SUMMARY
To test the gearbox model of HAMP signaling in the E. coli serine receptor, Tsr, we generated a series of amino acid replacements at each residue of the AS1 and AS2 helices. The residues most critical for Tsr function defined hydrophobic packing faces consistent with a 4-helix bundle. Suppression patterns of helix lesions conformed to the the predicted packing layers in the bundle. Although the properties and patterns of most AS1 and AS2 lesions were consistent with both proposed gearbox structures, some mutational features specifically indicate the functional importance of an x-da bundle over an alternative a-d bundle. These genetic data suggest that HAMP signaling could simply involve changes in the stability of its x-da bundle. We propose that Tsr HAMP controls output signals by modulating destabilizing phase clashes between the AS2 helices and the adjoining kinase control helices. Our model further proposes that chemoeffectors regulate HAMP bundle stability through a control cable connection between the transmembrane segments and AS1 helices. Attractant stimuli, which cause inward piston displacements in chemoreceptors, should reduce cable tension, thereby stabilizing the HAMP bundle. This study shows how transmembrane signaling and HAMP input-output control could occur without the helix rotations central to the gearbox model.
Keywords: chemotaxis, transmembrane signaling, piston model, gearbox model, conformational suppression
INTRODUCTION
Many transmembrane signaling proteins in prokaryotes contain a 50-residue HAMP motif, so called because it occurs in histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins (MCPs) and certain phosphatases. HAMP sequences have few conserved amino acid residues, but characteristic secondary structure features: two amphiphilic helices (AS1, AS2) joined by a non-helical connector (CTR) (Butler & Falke, 1998; Aravind & Ponting, 1999; Williams & Stewart, 1999). In transmembrane signaling proteins, the HAMP domain typically lies near the inner face of the cytoplasmic membrane, between the periplasmic sensing and cytoplasmic signaling portions of the molecule, where it plays a central role in converting stimulus-induced conformational changes into behavior-controlling output signals [see (Hazelbauer et al., 2008) for a recent review].
Chemoreceptors of the MCP family provide a tractable system for investigating the mechanism of HAMP-mediated input-output control. Native MCP molecules are homodimers of ~550-residue subunits organized into three functional modules (Fig. 1A): a transmembrane sensing module composed of a periplasmic ligand binding domain and four transmembrane helices, a signal-converting HAMP domain that adjoins one of the transmembrane helices in each subunit, and a 4-helix bundle kinase control module connected to the HAMP domain. The kinase control portion of MCP molecules is highly conserved in sequence (Alexander & Zhulin, 2007) and tertiary structure (Kim et al., 1999; Park et al., 2006; Pollard et al., 2009) and regulates the autophosphorylation activity of the histidine kinase, CheA. The kinase control module typically contains: 4-6 glutamic acid residues that are sites for adaptational modification (methylation and demethylation); a flexible bundle segment with a glycine hinge; and a hairpin tip that interacts with other MCP molecules, with CheA, and with CheW, a protein that couples CheA to receptor control [see (Hazelbauer et al., 2008) for a recent review].
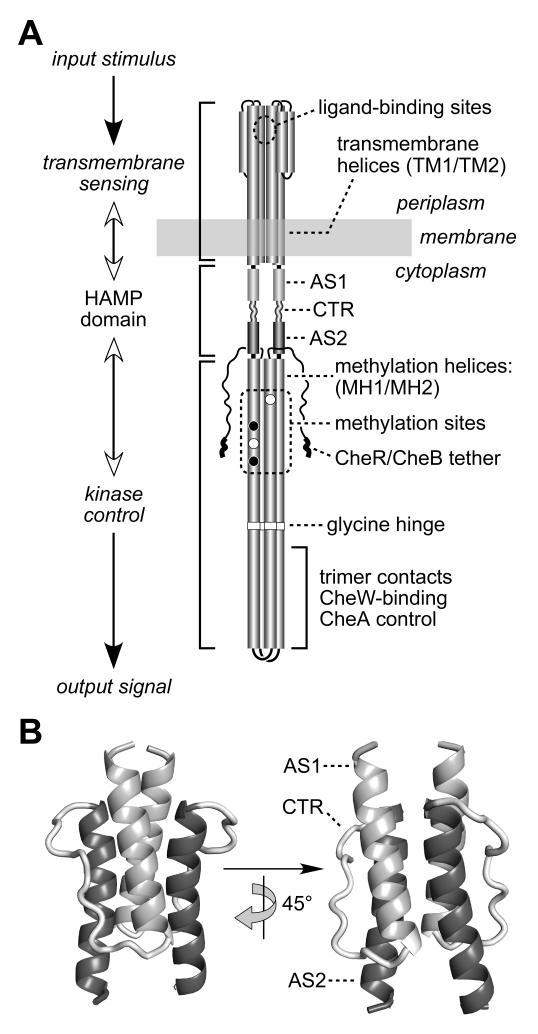
Fig. 1. Functional architecture of MCP molecules.
A) Schematic model of the Tsr homodimer. The CheR/CheB tether (NWETF pentapeptide) marks the C-terminus of each subunit. Helical segments are represented as thickened cylinders, whose relative lengths are approximately to scale. Each Tsr subunit has two transmembrane helices (TM1, TM2) and two methylation helices (MH1, MH2) containing four principal methylation sites; two (black circles) are synthesized as glutamines and subsequently deamidated to glutamates (white circles).
B) Modeled structure of the Tsr HAMP domain. Atomic coordinates were obtained by threading the Tsr HAMP sequence onto the Af1503 HAMP structure (Hulko et al., 2006). The AS1 helix is labeled at its N-terminus; the AS2 helix is labeled at its C-terminus; CTR is the connector segment joining the two helices.
E. coli has two amino acid-sensing MCPs, Tar (aspartate) and Tsr (serine) that have been extensively studied (Hazelbauer et al., 2008). Both receptors contain two symmetric, negatively cooperative ligand binding sites at the dimer interface (Lee et al., 1988; Milburn et al., 1991; Biemann & Koshland, 1994; Lin et al., 1994). In the unbound state, Tar and Tsr activate their associated CheA molecules (the kinase-on state). Binding of one attractant molecule produces an inward displacement of one of the transmembrane segments adjoining the HAMP domain (Falke & Hazelbauer, 2001). HAMP converts this piston-like movement into a conformational change of the kinase control region that deactivates CheA to elicit an appropriate chemotactic response. Locomotor behavior and CheA activity are subsequently reset to prestimulus values by a sensory adaptation system that adjusts the detection sensitivity and dynamic range of the MCPs through reversible, feedback-controlled covalent modifications. Methylation or demethylation of the adaptation sites interposed between HAMP and the kinase control region influences both kinase activity and positioning of the signaling transmembrane helix, demonstrating bidirectional conformational control through HAMP (Lai et al., 2006).
A recently determined nuclear magnetic resonance structure for an isolated HAMP domain from Af1503, a thermophile protein of unknown function, revealed a dimeric, parallel 4-helix bundle, with the connector segments encircling helices from the same subunit (Hulko et al., 2006) (Fig. 1B). The amino acid sequences of MCP HAMP domains readily thread onto the Af1503 HAMP structure and MCP HAMP domains exhibit structural features consistent with the Af1503 bundle (Swain & Falke, 2007; Ames et al., 2008; Doebber et al., 2008; Watts et al., 2008). However, the nature of HAMP signaling states and their role in input-output signaling are still poorly understood. The report of the Af1503 HAMP structure also presented a 2-state working model for HAMP, dubbed the gearbox model, in which the kinase-on and kinase-off output states corresponded to alternative HAMP bundle structures, interconvertible by concerted 26° counter-rotations of the HAMP helices (Hulko et al., 2006). To date, there has been little published evidence for or against the gearbox model.
