. Our observations imply that TRPC6 is upregulated in PASMC from all IPAH patients, however, NF-κB only upregulates TRPC6 expression in PASMC from patients who have a −254G allele.
Using PASMC from −254C/G and −254C/C individuals, we examined the regulation of TRPC6 expression by TNF-α. In the presence of TNF-α, the protein expression level of TRPC6 was not significantly different in the −254C/C IPAH PASMC infected with control adenovirus (Ad-null) and IκBα super-repressor (Ad-IκB), a nondegradable mutant dominant-negative IκBα variant that inhibits NF-κB activation and/or nuclear translocation of p65 and p50 (Supplementary Figure 1). However, in the −254C/G IPAH PASMC, the TRPC6 protein level was significantly reduced by IκBα super-repressor (Ad-IκB). These results suggest that NF-κB-mediated TRPC6 upregulation only occurs in PASMC from −254G allele patients.
Supplement Table 6. Demographics and hemodynamics of −254G and −254C IPAH patients with isolated PASMC.
Supplement Figure 1. The −254G allele regulates NF-κB-mediated protein TRPC6 upregulation. Representative gels depict data from nuclear (A) and cytoplasmic (B) protein fractions from TNF-treated PASMC isolated from heterozygous (−254C/G: top) and homozygous (−254C/C: bottom) IPAH patients. Both fractions were treated with (Ad-IkB) or without (Ad-null) IκBα super-repressor. Left. IκBα treatment inhibits significantly decreases nuclear translocation of the p65 and p50 in both −254C/G and −254C/C PASMC. Right. TRPC6 expression is evaluated in Ad-null and Ad-IκB treated cells. IκB expression is enhanced by Ad-IκB in both −254C/G and −254C/G cell types. However, only the −254C/G IPAH cells exhibit decreased TRPC6 protein expression. Data for the −254C/G patient is reproduced from Figure 7.
Acknowledgments
We thank Dr J.A. Kozak for helpful discussions and advice on the electrophysiological recording of TRPC6 currents and Drs H. Gall, S.S. Pullamsetti, H.T. Mueller, and T. Stoecker for technical assistance.
Footnotes
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/cgi/content/full/CIRCULATIONAHA.108.782458/DC1.
Sources of Funding
This work was supported in part by the American Heart Association (0630117N to Dr Yu) and the National Institutes of Health (HL64945, HL66012, and HL54043 to Dr Yuan; NS14609 to Dr Cahalan).
Disclosures
None.
References
- 1.Runo JR, Loyd JE. Primary pulmonary hypertension. Lancet. 2003;361:1533–1544. doi: 10.1016/S0140-6736(03)13167-4. [DOI] [PubMed] [Google Scholar]
- 2.Farber HW, Loscalzo J. Pulmonary arterial hypertension. N Engl J Med. 2004;351:1655–1665. doi: 10.1056/NEJMra035488. [DOI] [PubMed] [Google Scholar]
- 3.Newman JH, Fanburg BL, Archer SL, Badesch DB, Barst RJ, Garcia JGN, Kao PN, Knowles JA, Loyd JE, McGoon MD, Morse JH, Nichols WC, Rabinovitch M, Rodman DM, Troy Stevens T, Tuder RM, Voelkel NF, Gail DB. Pulmonary arterial hypertension: future directions: report of a National Heart, Lung and Blood Institute/Office of Rare Diseases workshop. Circulation. 2004;109:2947–2952. doi: 10.1161/01.CIR.0000132476.87231.6F. [DOI] [PubMed] [Google Scholar]
- 4.Robbins IM, Barst RJ, Rubin LJ, Gaine SP, Price PV, Morrow JD, Christman BW. Increased levels of prostaglandin D2 suggest macrophage activation in patients with primary pulmonary hypertension. Chest. 2001;120:1639–1644. doi: 10.1378/chest.120.5.1639. [DOI] [PubMed] [Google Scholar]
