Abstract
Eukaryotic cells normally restrict genome duplication to once per cell division. In metazoa, re-replication of DNA during a single S-phase appears to be prevented solely by suppressing CDT1 activity, a protein required for loading the replicative MCM DNA helicase. However, siRNA suppression of geminin (a specific inhibitor of CDT1) arrested proliferation only of cells derived from malignant cancers by inducing DNA re-replication and DNA damage that spontaneously triggered apoptosis. None of these effects were detected either in cells derived from normal human tissues, or in cells immortalized by a viral oncogene. To induce these effects in non-cancer cells required suppression of both geminin and cyclin A, another cell cycle regulator. Therefore, initiating DNA replication in some cancer cells is limited solely by regulating the level of CDT1 activity with geminin, whereas non-cancer cells contain additional safeguards that prevent DNA re-replication. These results demonstrate that inhibition of geminin activity could be used to selectively kill cancer cells without harming other cells.
Keywords: siRNA, Cyclin A, geminin, DNA re-replication, Cell Cycle
INTRODUCTION
DNA replication begins with assembly of pre-replication complexes (preRCs) at multiple sites throughout the genome as cells exit metaphase (1). PreRCs are then assembled into pre-initiation complexes that are subsequently activated by protein kinases to begin DNA synthesis (S-phase) (2). Once S-phase begins, further assembly of preRCs is prevented by phosphorylation, ubiquitination and degradation of preRC proteins ORC1, CDC6 and CDT1, and by geminin, a specific protein inhibitor of CDT1 activity unique to metazoa (3, 4).
Eukaryotic cell division is regulated through multiple convergent pathways to restrict genome duplication to once and only once each time a cell divides (3, 4). When these pathways are subverted, random reinitiation of DNA replication occurs throughout the genome prior to mitosis, an event referred to as DNA re-replication. This results in cells with greater than 4N DNA content that are sensitive to drugs that inactivate DNA damage response pathways (5–7). Therefore, if DNA re-replication could be induced selectively in cancer cells, cancer cells could be killed without harming normal cells.
An effective cancer therapy must target cancer cells without harming non-cancer cells. Over expression of geminin in cancer cells does not prevent their proliferation (8–10), but inhibition of preRC assembly either by over-expressing a non-degradable form of geminin (9,10), or by suppressing expression of Orc2, Cdc6, or Mcm2 genes induces apoptosis. However, these treatments also arrest non-cancer cells in G1-phase (10–12). Cancer cells induce apoptosis under these conditions by entering S-phase with too few licensed replication origins, a situation that increases the frequency of stalled replication forks and thereby triggers the cell’s DNA damage response (13). Unfortunately, non-cancer cells that are arrested in G1-phase by preventing preRC assembly will also eventually undergo apoptosis, because only cells arrested in a quiescent G0 state are stable for prolonged periods of time. PreRC assembly occurs during the mitotic anaphase to G1-phase transition (1, 14). When the cell cycle is arrested prior to phosphorylation of the retinoblastoma tumor suppressor protein (Rb) and prior to the ‘restriction point’ at which entry into S-phase becomes independent of serum mitogens, they undergo apoptosis, although perhaps not as quickly as cancer cells (15, 16). This difference between cancer and non-cancer cells arises from the fact that cancer cells can pass through the restriction point (G1/S-phase checkpoint) under conditions where non-cancer cells cannot (17). Thus, inhibition of origin licensing simply induces apoptosis more quickly in cancer cells than in non-cancer cells.
Here we took the opposite tack in an effort to determine the feasibility of a cancer therapy based on the spontaneous induction of apoptosis in response to re-licensing replication origins during S-phase. From yeast to mammals, re-licensing of replication origins during cell proliferation is prevented either by modification or degradation of various preRC proteins (3, 13, 18). However, numerous results suggest that the primary regulatory target in most, perhaps all, metazoan cells is CDT1, a preRC protein that is required for loading the MCM DNA helicase onto the replication origin. Over-expression of CDT1 induces DNA re-replication in cells from mammals (19, 20), flies (21), frogs 22–25 and plants (26), and siRNA depletion of geminin induces DNA re-replication in cells from flies (27) and mammals (5–7, 28). Moreover, deletion of the geminin gene results in DNA re-replication in preimplantation mouse embryos (29, 30). These results make a compelling case that CDT1 activity is the rate limiting component for initiation of DNA replication in metazoa.