To explore the in vivo structure and function of a HAMP domain in a canonical chemoreceptor, and to experimentally address the gearbox model, we surveyed each residue in the AS1 and AS2 helices of Tsr to determine: (i) whether functionally critical residues corresponded to those predicted to participate in structure-stabilizing packing interactions in the 4-helix HAMP bundle; (ii) whether function-disrupting lesions could be suppressed by compensatory structural changes in predicted partner helix residues; and (iii) whether the patterns of mutations and suppression effects were consistent with the gearbox model. Our findings indicate that the Tsr HAMP domain has a functionally important in vivo structure that closely resembles the 4-helix bundle of Af1503 HAMP. However, that bundle may be the only functionally important Tsr-HAMP structure; our genetic evidence provided no strong support for the alternative bundle structure proposed in the gearbox model. Accordingly, we propose that HAMP signaling could simply involve modulated stability changes in the HAMP bundle. To stimulate further work in the field, we present possible mechanisms for the control interactions between Tsr HAMP and its adjoining input and output domains.
RESULTS
Mutational scan of the Tsr-HAMP helices
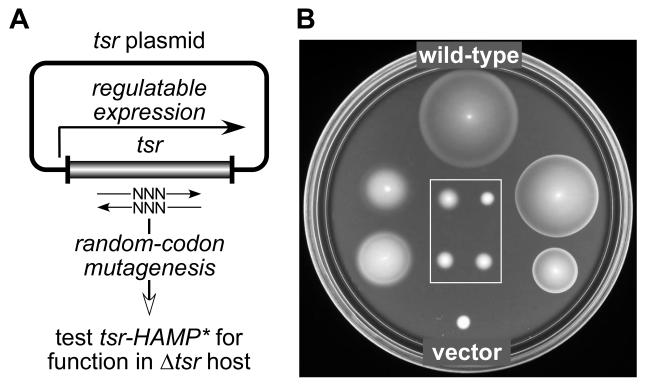
To identify the AS1 and AS2 residues most critical for Tsr function in vivo, we carried out a codon-by-codon mutagenesis of the tsr regions encoding residues K215-A233 (encompassing AS1) and E248-R265 (encompassing AS2) in two regulatable tsr expression plasmids, pRR53 and pPA114 (Fig. 2A). At optimal inducer concentrations (100 μM IPTG or 0.6 μM sodium salicylate, respectively), pRR53 and pPA114 confer wild-type Tsr function to UU1250, a receptorless host strain carrying deletions of all five E. coli MCP-family genes (tsr, tar, tap, trg, aer). Candidate mutant plasmids were tested in strain UU1250 for Tsr function at optimal induction conditions on tryptone soft agar plates. On the basis of these initial chemotaxis tests, each mutant plasmid was assigned to one of three functional categories: Tsr+ (colony size and morphology very similar to wild-type); Tsr± (reduced colony size and/or mutant ring morphology; see examples in Fig. 2B); and Tsr− (complete loss of function, comparable to the vector control phenotype; see boxed examples in Fig. 2B).
Fig. 2. Isolation of Tsr-HAMP mutants.
A) Strategy for constructing mutations. Mutations were created by randomizing the sequences of individual HAMP codons in plasmids carrying the tsr coding region under inducible expression control. Candidate plasmids were tested for Tsr function in UU1250, a receptorless host that lacks tsr and other MCP family receptors.
B) Examples of mutant Tsr-HAMP phenotypes. UU1250 transformants from (A) were tested for Tsr function on tryptone soft agar plates containing 12.5 μg/ml chloramphenicol and 0.6 μM sodium salicylate and incubated at 30°C for 7 hours. The wild-type control was pPA114; the vector control was pKG116. Eight mutant examples are shown: The two on the left (I229Y, M249S) and the two on the right (I226M, S255E) exhibit reduced or impaired Tsr function (i.e., Tsr±). The four boxed mutants have null defects (i.e., Tsr−).
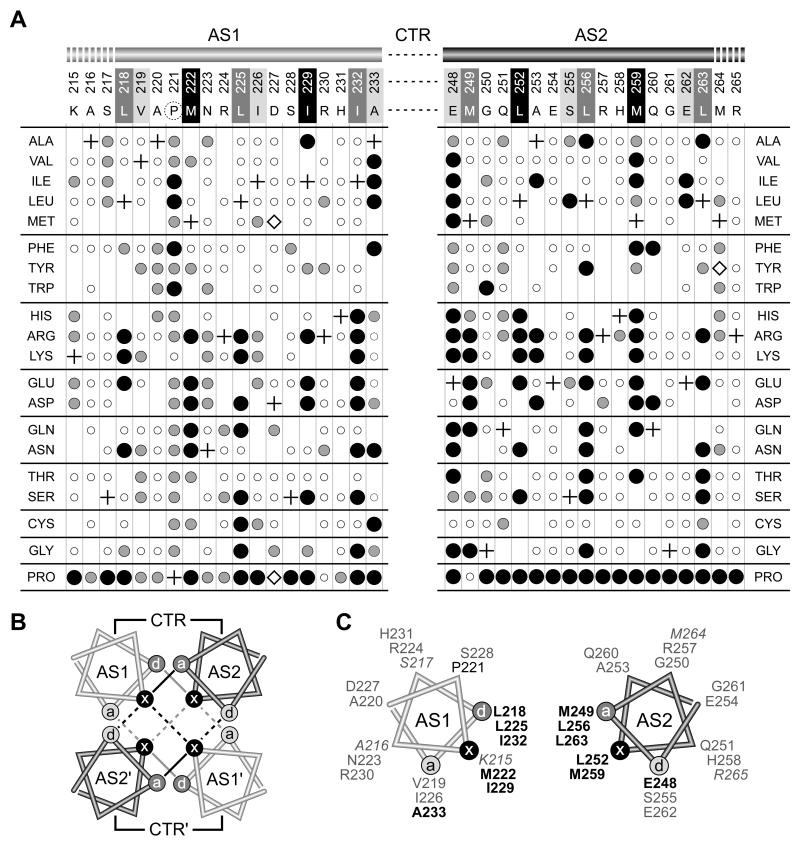
We obtained about 13 mutant amino acids at each of the residues surveyed, sufficient to identify functionally critical helix residues and their important side-chain characteristics (Fig. 3). We define important residues as those at which a majority of amino acid replacements demonstrably impaired Tsr function. Among those, we consider particularly critical residues to be the ones at which a majority of the deleterious replacements produced a complete loss-of-function (null) phenotype. By these criteria, AS1 has one important (P221) and six critical (L218, M222, L225, I229, I232, A233) residues; AS2 has six critical residues (E248, M249, L252, L256, M259, L263) (Fig. 3A).
Fig. 3. Mutant phenotypes and packing faces of AS1 and AS2 helices in Tsr HAMP.
A) AS1 and AS2 missense mutants. Residue numbers and wild-type amino acids of Tsr HAMP are shown at the top of the figure. In the x-da packing notation, critical d positions in AS1 and critical a positions in AS2 are shaded dark gray. Noncritical a positions in AS1 and noncritical d positions in AS2 are shaded light gray. Residues at x positions in both helices are shaded black. The dashed circle around P221 indicates that this residue is less critical, but also important for function. Missense changes obtained at each residue are indicated by circles below the corresponding wild-type residue: small, open circles = Tsr+; intermediate-sized gray circles = Tsr±; large black circles = Tsr−. Plus symbols identify the wild-type residues at each position. Where no symbol is given, that particular replacement was not isolated. Mutants denoted by open diamonds expressed Tsr at less than 25% of the wild-type level; all other mutants expressed Tsr at 50% or more of the wild-type level (Table S1).
B) Helix packing in the x-da 4-helix HAMP bundle. Helical wheels, viewed toward the C-terminus from the N-terminal side, show the x, d, and a positions of the bundle-stabilizing residues (Hulko et al., 2006). Two layers of hydrophobic packing interactions are indicated by black (upper layer) and gray (lower layer) lines. Interactions that stabilize the intrasubunit interface are indicated by solid lines; interactions that stabilize the intersubunit interface are indicated by dashed lines.