- 5.Yuan JX-J, Aldinger AM, Juhaszova M, Wang J, Conte JV, Jr, Gaine SP, Orens JB, Rubin LJ. Dysfunctional voltage-gated K+ channels in pulmonary artery smooth muscle cells of patients with primary pulmonary hypertension. Circulation. 1998;98:1400–1406. doi: 10.1161/01.cir.98.14.1400. [DOI] [PubMed] [Google Scholar]
- 6.Eddahibi S, Humbert M, Fadel E, Raffestin B, Darmon M, Capron F, Simonneau G, Dartevelle P, Hamon M, Adnot S. Serotonin transporter overexpression is responsible for pulmonary artery smooth muscle hyperplasia in primary pulmonary hypertension. J Clin Invest. 2001;108:1141–1150. doi: 10.1172/JCI12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane KB, Machado RD, Pauciulo MW, Thomson JR, Phillips JA, III, Loyd JE, Nichols WC, Trembath RC, for the IPPH Consortium Heterozygous germline mutations in BMPR2, encoding a TGF-β receptor, cause familial primary pulmonary hypertension. Nat Genet. 2000;26:81–84. doi: 10.1038/79226. [DOI] [PubMed] [Google Scholar]
- 8.Morse JH, Jones AC, Barst RJ, Hodge SE, Wilhelmsen KC, Nygaard TG. Mapping of familial primary pulmonary hypertension locus (PPH1)to chromosome 2q31-q32. Circulation. 1997;95:2603–2606. doi: 10.1161/01.cir.95.12.2603. [DOI] [PubMed] [Google Scholar]
- 9.Newman JH, Wheeler L, Lane KB, Loyd E, Gaddipati R, Phillips JA, III, Loyd JE. Mutation in the gene for bone morphogenetic protein receptor II as a cause of primary pulmonary hypertension in large kindred. N Engl J Med. 2001;345:319–324. doi: 10.1056/NEJM200108023450502. [DOI] [PubMed] [Google Scholar]
- 10.Stenmark KR, Mecham RP. Cellular and molecular mechanisms of pulmonary vascular remodeling. Annu Rev Physiol. 1997;59:89–144. doi: 10.1146/annurev.physiol.59.1.89. [DOI] [PubMed] [Google Scholar]
- 11.Rich S, Kaufmann E, Levy PS. The effect of high doses of calcium-channel blockers on survival in primary pulmonary hypertension. N Engl J Med. 1992;327:76–81. doi: 10.1056/NEJM199207093270203. [DOI] [PubMed] [Google Scholar]
- 12.Golovina VA, Platoshyn O, Bailey CL, Wang J, Limsuwan A, Sweeney M, Rubin LJ, Yuan JX-J. Upregulated TRP and enhanced capacitative Ca2+ entry in human pulmonary artery myocytes during proliferation. Am J Physiol Heart Circ Physiol. 2001;280:H746–H755. doi: 10.1152/ajpheart.2001.280.2.H746. [DOI] [PubMed] [Google Scholar]
- 13.Yu Y, Sweeney M, Zhang S, Platoshyn O, Landsberg J, Rothman A, Yuan JX-J. PDGF stimulates pulmonary vascular smooth muscle cell proliferation by upregulating TRPC6 expression. Am J Physiol Cell Physiol. 2003;284:C316–C330. doi: 10.1152/ajpcell.00125.2002. [DOI] [PubMed] [Google Scholar]
- 14.Lin M-J, Leung GPH, Zhang W-M, Yang X-R, Yip K-P, Tse C-M, Sham JSK. Chronic hypoxia-induced upregulation of store-operated and receptor-operated Ca2+ channels in pulmonary arterial smooth muscle cells: a novel mechanism of hypoxic pulmonary hypertension. Circ Res. 2004;95:496–505. doi: 10.1161/01.RES.0000138952.16382.ad. [DOI] [PubMed] [Google Scholar]