There are two ways to increase CDT1 activity in mammalian cells: increase the amount of CDT1 protein or inactivate geminin. Over-expression of CDT1 in mammalian cells has been reported to induce DNA re-replication only in cells that lack p53 activity (19, 20), but other studies conclude that over-expression of CDT1 does not induce significant DNA re-replication in mammalian cells as long as the ATR checkpoint is intact (31). Moreover, DNA lesions accumulate in primary cells even when CDT1 over-expression failed to induce re-replication, making all cells with excess CDT1 sensitive to genotoxic stress (31). Thus, over-expression of CDT1 does not appear to be a useful approach to cancer therapy. Therefore, we considered the alternative approach of increasing CDT1 activity by reducing geminin levels.
Remarkably, the response of mammalian cells to geminin depletion was entirely cell-type dependent. Depletion of geminin in cells derived from malignant cancers resulted in arrest of cell proliferation, DNA re-replication and apoptosis without addition of inhibitors of the DNA damage response pathway. Depletion of geminin in immortalized cells or cells derived from normal tissues did not arrest their proliferation and did not induce DNA re-replication. Moreover, normal cells neither accumulated DNA damage nor expressed DNA damage control genes. Normal cells responded to geminin depletion like cancer cells only when they were depleted of both geminin and cyclin A, a protein required for cell cycle dependent CDK phosphorylation of preRC proteins. These and other results demonstrate that regulation of CDT1 activity by geminin is rate limiting for initiation of DNA replication only in certain cancer cells, and provide clear evidence for the supposition that suppression of geminin activity could be used to selectively kill cancer cells under conditions that do not affect growth and proliferation of normal cells.
MATERIALS AND METHODS
Cells And Cell Culture
The human cells listed in Table SI were grown in media recommended by the providers. Primary dermal fibroblasts D-1 and primary epidermal keratinocytes K-1 were provided by Dr. Jonathon Vogel (NCI/NIH). 786-O was provided by Dr. Marston Linehan (NCI/NIH). D-1 cells were maintained in DMEM medium supplemented with 10% FBS. K-1 were maintained in Keratinocyte Medium (ScienCell, Cat. No. 2101). MCF10A and H1299 cells were kindly provided by Dr. Zong Guo (University of Pittsburgh Cancer Institute). H1299 cells were maintained in DMEM medium supplemented with 10% FBS, and MCF10A cells were maintained MCF10A breast cells were grown in a serum-free MEGM (Mammary Epithelial Growth Medium) supplemented with a MEGM bullet kit (Lonza, cc3150).
siRNA
Short interfering oligoribonucleotides (Dharmacon) for luciferase (siGL2) and cyclin A (siCCNA) were as previously described (6,7)}. Three siRNAs targeted against geminin RNA were siGEM (UGCCAACUCUGGAAUCAAA) (7), siGEM2 (AACUUCCAGCCCUGGGGUUAU) (28), siGEM3 (AAUCACUGGAUAAUCAGGAAU). Transfections were performed with 20 – 50 nM siRNA oligonucleotide duplexes using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. In brief, cells 50%–60% confluent were transfected twice (0 hours and 24 hours later) for 4 hours per transfection before adding fresh medium and then harvested 48 hours after the first transfection, unless otherwise indicated.
FACS Analysis
Fluorescence activated cell sorting was done on cells that were collected by trypsinization, fixed with 70% ethanol overnight at 4°C, then collected by centrifugation, stained in 50 ng propidium iodide/ml, 0.05% NP-40, and 10 µg RNase A/ml, and then analyzed with a Becton Dickinson FACSCalibur using Cellquest software.
Protein Detection
Western immuno-blotting was done using Rabbit anti-geminin Geminin (FL-209, Santa Cruz Biotechnology), rabbit anti-actin (A2066, Sigma), rabbit anti-GAPDH (G9545, Sigma), Mouse anti-p53 (Sigma), rabbit anti-p53 phosphorylated at serine15 (Cell Signaling), rabbit antip-21 (C-19, Santa Cruz), rabbit anti-cyclin A (H432, Santa Cruz) were used for immuno-blotting. Rabbit anti-HsCDT1 antibody was a gift from Dr. Anindya Dutta. Cells were washed twice in PBS and then sonicated in RIPA buffer. Proteins were fractionated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and incubated with the indicated antibodies. For immuno-fluorescence cells were fixed with 2% paraformaldehyde in PBS for 10 min, permeabilized with 0.2% Triton X-100 for 10 min at room temperature, and then washed and mounted with solution containing 4',6'-diamidino-2-phenylindole (DAPI) (Vector Laboratories).