C) One subunit of the x-da bundle. The AS1 and AS2 residues important or critical (bold) for function are labeled in black type; noncritical residues are labeled in gray type. Italic labels indicate residues on the N-terminal (K215-S217) or C-terminal (M264-R265) side of the HAMP bundle that may serve as transmission links to adjacent domains (see Discussion).
Residues P221 (AS1) and E248 (AS2) are conserved, defining features of HAMP domains (Aravind & Ponting, 1999; Williams & Stewart, 1999). We generated additional mutations at these two positions by site-directed mutagenesis to obtain a complete set of amino acid replacements for each. Despite its presence in nearly every HAMP domain, P221 is not a highly critical residue in Tsr-HAMP, although most replacements at this position did impair Tsr function to some extent (Fig. 3A). E248 proved much less tolerant of amino acid replacements. Only cysteine and aspartate preserved wild-type function, whereas most other E248 replacement mutants exhibited a null phenotype (Fig. 3A).
We also made site-directed mutations at some HAMP codons to obtain proline replacements at every AS1 and AS2 residue. AS2 was quite sensitive to proline replacements; nearly all positions yielded a null defect (17/18 Tsr−; Fig. 3A), consistent with the premise that this HAMP segment has helical character in vivo. AS1 also proved sensitive to proline replacements, but replacements at some AS1 residues did not cause a complete loss of function (11/18 Tsr−; 6/18 Tsr±). We note that four of the six partially functional AS1 proline mutants had replacements close to P221, the naturally-occurring AS1 proline that is optimal for HAMP function (see above). We conclude that AS1 also has helical character in vivo, but its structural stability may be less critical to HAMP function than that of AS2.
In all, we characterized 51 AS1 and 80 AS2 missense mutants with complete loss-of-function Tsr phenotypes. The remainder of this report deals exclusively with these Tsr-HAMP* null mutants.
Critical AS1 and AS2 residues correspond to packing faces in the HAMP 4-helix bundle
The Af1503 HAMP structure exhibits “complementary x-da” helix packing interactions in which three residue positions along the hydrophobic face of each helix contribute to the stability of the 4-helix bundle (Hulko et al., 2006) (Fig. 3B). Note that the residue designations in the x-da arrangement differ from those in the a-d heptad repeat in many coiled-coils. To identify corresponding residues in the two packing arrangements (in Figs. 3 and 7), positions critical to both arrangements are shown with black and dark gray shading and less critical “edge” residues with light gray shading. In x-da packing, the side-chains of symmetry-related x residues face one another in “knob-to-knob” orientation, whereas the side-chains of the flanking d and a residues lie one helical turn above or below and pack against the x residues in the other helix (Fig. 3B). In this packing arrangement, some a/d residues (dark gray) contribute to intrasubunit interactions, whereas others (light gray) contribute to intersubunit interactions. Note, however, that the x residues stabilize both the intra- and intersubunit helix interfaces (Fig. 3B). In Tsr HAMP, all but one of the functionally critical residues in each helix fell at two packing positions (x and d in AS1 and a and x in AS2; see Fig. 3C), indicating that those two positions play predominant roles in stabilizing the Tsr-HAMP structure. These mutational features are consistent with a four-helix bundle structure for Tsr HAMP. If that bundle has an x-da arrangement, then the mutation patterns suggest that packing interactions within each subunit (involving the dark gray a and d positions in Fig. 3C) are more critical to function than are those at the interface between subunits (involving the light gray a and d positions in Fig. 3C).
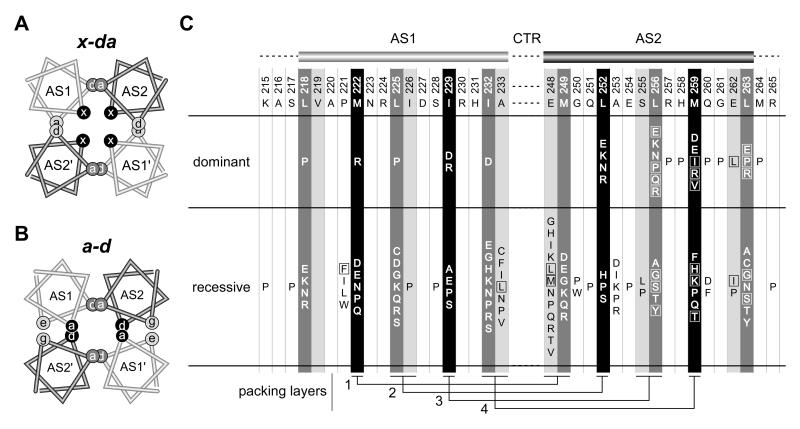
Fig. 7. Bundle arrangements and null lesions in the Tsr-HAMP AS1 and AS2 helices.
A) Helical wheel cross-section of the x-da packing arrangement. This view is similar to that shown in Figure 3B, but the helices have been moved closer together and overlapped to emphasize the layers and packing interactions. Residues at overlapping a and d positions contribute to intrasubunit (dark gray) or intersubunit (light gray) packing interactions; residues at the x positions (black) stabilize both the intra- and intersubunit interface, as summarized in Fig. 3B and 5A. B)
B) Helical wheel cross-section of a possible a-d bundle. This HAMP packing arrangement has not been observed experimentally, but was suggested to be the functional alternative to the x-da signaling state (Hulko et al., 2006). Overlapping sites indicate packing layers and residue interactions. As in the x-da arrangement, interactions between dark gray a/d residue positions stabilize the intrasubunit interface. However, the black a/d residue positions that stabilize the intersubunit interface are x residues in the x-da arrangement of (A). The e (AS1) and g (AS2) positions (light gray shading) correspond to the noncritical a (AS1) and d (AS2) residues in the x-da arrangement.
C) Summary of null (Tsr−) lesions in the AS1 and AS2 helices. Wild-type amino acids and residue numbers are given below the AS1 and AS2 cartoons. Shading of the residue positions corresponds to that of the x, d, and a positions in the HAMP bundles of (A) and (B). Boxes enclose amino acid replacements that caused moderate to severe jamming behavior (E2 or E3 classification, see Table S1). Residue interactions in the four packing layers are shown at the bottom of the figure.
Expression level and stability of Tsr-HAMP* proteins
Owing to their loss-of-function nature, we cannot be certain that Tsr-HAMP* null mutant proteins retain native or near-native structures. However, at optimal induction conditions for wild-type Tsr, all but three of the null mutants (D227M, D227P and M264Y) exhibited a steady-state Tsr protein level above 50% of wild-type (Fig. 3A; Table S1), which indicates that the vast majority of null mutant proteins have essentially wild-type intracellular stabilities, and perhaps mainly native structures, as well.