- 15.Dietrich A, Chubanov V, Kalwa H, Rost BR, Gudermann T. Cation channels of the transient receptor potential superfamily: their role in physiological and pathophysiological processes of smooth muscle cells. Pharmacol Ther. 2006;112:744–760. doi: 10.1016/j.pharmthera.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Yu Y, Fantozzi I, Remillard CV, Landsberg JW, Kunichika N, Platoshyn O, Tigno DD, Thistlethwaite PA, Rubin LJ, Yuan JX-J. Enhanced expression of transient receptor potential channels in idiopathic pulmonary arterial hypertension. Proc Natl Acad Sci U S A. 2004;101:13861–13866. doi: 10.1073/pnas.0405908101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nilius B, Owsianik G, Voets T, Peters JA. Transient receptor potential cation channels in disease. Physiol Rev. 2007;87:165–217. doi: 10.1152/physrev.00021.2006. [DOI] [PubMed] [Google Scholar]
- 18.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marasa BS, Rao JN, Zou T, Liu L, Keledjian KM, Zhang A-H, Xiao L, Chen J, Turner DJ, Wang J-Y. Induced TRPC1 expression sensitizes intestinal epithelial cells to apoptosis by inhibiting NF-κB activation through Ca2+ influx. Biochem J. 2006;397:77–87. doi: 10.1042/BJ20060124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Saffen D. Muscarinic acetylcholine receptor regulation of TRP6 Ca2+ channel isoforms: molecular structures and functional characterization. J Biol Chem. 2001;276:13331–13339. doi: 10.1074/jbc.M008914200. [DOI] [PubMed] [Google Scholar]
- 21.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 22.Clapham DE, Runnels LW, Strübing C. The TRP ion channel family. Nat Rev Neurosci. 2001;2:387–396. doi: 10.1038/35077544. [DOI] [PubMed] [Google Scholar]
- 23.Inoue R, Jensen LJ, Shi J, Morita H, Nishida M, Honda A, Ito Y. Transient receptor potential channels in cardiovascular function and disease. Circ Res. 2006;99:119–131. doi: 10.1161/01.RES.0000233356.10630.8a. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich A, Mederos y Schnitzler M, Gollasch M, Gross V, Storch U, Dubrovska G, Lauterbach B, Herz U, Obst M, Essin K, Renz H, Luft FC, Gudermann T, Birnbaumer L. Increased vascular smooth muscle contractility in TRPC6−/− mice. Mol Cell Biol. 2005;25:6980–6989. doi: 10.1128/MCB.25.16.6980-6989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuwahara K, Wang Y, McAnally J, Richardson JA, Bassel-Duby R, Hill JA, Olson EN. TRPC6 fulfills a calcineurin signaling circuit during pathologic cardiac remodeling. J Clin Invest. 2006;116:3114–3126. doi: 10.1172/JCI27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Weigand L, Lu W, Sylvester JT, Semenza GL, Shimoda LA. Hypoxia inducible factor 1 mediates hypoxia-induced TRPC expression and elevated intracellular Ca2+ in pulmonary arterial smooth muscle cells. Circ Res. 2006;98:1528–1537. doi: 10.1161/01.RES.0000227551.68124.98. [DOI] [PubMed] [Google Scholar]
- 27.Glasgow JN, Wood T, Perez-Polo JR. Identification and characterization of nuclear factor κB binding sites in the murine bcl-X promoter. J Neurochem. 2000;75:1377–1389. doi: 10.1046/j.1471-4159.2000.0751377.x. [DOI] [PubMed] [Google Scholar]
- 28.Baldwin ASJ. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 29.Karin M, Greten FR. NF-κB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 30.Dorfmuller P, Perros F, Balabanian K, Humbert M. Inflammation in pulmonary arterial hypertension. Eur Respir J. 2003;22:358–363. doi: 10.1183/09031936.03.00038903. [DOI] [PubMed] [Google Scholar]