TUNEL Assay
TUNEL assays were performed according to manufacturer’s instructions (Roche manual, Cat. No. 11684795910). Briefly, cells were washed three times in PBS, and fixed in freshly prepared 4% paraformaldehyde in PBS for 1 hour at room temperature, and then washed with PBS and permeabilized in 0.1% Triton X-100 in 0.1% sodium citrate. Cells were then incubated with TUNEL reaction mixture (Roche) for 60 min at 37°C in a humidified atmosphere in the dark. After washing twice with PBS, cells were mounted in a DAPI solution (Vector Laboratories). A positive control run in parallel consisted of permeabilized cells that had been incubated with DNase I (1 U/µl) for 10 min at room temperature to introduce DNA strand breaks prior to the TUNEL assay.
RESULTS
Geminin Depletion Selectively Induced DNA Re-Replication In Malignant Cancer Cells
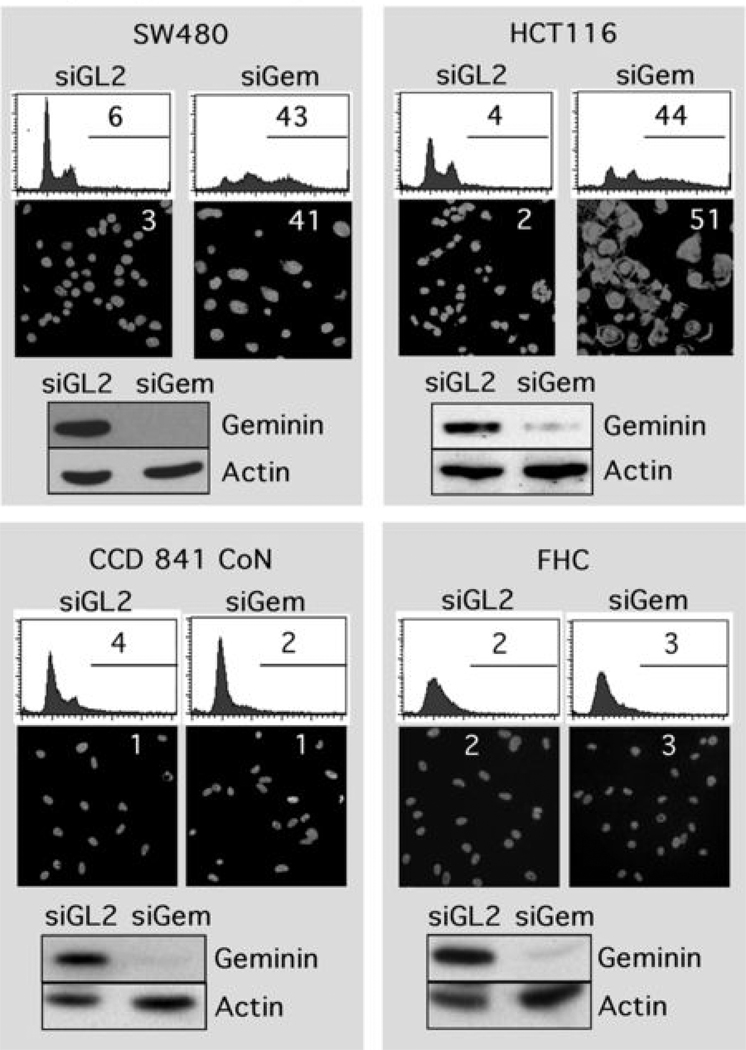
Previous studies (7) have shown that siRNA targeted against geminin (siGem) can induce DNA re-replication in HCT116 cells derived from a colorectal carcinoma, as evidenced by increased DNA content and the appearance of giant nuclei. To determine whether or not this result was unique to these cells, their response to siGem was compared with the response of three other colon cancer cells (SW480, COLO 320DM, DLD-1, Table SI). In each case, geminin was reduced at least 10-fold by siGem (Fig. 1, Fig. S3). The fraction of cells with greater than 4N DNA increased 7 to 11-fold within 2 days of siGem transfection, as judged by fluorescence activated cell sorting (FACS) analysis, and the number of giant nuclei detected by microscopy increased 14 to 25-fold (Fig. 1, Fig. S3). In total, from 30% to 50% the cells in each population underwent DNA re-replication, a proportion equivalent to the population of cells in S and G2/M phases of the cell cycle (those phases in which geminin is present). In contrast, transfection with siRNA targeted against firefly luciferase (siGL2), a gene not encoded by mammals, did not affect these parameters.
Figure 1.