Dominance and asymmetry tests of Tsr-HAMP* lesions
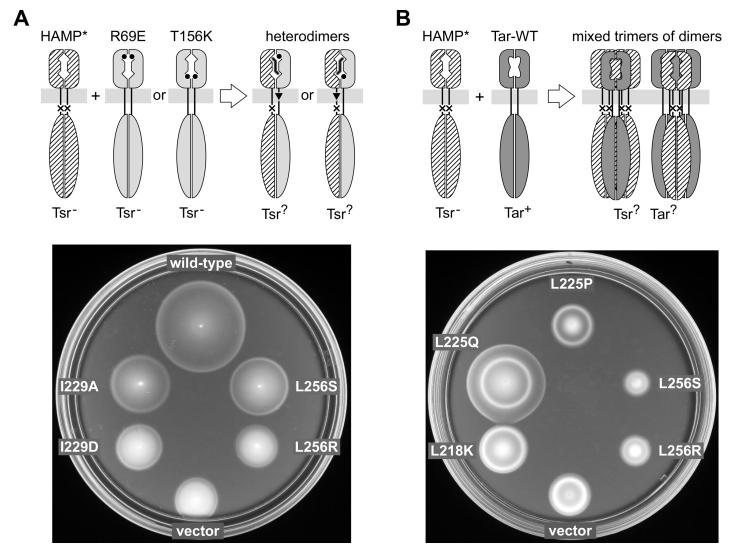
To assess the extent of structural perturbation in Tsr-HAMP* null mutants, we tested their signaling properties in heterodimers that contained one wild-type (HAMP+) subunit. These tests were done by expressing plasmid-encoded Tsr-HAMP* subunits in tsr mutant hosts that had a recessive serine-binding defect (R69E or T156K) (Ames et al., 2008). Tsr heterodimers containing either the R69E or T156K lesion in one subunit have one functional serine-binding site, but the subsequent piston displacement takes different routes (Fig. 4A). In R69E heterodimers, ligand-induced piston motions occur in the R69E subunit, whereas in T156K heterodimers, the piston displacement occurs in the opposite, HAMP* subunit (Yang et al., 1993). Because the mutant homodimers cannot support Tsr function, positive tests identify recessive lesions and negative tests identify dominant lesions. We assume that recessive HAMP lesions have relatively modest structural changes, whereas dominant HAMP lesions must have more drastic structural changes that perturb HAMP structure even in association with a wild-type HAMP subunit. Examples of recessive and dominant Tsr-HAMP* behaviors are shown in Fig. 4A; test results for all Tsr-HAMP* nulls are summarized in Table S1 (and in Fig. 7C, to be discussed in more detail below). Of 128 Tsr-HAMP* null mutants tested, 30 had dominant defects, the remainder were recessive. However, none of the mutants exhibited functional asymmetry in these tests, for example by blocking signal transmission in the T156K heterodimer, but not in the R69E heterodimer. The symmetric behavior of all HAMP* null mutants in these tests implies that AS1 and AS2 lesions disrupt Tsr function by interfering with symmetric structural interactions between the two HAMP subunits in a Tsr dimer. Thus, these test results are also consistent with a 4-helix bundle HAMP structure.
Fig. 4. Functional tests of Tsr-HAMP mutations.
A) Dominance and asymmetry test. Upper: Plasmid-encoded mutant Tsr-HAMP (HAMP*) subunits were expressed in a host strain with chromosomally-encoded Tsr-R69E or Tsr-T156K subunits, which have lesions at the serine-binding site. The resulting heterodimers have one functional binding site, which produces ligand-induced piston motions through the wild-type HAMP subunit (in the case of R69E) or through the mutant HAMP subunit (in the case of T156K). Lower: Examples of dominant and recessive Tsr-HAMP defects. UU2378 (tsr-T156K) cells containing pPA114 derivatives were tested for Tsr function on tryptone soft agar plates containing 12.5 μg/ml chloramphenicol and 0.6 μM sodium salicylate and incubated at 30°C for 7.5 hours. The wild-type control was pPA114; the vector control was pKG116. Tsr-HAMP mutants I229A and L256S are recessive; I229D and L256R are dominant.
B) Jamming and rescue test. Upper: Plasmid-encoded Tsr-HAMP* molecules were expressed in a tar+ Δtsr host strain. Tar and Tsr subunits do not form heterodimers, but do form mixed trimers of dimers, in which some Tsr defects regain function (rescue) and other Tsr defects spoil Tar function (jamming). Lower: Examples of rescuable and jamming Tsr-HAMP defects. UU1623 cells containing pPA114 derivatives were tested for Tsr function on tryptone soft agar plates containing 12.5 μg/ml chloramphenicol and 0.6 μM sodium salicylate and incubated at 32.5°C for 9 hours. The vector control was pKG116. L256S and L256R jam Tar; L225Q is rescuable (and does not jam Tar); L218K and L225P are neither rescuable nor jamming.
Rescue and jamming tests of Tsr-HAMP* lesions
To assess the extent of functional perturbation in Tsr-HAMP* null mutants, we tested their signaling properties in cells containing wild-type Tar molecules, the aspartate chemoreceptor. These tests were done by expressing plasmid-encoded Tsr-HAMP* subunits in a tar+, but otherwise receptorless, host and assessing its Tsr and Tar function on tryptone soft agar plates (Ames et al., 2008). Tsr and Tar subunits do not form heterodimers (Milligan & Koshland, 1988) but Tsr and Tar homodimers can form mixed trimer of dimer signaling teams (Fig. 4B) (Ames et al., 2002; Studdert & Parkinson, 2004; Ames & Parkinson, 2006). Some Tsr* defects are functionally rescued by wild-type Tar team partners, other Tsr* defects can block or jam Tar function in the mixed teams. We assume that rescuable Tsr* mutants have relatively modest functional defects, whereas jamming Tsr* mutants have relatively severe defects that prevent team function, despite the presence of wild-type members. Examples of rescuable and jamming Tsr-HAMP* behaviors are shown in Fig. 4B; test results for all Tsr-HAMP* nulls are summarized in Table S1. (Jamming results are also summarized in Fig. 7C, to be discussed in more detail below.) Among 128 null mutants, 24 exhibited jamming properties and 51 others had rescuable defects. Thus, at least 75/128 HAMP* null proteins must be able to form mixed trimers of dimers with wild-type Tar molecules. We did not directly test the non-rescuable and non-jamming HAMP* mutants for trimer-of-dimer formation, but some of those mutant receptors are probably able to form mixed trimers with Tar, as well, even though they cannot influence the signaling properties of the mixed teams (Ames et al., 2002; Ames & Parkinson, 2006; Mowery et al., 2008).
Compensatory interactions between mutant AS1 and AS2 helices
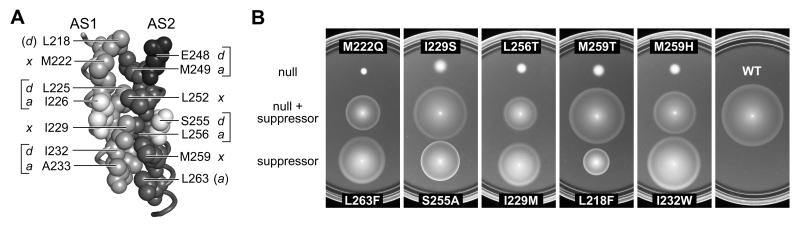
The hydrophobic residues that stabilize the 4-helix HAMP bundle interact in four packing layers (Hulko et al., 2006) (Fig. 5A). In addition to inter-subunit interactions between helix residues, within each subunit the x residues in one HAMP helix pack against residues at the d and a positions in the same layer of the other HAMP helix: M222 (x) / E248 (d) / M249 (a) [layer 1]; L252 (x) / L225 (d) / I226 (a) [layer 2]; I229 (x) / S255 (d) / L256 (a) [layer 3]; and M259 (x) / I232 (d) / A233 (a) [layer 4]; (Fig. 5A). These structural features suggest that deleterious amino acid replacements at a critical residue in one HAMP helix could act by destabilizing helix packing interactions. Accordingly, null defects in a HAMP helix might be suppressible by a compensatory change at a packing partner residue in the other HAMP helix. To test this possibility, we chose amino acid replacements in AS1 or AS2 that caused null phenotypes and scanned potential packing partner residues in the other helix for amino acid changes that could restore some degree of signaling function. To survey a full spectrum of amino acid replacements at potential packing partners, we targeted individual residues in each Tsr-HAMP null mutant for all-codon mutagenesis and tested pools of several hundred mutant plasmids for ones with enhanced Tsr function.
Fig. 5. Tsr-HAMP packing layers and example suppression pairs.
A) Side view of AS1-AS2 packing interactions in one subunit of the modeled 4-helix Tsr-HAMP bundle. Space-filled residues compose the packing interface. Critical AS1 residues are shown with light gray atoms); critical AS2 residues with either dark gray or black (E248) atoms. Noncritical residues at the packing interface (I226, S255) have white atoms. Brackets denote the four packing layers (x in one helix; a, d in the other). L218 (d) and L263 (a) participate in packing interactions at the top and bottom of the bundle, respectively.