- 31.Cool CD, Rai PR, Yeager ME, Hernandez-Saavedra D, Serls AE, Bull TM, Geraci MW, Brown KK, Routes JM, Tuder RM, Voelkel NF. Expression of human herpesvirus 8 in primary pulmonary hypertension. N Engl J Med. 2003;349:1113–1122. doi: 10.1056/NEJMoa035115. [DOI] [PubMed] [Google Scholar]
- 32.Rubin LJ. Primary pulmonary hypertension. N Engl J Med. 1997;336:111–117. doi: 10.1056/NEJM199701093360207. [DOI] [PubMed] [Google Scholar]
- 33.Trembath RC, Thomson JR, Machado RD, Morgan NV, Atkinson C, Winship I, Simonneau G, Galie N, Loyd JE, Humbert M, Nichols WC, Berg J, Manes A, McGaughran J, Pauciulo M, Wheeler L, Morrell NW. Clinical and molecular genetic features of pulmonary hypertension in patients with hereditary hemorrhagic telangiectasia. N Engl J Med. 2001;345:325–334. doi: 10.1056/NEJM200108023450503. [DOI] [PubMed] [Google Scholar]
- 34.Long L, MacLean MR, Jeffery TK, Morecroft I, Yang X, Rudarakanchana N, Southwood M, James V, Trembath RC, Morrell NW. Serotonin increases susceptibility to pulmonary hypertension in BMPR2-deficient mice. Circ Res. 2006;98:818–827. doi: 10.1161/01.RES.0000215809.47923.fd. [DOI] [PubMed] [Google Scholar]
- 35.Song Y, Jones JE, Beppu H, Keaney JF, Jr, Loscalzo J, Zhang Y-Y. Increased susceptibility to pulmonary hypertension in heterozygous BMPR2-mutant mice. Circulation. 2005;112:553–562. doi: 10.1161/CIRCULATIONAHA.104.492488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellicelli AM, Palmieri F, Cicalini S, Petrosillo N. Pathogenesis of HIV-related pulmonary hypertension. Ann N Y Acad Sci. 2001;946:82–94. doi: 10.1111/j.1749-6632.2001.tb03904.x. [DOI] [PubMed] [Google Scholar]
- 37.Humbert M, Monti G, Brenot F, Sitbon O, Portier A, Grangeot-Keros L, Durous P, Galanaud P, Simonneau G, Emilie D. Increased interleukin-1 and interleukin-6 serum concentrations in severe primary pulmonary hypertension. Am J Respir Crit Care Med. 1995;151:1628–1631. doi: 10.1164/ajrccm.151.5.7735624. [DOI] [PubMed] [Google Scholar]
- 38.Abenhaim L, Moride Y, Brenot F, Rich S, Benichou J, Kurz X, Higenbottam T, Oakley C, Wouters E, Aubier M, Simonneau G, Bégaud B. Appetite-suppressant drugs and the risk of primary pulmonary hypertension. N Engl J Med. 1996;335:609–616. doi: 10.1056/NEJM199608293350901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL MATERIAL
Determination of pulmonary hemodynamics. A flow directed, balloon tipped Swan-Ganz catheter was positioned into right ventricle and then pulmonary artery via the internal jugular vein. Hemodynamic measurements, such as mean PAP, were obtained by a pressure transducer (Namic/Boston Scientific, Natick, MA) connected to a Mac-Laboratory 7000 hemodynamic and electrocardiographic monitoring system (GE Medical System, Waukesha, WI). Cardiac output (CO) was measured by a thermodilution technique, and PVR was calculated according to the measured mean PAP, pulmonary artery wedge pressure (Pwedge) and CO by the monitoring system.
Supplementary Table 1. Demographic and hemodynamic characteristics of normal subjects and IPAH patients
The blood samples (for DNA extraction) from IPAH patients were obtained from IPAH patients underwent right heart catheterization. The normal subjects (non-smokers) had no history of cardiopulmonary disease, drug abuse, and HIV/AIDS. The blood samples from normal subjects were collected by phlebotomists, while the blood samples from IPAH patients were collected during right heart catheterization via the catheter.