B) Examples of suppression quality. Derivatives of pPA114 carrying a Tsr-HAMP null mutation alone, a suppressor mutation alone, and the double mutant combination were tested for Tsr function in receptorless host strain UU1250. Transformant cells were placed on tryptone soft agar plates containing 12.5 μg/ml chloramphenicol and 0.6 μM sodium salicylate and incubated at 30°C for 7 hours.
Nine Tsr-HAMP null lesions proved suppressible; examples are shown in Fig. 5B and summarized in Fig. 6A. All suppressible defects were recessive, most (7/9) were non-jamming and functionally rescuable (Fig. 6A and Table S1). These properties suggest that suppressible lesions have relatively modest structure- and function-perturbing character. The suppressible null defects had polar amino acid replacements at one of the critical hydrophobic residues (AS1: L218N, M222Q, I229A/S; AS2: L256A/T, M259H/T, L263T). Except for one residue (S255A/E/G), the suppressors involved replacements at other critical hydrophobic residues (AS2: M249I/L/V/W, L256F/I/M; L263F; AS1: L218F; I229F/L/M; I232M/W; A233G). However, contrary to our initial expectations, the suppressor changes at hydrophobic residues did not involve polar amino acid replacements, but rather hydrophobic ones, often with different side-chain volumes than the wild-type residue. This finding implies that relatively modest changes in HAMP packing interactions can overcome the destabilizing effects of polar replacements at critical helix residues.
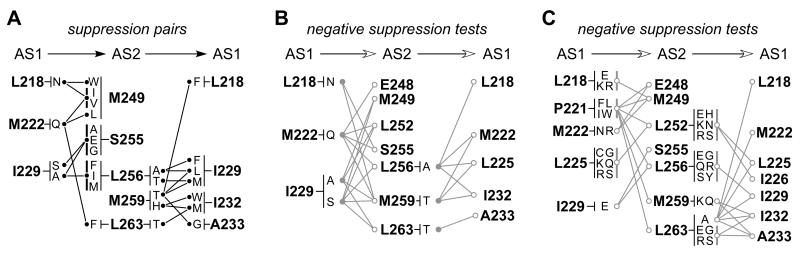
Fig. 6. Summary of AS1-AS2 suppression tests.
A) Positive suppression pairs. Horizontal arrows at the top point away from the helix with a starting null lesion and toward the targeted helix. Thick vertical lines next to some amino acid replacements at suppressor residues indicate several different changes that suppressed the same null defect.
B) Negative suppression tests involving suppressible null alleles. This diagram indicates targeted residues that did not give rise to suppressors of null defects known to be suppressible from the tests in (A). Not all combinations of null and target residues were tested. Gray circles indicate previously-suppressible null alleles; open circles indicated targeted residues that yielded no suppressors.
C) Negative suppression tests of other null alleles. This diagram summarizes combinations of null defects and targeted residues that did not give rise to suppression effects. Not all combinations of null and target residues were tested. Thick vertical gray lines group multiple null lesions tested at some residues.
Of the 18 different amino acid replacements found as suppressors (see Fig. 6A), we had obtained 14 of them in our HAMP mutagenesis study (Fig. 3A). Ten of these single replacement mutants exhibited essentially wild-type Tsr function (see L263F, I229M, I232W in Fig. 5B), whereas the other four formed small, sharp-edged colonies on tryptone soft agar plates, indicating somewhat reduced Tsr function (see S255A and L218F in Fig. 5B). Interestingly, the mutations causing partial Tsr defects seemed to be the most effective suppressors, yielding large, wild-type colony phenotypes when combined with a suppressible null lesion (Fig. 5B).
To assess the residue position and side-chain specificity of these suppression effects, we tested the suppressible mutants, and other null defects at the same residues, in combination with a variety of target residues. Residue combinations that did not lead to a suppression effect are summarized in Fig. 6B and 6C. Most targeted residues failed to suppress the starting null defects. For example, although the M222Q null defect was suppressible by changes at M249 and L263 (Fig. 6A), it was not suppressible by replacements at E248, L252, S255, L256, or M259 (Fig. 6B). Similarly, the M259T null defect was suppressible by changes at L218, I229, and A233 (Fig. 6A), but not by any amino acid replacement at M222, L225 or I232 (Fig. 6B). Moreover, other null defects at these same residues (M222N, M222R; M259K, M259Q) were not suppressible by any amino acid changes at the targeted suppressor residues (Fig. 6C). In all, 112 combinations of null mutations and target residues were tested for suppression effects. Only 6/61 AS1 null combinations and 7/51 AS2 null combinations yielded suppressors. This high degree of residue and side-chain specificity is consistent with restoration of function through compensatory structural changes, presumably ones that interact to stabilize the native HAMP structure. Most of the positive suppression pairs conformed to the packing layers of the x-da four-helix bundle (see Fig. 9A, to be discussed in more detail below), providing additional support for this HAMP structure in Tsr.
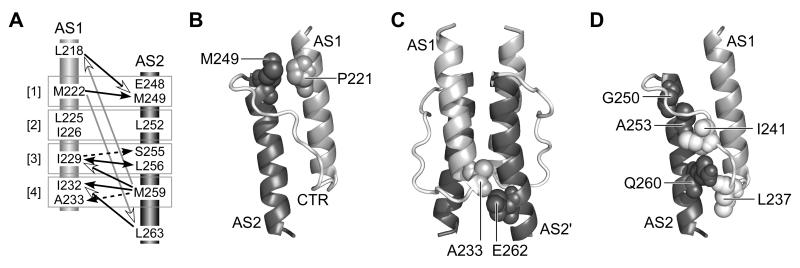
Fig. 9. Structural features of the Tsr-HAMP bundle.
A) Summary of AS1-AS2 suppression effects. Arrows point from the null residue to the suppressor residue. Numbers in brackets identify the packing layers. In the context of the x-da bundle structure, suppression effects that should primarily influence the stability of intrasubunit packing are indicated by black lines and arrowheads; effects that should primarily influence the stability of intersubunit packing are indicated by dashed black lines and arrowheads. Suppression effects involving the bundle-capping residues (L218, L263) that could influence both intra- and intersubunit interactions are indicated by open arrowheads: black lines for adjacent layer interactions and gray lines for long-range interactions.
B) Relative positions of P221 and M249 residues in the modeled Tsr-HAMP structure. Only one subunit is shown; the other subunit would be behind these helices.
C) Relative positions of A233 and E262 residues in the modeled Tsr-HAMP structure. The residues abut one another at the subunit interface.
D) Relative positions of AS2 and critical connector residues in the modeled Tsr-HAMP structure. One subunit is shown, as in (B). The side-chains of connector residues L237 and I241 (white, space-filled) project into the AS1-AS2 cleft of the same subunit. Amino acid replacements at AS2 residues G250, A253, and Q260 (dark gray, space-filled) may disrupt HAMP function by displacing the critical connector residues.
DISCUSSION
The 4-helix HAMP bundle
The structure of the Af1503 HAMP domain has provided important clues to the mechanism(s) of HAMP signaling (Hulko et al., 2006). The Af1503 HAMP domain is dimeric, with the AS1 and AS2 helices of both subunits organized in a parallel 4-helix bundle (Hulko et al., 2006). Because chemoreceptors of the MCP family and other HAMP-containing proteins have a dimeric native structure, many HAMP domains could have a 4-helix bundle structure. There is growing support for this idea. Cysteine-scanning (Butler & Falke, 1998) and disulfide mapping (Swain & Falke, 2007) studies have provided in vitro evidence for a 4-helix HAMP bundle in Tar, the aspartate chemoreceptor. Similar in vivo studies of Aer, an MCP-like aerosensor, have also indicated a 4-helix bundle HAMP structure (Watts et al., 2008). In HtrII, an MCP-like transducer that interacts with the sensory rhodopsin photoreceptor of Natronomonas pharaonis, HAMP signaling is accompanied by changes in the diffusion coefficients of AS1 and AS2 (Inoue et al., 2008) and by conversion from a “dynamic” state to a “compact“ structure (Doebber et al., 2008). Both results are at least consistent with a 4-helix bundle.