Identification of SNP in TRPC6 gene promoter region. To identify SNPs in the 5′- promoter region of TRPC6, approximately a 2000-bp size of TRPC6 5'-regulatory region sequence and total TRPC6 gene sequence were obtained from the human chromosome 11 clone: RP11−223E3 (AP003080) in human genomic database. Six paired amplification PCR primers (Supplementary Table 2) were designed to amplify six overlapping DNA segments spanning 2000 bp upstream and 110 bp downstream of the transcriptional start site of human TRPC6.
PCR amplification of genomic DNA encoding the putative TRPC6 promoter region was performed by a GeneAmp PCR System (Perkin Elmer) using a Platinum PCR Supermix Kit (Invitrogen). The PCR products were detected by agarose gel electrophoresis, purified with a Gel extraction kit (Qiagen), and sequenced. The sequencing data were analyzed by Chromas software and compared with known SNPs deposited in the NCBI SNP databank. Vertebrate transcription factor binding sites within the 5’-regulatory region of human TRPC6 were identified by Dragon TF association miner (http://research.i2r.a-star.edu.sg/DRAGON/TFAM/v1.html).
Supplementary Table 2. PCR Primers used for amplifying six overlapping DNA segments of the human TRPC6 gene
Supplementary Table 3. Allele frequency of the TRPC6 promoter SNPs sorted by cohort
Preparation of cytoplasmic and nuclear extracts from PASMCs. Cytosolic and nuclear extracts from cultured PASMCs were collected using a modified protocol. Briefly, cells were washed with PBS and incubated in cytoplasmic extraction buffer [10 mM Tris-HCl (pH 7.9), 60 mM KCl, 1 mM EDTA, 0.4% NP-40, 1mM DTT and the protease inhibitor cocktail (Complete Mini, Roche Diagnostics)] for 10 min on ice, the samples were then scraped and collected. After centrifuged at 1000× g for 5 min, extracted soluble material representing the cytoplasmic fraction was isolated. The pellet was further extracted in the nuclear extraction buffer [50 mM Tris-HCl (pH 8.0), 410 mM NaCl, 1.5 mM MgCl2, 25% glycerol] for 10 min on ice to isolate the nuclear fraction. Extracted soluble material representing the nuclear fraction was isolated after centrifuged at 16000× g for 10 min at 4°C. The paired cytoplasmic and nuclear fractions from PASMCs were stored at −20°C before use.
RNA isolation and RT-PCR. Total RNA was isolated from PASMC using TRIzol reagent (Invitrogen). Two μg of RNA was used to synthesize first-strand cDNA using the Super-ScriptJ First-Strand Synthesis System (Invitrogen). PCR was performed by a GeneAmp PCR System (Perkin–Elmer) using the Platinum PCR Supermix (Invitrogen). The sense and antisense primers are specifically designed from the coding regions of TRPC genes (TRPC4 and TRPC6) and GAPDH (Supplementary Table 4).
Supplementary Table 4. Oligonucleotide sequences of primers used for RT-PCR*
Western blot analysis. The cytoplasmic extract (10 μg of protein), the nuclear extract (20 μg of protein) or total cell lysates (30−50 μg of protein) were electrophoresed on NuPAGE® 4−12% Bis-Tris gels (Invitrogen) and transferred to a nitrocellulose membrane. Polyclonal antibodies against p50, p65 (Upstate), IκBα (Cell Signaling) and TRPC6 (Chemicon); monoclonal antibodies against p84, histone H2, and GAPDH were used as primary antibodies. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG was used as secondary antibodies. The bound antibody was detected using an enhanced chemiluminescence detection system (Amersham).
Immunofluorescence staining. PASMCs on cover slips were fixed in 4% paraformaldehyde/PBS, permeabilized and exposed to rabbit anti-TRPC6 antibody (Chemicon). FITC-conjugated goat anti-rabbit IgG was used to detect TRPC6. HA epitope tag was stained by TRITC-conjugated anti-HA mouse antibody. The cells on cover slips were visualized using a confocal laser scanning microscope (LSM510 META, Zeiss).