Our Tsr-HAMP suppression studies are also consistent with a 4-helix bundle organization. The Af1503 HAMP bundle is stabilized by four layers of hydrophobic packing interactions (Hulko et al., 2006). We tested this structural feature by identifying compensatory changes that could restore function to AS1 and AS2 mutants with null lesions at predicted packing residues (Fig. 9A). Most suppression effects occurred between residues in the same (5/10) or adjacent (3/10) packing layers. Moreover, two of the adjacent layer effects (L218/M249 and L263/I232) involved the bundle-capping residues (L218, L263), which could have special structural roles. The remaining two suppression effects (L218/M259 and M222/L263) occurred across multiple layers, but both of these long-range interactions involved the bundle-capping residues, as well. All suppression effects were highly specific with respect to both residue position and side-chain character, consistent with restoration of function via compensatory conformational changes. We conclude that the suppression analysis reflects structural interactions between the AS1 and AS2 helices that are consistent with a 4-helix bundle.
Mutational evidence for an x-da packing arrangement in the Tsr-HAMP bundle
The experimentally-determined Af1503 HAMP structure exhibits a “complementary x-da” packing arrangement of the helices in the bundle (Hulko et al., 2006). Hulko et al. suggested that a more conventional “a-d” packing arrangement could represent an alternative HAMP signaling conformation (Hulko et al., 2006). The two bundle structures are related by a concerted 26° counter-rotation of each of the four helices, analogous to meshed gears in a transmission (the “gearbox”; see Fig. 7A&B). In the x-da structure, three residue positions contribute to bundle stability (Fig. 7A). The x positions are structurally distinct from the flanking a and d positions; their side-chains project into the bundle core and interact with their counterparts in the opposing subunit (Fig. 7A). In contrast, the two critical hydrophobic positions in the a-d arrangement (dark gray shading in Fig. 7B) contribute more or less equally to bundle integrity. The d position in AS1 and the a position in AS2 correspond to critical a and d residues in the x-da arrangement (dark gray shading in Fig. 7A). However, in the a-d arrangement, the a positions in AS1 and the d positions in AS2 (black shading in Fig. 7B) correspond to x residues in the x-da arrangement (black shading in Fig. 7A). Note that the noncritical a/d positions in the x-da arrangement (light gray shading in Fig. 7A) correspond to g or e “edge” residues in the a-d arrangement (light gray shading in Fig. 7B). It follows that in the a-d packing arrangement, x and a/d residues should have comparable mutational sensitivities, whereas in the x-da arrangement, they might have different mutational properties that reflect their different structural environments.
The dominance properties of proline and arginine replacements at critical AS1 residues are more consistent with an x-da bundle organization than with an alternative a-d arrangement. Arginine replacements at the AS1 x positions (black shading in Fig. 7C) caused dominant defects, whereas at the critical AS1 d positions (dark gray shading in Fig. 7C), arginine replacements caused recessive defects. The more severe structural consequences of arginine replacements at x positions are consistent with their core location in the x-da structure, which should not have enough space to accommodate a large polar residue. In contrast, proline replacements were dominant at 2/3 critical AS1 d positions (L218, L225), but recessive at the two x positions in AS1 (Fig. 7C). These differences also distinguish the x and d residue positions in AS1. Perhaps prolines are better tolerated at x positions because of their relatively small volume and the fact that hydrophobicity may be less critical to bundle stability at fully buried positions (Hulko et al., 2006). The most parsimonious conclusion to draw from this evidence is that the x-da packing arrangement is the only functionally important 4-helix HAMP bundle in Tsr.
Control linkage between the HAMP and methylation bundles
The mutation patterns of Tsr-HAMP helices suggest that AS2 has a more central role than AS1 in HAMP signaling (Fig. 7C). (i) The preponderance of dominant (24/30) and jamming (22/24) mutations occurred in AS2, indicating that AS2 lesions have more drastic structural and functional consequences than do AS1 lesions, which were mostly recessive and non-jamming. (ii) Residues 256-264, spanning the C-terminal region of AS2, were also especially sensitive to proline replacements, with 7/11 replacements causing dominant defects. Nowhere else in HAMP did proline replacements at noncritical residue positions cause dominant behavior (Fig. 7C). The heightened sensitivity to proline replacements probably reflects the importance of helix strength at the AS2/MH1 junction.
The AS2 helices connect to the MH1/MH1′ methylation helices of the kinase control domain, a long 4-helix bundle of two interwound, antiparallel coiled coils (Bass & Falke, 1999; Kim et al., 1999; Park et al., 2006; Alexander & Zhulin, 2007). Stimulus signals transmitted through HAMP impinge on the MH1 and MH1′ segments to modulate output signals from the kinase control bundle. Conversely, the modification states of the methylation helices in the kinase control bundle impinge on HAMP to modulate the transmembrane helices and the stimulus-sensing portion of the receptor molecule (Lai et al., 2006). The heptad phasing of the AS2 and MH1 helices is out-of-register by +4 (or −3) residues, a so-called “stutter” (Brown et al., 1996; Lupas, 1996) (compare the a heptad positions in AS2 and MH1 in Fig. 8A). Stutters destabilize coiled-coil pairing interactions, producing an underwound or kinked structure (Brown et al., 1996; Lupas, 1996). The AS2 helices emerge from the HAMP bundle in divergent directions (Hulko et al., 2006), ostensibly in an x-da phase arrangement, so their structural interactions with the adjoining MH1 helices might have even more drastic destabilizing consequences (Fig. 8A). Thus, the HAMP and methylation bundles appear to be oppositionally coupled through an AS2/MH1 phase clash: stability of one structure should promote instability of the other. We suggest that the interplay between these conformational signals is the essence of HAMP signaling transactions and propose a new mechanistic model of HAMP action based on the dynamic properties of the x-da bundle.
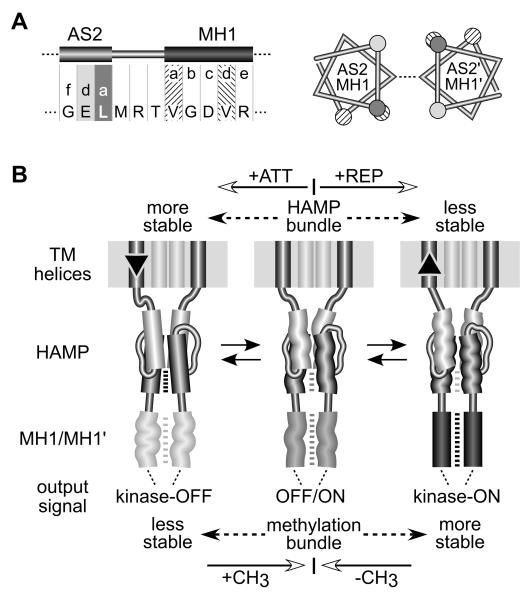
Fig. 8. Mechanistic model of HAMP signaling.
A) Junction of the AS2 helix and the first methylation helix (MH1) of the kinase control domain. Residue positions for AS2 are shaded as in Figures 3 and 7. The N-terminal (MH1) and C-terminal (MH2) methylation helices form a 4-helix coiled-coil in the Tsr dimer (Kim et al., 1999) (see Figure 1), with packing interactions mediated by a-d heptad repeats (Kim et al., 1999; Alexander & Zhulin, 2007). Left: The heptad packing residues of MH1 are four residues out of phase with those of AS2. Right: Helical wheels of the Tsr subunits at the AS2/MH1 junction, showing the alignment of residues in the AS2 helices as they emerge from a stable x-da bundle. The x positions (not highlighted) are connected by a dashed line. The a and d positions of the MH1 helices are nearly 180° out-of-phase, which should destabilize the methylation helices and packing interactions.
B) A modulated dynamics model of HAMP signaling. The model proposes that the x-da HAMP bundle is important to both the kinase-activating and kinase-deactivating signaling states because it controls the range of inter-subunit motions of the kinase control domain in the Tsr dimer. However, the dynamic states of the x-da bundle and methylation helices are oppositionally coupled through an out-of-register helical phase relationship; see (A). An unstable x-da bundle allows inter-subunit interactions of the methylation helices. Conversely, a stable x-da bundle destabilizes the methylation helix bundle. The structural interactions between AS2 and MH1 helices are bi-directional. Changes in methylation state modulate stability of the methylation region, which in turn impacts stability of the HAMP bundle. Importantly, this model predicts that attractant stimuli suppress CheA activity by enhancing stability of the HAMP bundle, whereas repellent stimuli should destabilize the bundle. These stimulus-induced changes in HAMP stability could be triggered by small vertical displacements of the transmembrane segments (TM2) adjoining the AS1 helices. The TM2/AS1 connection could act like a control cable to adjust structural tension on the HAMP domain, altering its dynamic properties. See text for additional explanation.
A dynamic bundle model of HAMP signaling
There is growing recognition that signaling proteins have dynamic structures that are central to their signaling activities (Smock & Gierasch, 2009). The signaling roles of HAMP domains may be best understood in terms of their dynamic behavior rather than a few discrete conformational states. The most stable HAMP conformation could be the 4-helix x-da bundle; the least stable would be a fully denatured bundle. Presumably, HAMP domains have been evolutionarily tuned to operate over some portion of this dynamics range, oscillating between bundle conformations of different stabilities. In our view, HAMP signal output is related to its average bundle stability, reflecting its current conformational space. Stimulus inputs and methylation changes modulate HAMP stability to initiate and terminate signaling responses (Fig. 8B).
The opposed structural coupling between the HAMP and kinase control domains establishes the signaling logic. Chemoreceptors with low methylation states produce low kinase activity (Li & Weis, 2000; Bornhorst & Falke, 2001; Sourjik & Berg, 2002). Their methylation regions are probably more dynamic than those of highly-methylated, CheA-activating receptors (Kim et al., 2002; Starrett & Falke, 2005). Accordingly, stabilization of the HAMP bundle should lower CheA activity; destabilization should increase CheA activity (Fig. 8B). Subsequent methylation or demethylation changes, produced by the sensory adaptation enzymes, would counter these behavioral responses by increasing or decreasing the structural stability of the methylation bundle to offset the HAMP dynamic effects. It follows that attractant stimuli elicit kinase-off responses by stabilizing the HAMP bundle. Because attractants are known to cause an inward piston displacement of the TM2 helices (Falke & Hazelbauer, 2001), stimulus control might involve tension changes in the TM2/AS1 linkage, with no need for the helix rotations implicit in the gearbox model. We suggest that the TM2/AS1 links function as control cables, adjusting structural tension on the HAMP bundle to manipulate its stability (Fig. 8B).
Mutational evidence for the dynamic bundle model
The AS1-AS2 conformational suppression effects are more consistent with a dynamic bundle model than a conventional two-state signaling model. In both models, most null lesions should affect function by damaging a signaling structure. The dynamic model predicts that suppressors should restore function by returning HAMP stability to the physiological operating range. In contrast, two-state models predict that some suppressors could restore function by damaging the alternative signaling structure, producing a workable balance between the two compromised states. The structural specificity of the suppression effects argues against this two-state scenario (Fig. 9A). If packing interactions in the wild-type HAMP bundle confer optimal dynamic behavior, few residue changes outside the null lesion layer would be expected to enhance bundle stability. However, changes within the mutant layer, which is already destabilized, could introduce novel residues that work well in combination with the null replacement, for example, by better filling the altered packing volume or by accommodating water molecules that hydrogen bond to polar groups in the null mutant residue. Thus, null lesions that introduced polar residues at x positions (M222Q, I229A/S, L256A/T, and M259H) were suppressible by changes that altered the side chain volume or packing configurations of their major interaction partner in the same layer (M249I/L/V, L256F/I/M, I229F/L/M and I232M/W, respectively) (Fig. 9A). Two-state models predict a wider assortment of suppressors that need not be confined to the vicinity of the original lesion.
The long-range suppression effects (M222Q-L263F; M259T-L218F) involving the bundle-capping residues are interesting exceptions to the general rule of layer-specific suppression, but also understandable in terms of compensatory effects on bundle stability. M222 and M259 are the endmost x positions in the bundle. The M222Q and M259T lesions introduce polar groups that should destabilize one end of the bundle, conceivably creating stresses that could be offset by enhanced packing forces at the other end of the bundle. The bundle-capping location is open and better able to accommodate increased side-chain size. Notably, only phenylalanine replacements at L218 and L263 served as long-range suppressors. Phenylalanine is bulkier than leucine and could conceivably create additional bundle-stabilizing forces through stacking interactions.
Bundle stability changes can also account for the few (non-proline) null lesions found at non-critical AS1 and AS2 residues (Fig. 7C). For example, large hydrophobic replacements at P221 disrupted function. In the modeled Tsr-HAMP structure, P221 lies next to M249, a critical a residue in AS2 (Fig. 9B). Conceivably, bulky hydrophobics at P221 influence bundle stability through steric and hydrophobic interactions with M249. Similarly, several hydrophobic replacements at E262 (AS2), which borders on A233, a critical AS1 residue, disrupted function (Fig. 9C). Finally, a few replacements at AS2 residues G250, A253, and Q260, including large hydrophobic, acidic and basic amino acids disrupted function (Fig. 7C). These null lesions might influence bundle stability through nonspecific steric or charge clashes with the connector, which contains several bundle-stabilizing residues (L237 and I241) (Ames et al., 2008). Bulky or charged replacements at AS2 positions that face the connector could weaken those stabilizing connector interactions (Fig. 9D).
Output signals of mutant HAMP domains
HAMP lesions that abrogate chemoreceptor signaling function most likely destabilize the bundle, driving it far enough outside the physiological operating range to prevent effective compensation by the sensory adaptation system. Our current version of the dynamic bundle model does not attempt to define the relationship between HAMP stability and kinase activity outside the normal operating limits. This issue is beyond the scope of the present work, but will be addressed in detail in a follow-up study. However, based on the output patterns of Tsr-HAMP connector mutants, it appears that a very unstable or unstructured HAMP domain can also lock the receptor in a kinase-off mode (Ames et al., 2008). If so, then this kinase-off state is fundamentally different from the attractant-induced signaling state proposed in our model, even though their outputs are the same. This raises a caution flag about drawing mechanistic conclusions from the signal outputs of HAMP mutants in any signaling protein: The relationship between kinase activity and HAMP stability may not be a simple monotonic one.
Generality of the dynamic bundle model
The bundle stability model may also apply to the HAMP domains of signaling proteins other than chemoreceptors. There is evidence for a dynamic or unstructured HAMP state in several other systems (Bordignon et al., 2005; Kishii et al., 2007; Doebber et al., 2008) and the paucity of high-resolution structures implies that HAMP domains could be highly dynamic under physiological conditions. Indeed, the Af1503 HAMP bundle may have been unusually stable because it was examined well below its normal operating temperature (Hulko et al., 2006).
Despite the lack of HAMP structural information in many signaling proteins of interest, it should be possible to test key predictions of the dynamic bundle model by judicious mutant constructions guided by the Tsr-HAMP mutation data. For example: (i) arginine and proline replacements should have opposing dominance effects at x and a/d positions; and (ii) the C-terminal (output) end of AS2 should be sensitive to more kinds of amino acid replacements than any other parts of HAMP. These sorts of genetic approaches should help to elucidate structure-function relationships in HAMP domains under in vivo operating conditions.