Generation of recombinant adenovirus carrying human IκBαSR. The IκBα-super repressor (mutant IκBα, IκBSR) cDNA (S32A/S36A) was cloned into the pShuttle by digesting the pCMV-IκBαM with BamHI/HindIII and ligating the resulting fragments into the Xba1 site of the pShuttle vector. The pAdeno-X/IκBSR (Ad-IκB) was constructed by digesting pShuttle constructs with PI-SceI/I-CeuI and ligating the resulting fragments into the PI-SceI/I-CeuI sites of the pAdeno-X adenoviral vector. Recombinant adenoviral plasmids were packaged into infectious adenoviral particles by transfecting HEK-293 cells using LipofectAMINE PLUS reagent. The adenoviral particles were propagated in HEK-293 cells and purified on CsCl ultracentrifugation. Titers of the adenoviral stock were determined by standard plaque assay. Recombinant adenoviruses were screened for expression of the introduced gene by Western blot. pAdeno-X, which was the recombinant replication-incompetent adenovirus carrying no IκBSR cDNA insert, was used as a control adenovirus (Ad-null).
Generation of recombinant adenovirus carrying human TRPC6 specific siRNA. Recombinant adenovirus carrying siRNA targeting on human TRPC6 was generated using an Invitrogen Expression Kit. We also constructed DNA-based vector (pSilencer 2.0-U6-TRPC6) that expressed TRPC6 siRNA targeting on AAGGTCTTTATGCAATTGCTG (nucleotide 2214−2234 of TRPC6 mRNA, NM_004621). The siRNA expression DNA cassettes, including RNA Polymerase III promoter U6, a hairpin siRNA target sequence and terminator, were subcloned into pENTR1A entry vector (Invitrogen). A scrambled control vector and GAPDH specific siRNA vector were also constructed. After efficient recombination of the entry vector into the promoterless pAd/PL-DEST Gateway® vectors, followed by viral production and transduction. The adenoviral particles for TRPC6-siRNA, GAPDH-siRNA and scramble control were plaque-purified and amplified in 293A cells. The titer of the adenoviral stock was measured by spectrophotometer. For adenovirus infection experiments, cells were infected with the appropriate virus in SMBM containing 0.2% FBS for 4 hrs and used for experiments 24 to 48 hours after adenoviral infection.
Cytosolic [Ca2+] measurement. Cells were loaded with fura-2/AM (3 μM) in culture media for 30 min, then mounted in a perfusion chamber on the stage of a Zeiss Axiovert 35 microscope equipped with a 40× plan-Neofluar objective (NA 1.30 oil). Cells were then washed for 40 min by Ringer's solution to remove extracellular dye and allow intracellular esterases to cleave cytosolic fura 2-AM into active fura 2. Alternating 340 or 380 nm light was controlled by a wave length switcher (Lambda DG-4, Sutter Instruments) to elicit fura-2 fluorescence. A 400 nm dichroic mirror and 480 nm long-pass emission filter (Chroma) supplied light to a Photometrics CoolSNAP HQ CCD camera (Roper Scientific). Images were acquired with Metafluor software (Universal Imaging Corp.). [Ca2+]cyt was calculated from fura-2 fluorescent emission excited at 340 and 380 nm (F340/F380) using the ratio method based on the following equation: [Ca2+]cyt = Kd × (Sf2/Sb2) × (R − Rmin)/(Rmax − R), where Kd (225 nM) is the dissociation constant for Ca2+, Sf2 and Sb2 are emission fluorescence values at 380-nm excitation, R is the measured fluorescence ratio, and Rmin and Rmax are minimal and maximal ratios, respectively. The [Ca2+]cyt in each condition was determined as the average from 10 to 20 cells, and experiments were repeated at least four times.
Electrophysiological measurements. PASMCs were cultured on cover slips and mounted in a perfusable recording chamber on a Zeiss inverted microscope immediately before use. Membrane currents and capacitance were recorded with an EPC-9 amplifier coupled to ITC16 interface using whole-cell patch-clamp technique. Patch pipettes (2−2.5 MΩ) were pulled on a Sutter electrode puller using borosilicate glass capillary tubes, and fire polished on a Narishige microforge. Na+-containing Ringer solution was used as the extracellular (bath) solution and contained (in mM): NaCl 160, KCl 4, CaCl2 2, MgCl2 1, and HEPES 10 (pH 7.4). The pipette (intracellular) solution contained (in mM): cesium aspartate (CsAsp) 145, MgCl2 2, CaCl2 0.3, EGTA 10, and HPES 10 (pH 7.2 and pCa 8). Voltage ramps ranging from −100 mV to +100 mV were applied from a holding potential of 0 mV (which inactivates voltage-gated Na+ and Ca2+ channels). All experiments were performed at room temperature (24°C). The TRPC6 currents were activated by application of 100 μM OAG for 10−15 min.
The −254(G-to-C) SNP in normal subjects and IPAH patients in African Americans. In addition to Caucasian patients, the −254(C→G) SNP in the TRPC6 gene was also found in two African American IPAH patients; the frequency of the −254G allele in African American patients is also significantly higher than in African American controls. As shown in Supplementary Table 5, the allele frequency of the −254(C→G) SNP is 18.8% in African American IPAH patients (n=8; vs. 1.3% in African American normal controls, n=38). One of the two African American IPAH patients is a homozygote (−254G/G), and the other is a heterozygote (−254C/G). These data imply that the difference is not limited in Caucasians.
Supplement Table 5. The −254G allele frequency is significantly higher than in IPAH patients than in normal subjects in African Americans
Differential regulation of NF-κB expression in −254C/G and −254C/C IPAH patient PASMC. We have isolated PASMC from 4 IPAH patients, three of whom have a −254C/G genotype and one has the −254C/C genotype. Despite strenuous efforts, we have been unable to get tissue from a −254G/G allele patient prior to publication of the current study. Patient demographics and hemodynamics are reported in Supplement Table 6. Our observations imply that TRPC6 is upregulated in PASMC from all IPAH patients, however, NF-κB only upregulates TRPC6 expression in PASMC from patients who have a −254G allele.
Using PASMC from −254C/G and −254C/C individuals, we examined the regulation of TRPC6 expression by TNF-α. In the presence of TNF-α, the protein expression level of TRPC6 was not significantly different in the −254C/C IPAH PASMC infected with control adenovirus (Ad-null) and IκBα super-repressor (Ad-IκB), a nondegradable mutant dominant-negative IκBα variant that inhibits NF-κB activation and/or nuclear translocation of p65 and p50 (Supplementary Figure 1). However, in the −254C/G IPAH PASMC, the TRPC6 protein level was significantly reduced by IκBα super-repressor (Ad-IκB). These results suggest that NF-κB-mediated TRPC6 upregulation only occurs in PASMC from −254G allele patients.
Supplement Table 6. Demographics and hemodynamics of −254G and −254C IPAH patients with isolated PASMC.
Supplement Figure 1. The −254G allele regulates NF-κB-mediated protein TRPC6 upregulation. Representative gels depict data from nuclear (A) and cytoplasmic (B) protein fractions from TNF-treated PASMC isolated from heterozygous (−254C/G: top) and homozygous (−254C/C: bottom) IPAH patients. Both fractions were treated with (Ad-IkB) or without (Ad-null) IκBα super-repressor. Left. IκBα treatment inhibits significantly decreases nuclear translocation of the p65 and p50 in both −254C/G and −254C/C PASMC. Right. TRPC6 expression is evaluated in Ad-null and Ad-IκB treated cells. IκB expression is enhanced by Ad-IκB in both −254C/G and −254C/G cell types. However, only the −254C/G IPAH cells exhibit decreased TRPC6 protein expression. Data for the −254C/G patient is reproduced from Figure 7.