MATERIALS AND METHODS
Bacterial strains and plasmids
All strains were derivatives of E. coli K-12 strain RP437 (Parkinson & Houts, 1982) and contained the following markers relevant to this study: UU1250 [Δaer-1 Δtsr-7028 Δ(tar-tap)5201 Δtrg-100] (Ames et al., 2002); UU1535 [Δaer-1 Δ(tar-cheB)2234 Δtsr-7028 Δtrg-100] (Bibikov et al., 2004); UU1623 [Δtsr-7028 Δtap-3654 Δtrg-100] (Ames et al., 2008); UU2377 [tsr-R69E Δaer-1 Δ(tar-tap)5201 Δtrg-4543 ΔrecA-SstII/EcoRI; (Ames et al., 2008); and UU2378 [tsr-T156K Δaer-1 Δ(tar-tap)5201 Δtrg-4543 ΔrecA-SstII/EcoRI] (Ames et al., 2008). Plasmids used in this work were: pKG116, which confers chloramphenicol resistance and has a sodium salicylate-inducible expression/cloning site (Buron-Barral et al., 2006); pPA114, a relative of pKG116 that carries wild-type tsr under salicylate control (Ames et al., 2002); pRR48, which confers ampicillin resistance and has an expression/cloning site with a tac promoter and ideal (perfectly palindromic) lac operator under control of a plasmid-encoded lacI repressor, inducible by isopropyl-β-D-thiogalactopyranoside (IPTG) (Studdert & Parkinson, 2005); and pRR53, a derivative of pRR48 that carries wild-type tsr under IPTG control (Studdert & Parkinson, 2005). Plasmids pKG116 and pPA114 are compatible with plasmids pRR48 and pRR53; both types can stably co-exist in the same cell.
Directed mutagenesis
Plasmid mutations were generated by QuikChange™ mutagenesis, as previously described (Ames et al., 2002). For all-codon mutagenesis, we used oligonucleotide primer pools that were fully degenerate, i.e., with equal frequencies of the four bases A, C, G and T at all three base positions of the targeted codon. Mutations were verified by sequencing the entire tsr coding region in the mutant plasmid.
Chemotaxis assays
Host strains carrying Tsr expression plasmids were assessed for chemotactic ability on tryptone soft agar plates (Parkinson, 1976) containing appropriate antibiotics (chloramphenicol [12.5 μg/ml] or ampicillin [50 μg/ml]) and inducer (sodium salicylate or IPTG). Plates were incubated for 7-10 hours at 30°C or 32.5°C.
Expression levels of mutant Tsr proteins
The steady-state cellular levels of plasmid-encoded Tsr proteins were assessed in strain UU1535 as described (Ames & Parkinson, 2006).
Protein modeling and structural display
Atomic coordinates for the Tsr-HAMP domain were generated from the Af1503-HAMP coordinates (PDB ID 2ASW) by the Swiss-Model server (Schwede et al., 2003). Structure images were prepared with MacPyMOL software (http://www.pymol.org).
Supplementary Material
ACKNOWLEDGMENTS
Thanks to Tom Alber (U.C. Berkeley) for a most helpful discussion on coiled-coils and some useful reality checks. Thanks to Dave Blair (U. Utah) for constructive comments and creative ideas on an earlier version of the manuscript. Thanks to Bob Bourret (U. North Carolina), Joe Falke (U. Colorado), Mike Manson (Texas A & M U.), Patricia Mowery (Hobart & William Smith Colleges), and Joachim Schultz (U. Tuebingen) for comments on the manuscrript. Thanks to Andrei Lupas (Max Planck Institute - Tuebingen) for thought-provoking discussions. This work was supported by research grant GM19559 from the National Institute of General Medical Sciences. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute.
REFERENCES
- Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Parkinson JS. Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. USA. 2006;103:9292–9297. doi: 10.1073/pnas.0602135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Zhou Q, Parkinson JS. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleman JA, Stewart V. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 2003;185:89–97. doi: 10.1128/JB.185.1.89-97.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- Bass RB, Falke JJ. The aspartate receptor cytoplasmic domain: In situ chemical analysis of structure, mechanism and dynamics. Structure. 1999;7:829–840. doi: 10.1016/s0969-2126(99)80106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikov SI, Miller AC, Gosink KK, Parkinson JS. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 2004;186:3730–3737. doi: 10.1128/JB.186.12.3730-3737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemann HP, Koshland DE., Jr. Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochem. 1994;33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- Bordignon E, Klare JP, Doebber M, Wegener AA, Martell S, Engelhard M, Steinhoff HJ. Structural analysis of a HAMP domain: the linker region of the phototransducer in complex with sensory rhodopsin II. J. Biol. Chem. 2005;280:38767–38775. doi: 10.1074/jbc.M509391200. [DOI] [PubMed] [Google Scholar]
- Bornhorst JA, Falke JJ. Evidence that both ligand binding and covalent adaptation drive a two- state equilibrium in the aspartate receptor signaling complex. J Gen Physiol. 2001;118:693–710. doi: 10.1085/jgp.118.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Cohen C, Parry DA. Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Buron-Barral M, Gosink KK, Parkinson JS. Loss- and gain-of-function mutations in the F1-HAMP region of the Escherichia coli aerotaxis transducer Aer. J. Bacteriol. 2006;188:3477–3486. doi: 10.1128/JB.188.10.3477-3486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochem. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebber M, Bordignon E, Klare JP, Holterhues J, Martell S, Mennes N, Li L, Engelhard M, Steinhoff HJ. Salt-driven equilibrium between two conformations in the HAMP domain from Natronomonas pharaonis: the language of signal transfer? J. Biol. Chem. 2008;283:28691–28701. doi: 10.1074/jbc.M801931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends. Biochem. Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends. Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- Inoue K, Sasaki J, Spudich JL, Terazima M. Signal transmission through the HtrII transducer alters the interaction of two alpha-helices in the HAMP domain. J. Mol. Biol. 2008;376:963–970. doi: 10.1016/j.jmb.2007.12.026. [DOI] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Kim SH, Wang W, Kim KK. Dynamic and clustering model of bacterial chemotaxis receptors: Structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA. 2002;99:11611–11615. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishii R, Falzon L, Yoshida T, Kobayashi H, Inouye M. Structural and functional studies of the HAMP domain of EnvZ, an osmosensing transmembrane histidine kinase in Escherichia coli. J Biol Chem. 2007;282:26401–26408. doi: 10.1074/jbc.M701342200. [DOI] [PubMed] [Google Scholar]
- Lai WC, Beel BD, Hazelbauer GL. Adaptational modification and ligand occupancy have opposite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Mol. Microbiol. 2006;61:1081–1090. doi: 10.1111/j.1365-2958.2006.05296.x. [DOI] [PubMed] [Google Scholar]
- Lee L, Mizuno T, Imae Y. Thermosensing properties of Escherichia coli tsr mutants defective in serine chemoreception. J. Bacteriol. 1988;170:4769–4774. doi: 10.1128/jb.170.10.4769-4774.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- Lin LN, Li J, Brandts JF, Weis RM. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochem. 1994;33:6564–6570. doi: 10.1021/bi00187a025. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr., Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan DL, Koshland DE., Jr. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- Mowery P, Ostler JB, Parkinson JS. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 2008;190:8065–8074. doi: 10.1128/JB.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Parkinson JS. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 1976;126:758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard AM, Bilwes AM, Crane BR. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochem. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrett DJ, Falke JJ. Adaptation mechanism of the aspartate receptor: electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochem. 2005;44:1550–1560. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. USA. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochem. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts KJ, Johnson MS, Taylor BL. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. J. Bacteriol. 2008;190:2118–2127. doi: 10.1128/JB.01858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SB, Stewart V. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 1999;33:1093–1102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Park H, Inouye M. Ligand binding induces an asymmetrical transmembrane signal through a receptor dimer. J. Mol. Biol. 1993;232:493–498. doi: 10.1006/jmbi.1993.1405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.