Depletion of geminin induced DNA re-replication in colorectal cancer cells (SW480, HCT116), but not in normal colon cells (CCD 841 CoN, FHC). The indicated cells were transfected with siRNA against either firefly luciferase (siGL2) or human geminin (siGem). At 48 hours post-transfection, cells were harvested and stained either with propidium iodide to quantify their DNA content by fluorescence activated flow cytometry (FACS) analysis, or with DAPI to visualize their nuclei by fluorescence microscope. The percentage of cells is given with greater than 4N DNA content (FACS profiles) or with nuclei whose diameter is greater than twice that of nuclei in siGL2 treated cells. Geminin and actin proteins were detected by Western immuno-blotting.
To determine whether or not siGem had the same effect on non-carcinogenic colon cells, the experiment was repeated with cells derived from normal fetal colon tissue (CCD841 CoN, FHC cells, Table SI). Again siGem reduced the level of geminin protein in these cells at least 10-fold (Fig. 1). However, no change was detected either in the fraction of normal colon cells with greater than 4N DNA content, or in the fraction of normal colon cells with giant nuclei (Fig. 1), Thus, geminin depletion induced DNA re-replication in colorectal cancer cells, but not in their normal epithelial counterparts (summarized in Table I).
Table I.
Effect of siRNA against geminin on DNA re-replication in human cells.
| Tissue | Cell Type | Cell Name | DNA Re-replication (%) | ||
|---|---|---|---|---|---|
| siGL2 | siGem | siGem+siCcnA | |||
| Colon | Normal Epithelial | CCD-841 CoN | 4 | 2 | 29 |
| Normal Epithelial | FHC | 2 | 3 | ||
| Carcinoma | HCT-116 | 4 | 44 | 50 | |
| Adenocarcinoma | SW480 | 6 | 43 | ||
| Adenocarcinoma | COLO 320DM | 4 | 30 | ||
| Adenocarcinoma | DLD-1 | 3 | 28 | ||
| Breast | Normal Epithelial | AG11132 | 1 | 2 | 30 |
| Normal Epithelial | AG11134 | 6 | 5 | ||
| Normal Epithelial | MCF-10A | 4 | 4 | 35 | |
| Adenocarcinoma | MCF7 | 6 | 36 | ||
| Lung | Normal Fibroblast | WI-38 | 4 | 3 | |
| Carcinoma | H1299 | 2 | 33 | ||
| Kidney | Immortalized Epithelial | 293T | 3 | 4 | |
| Adenocarcinoma | 786-O | 1 | 28 | ||
| Bone | Immortalized Osteoblast | hFOB 1.19 | 5 | 3 | |
| Osteosarcoma | U-2OS | 2 | 42 | ||
| Brain | Glioblastoma | U-87 MG | 4 | 30 | |
| Glioblastoma | T98G | 2 | 39 | ||
| Skin | Normal Fibroblast | D-1 | 1 | 1 | 44 |
| Normal Fibroblast | K-1 | 3 | 4 | ||
| Melanoma | WM-266-4 | 2 | 2 | ||
| Melanoma | A375 | 1 | 2 | ||
| Cervix | Adenocarcinoma | HeLa | 2 | 2 | |
To determine whether or not similar results would be obtained with other tissues, the same experiment was carried out with epithelial and fibroblast cells from breast, lung, kidney, bone, brain, skin and cervix (Table SI). In each case, siGem reduced geminin levels by ~10-fold. Nevertheless, of the 10 cell types derived from normal tissues, and the 13 cell types derived from cancer tissues, only those cells derived from malignant cancers were induced to re-replicate their DNA upon geminin depletion (Table I; Figs. S1 – S5). Otherwise, no change was detected either in the fraction of cells with greater than 4N DNA, or in the fraction of giant nuclei. Surprisingly, two cells derived from normal tissues, but later immortalized by constitutive expression of SV40 large tumor antigen (293T, hGOB 1.19) also were not induced to re-replicate their DNA in response to geminin depletion. Since 293T cells are tumorigenic but non-metastatic (32), only malignant cancer cells appear to be sensitive to geminin depletion. However, two skin melanoma cells (WM-266-4, A375) as well as the frequently studied cervix adenocarcinoma HeLa cells also were resistant to induction of DNA re-replication in response to geminin depletion. In contrast, cells derived from breast adenocarcinoma (MCF7), lung carcinoma (H1299), kidney adenocarcinoma (786-O), osteosarcoma (U2OS), and brain glioblastoma (U87 MG; Y98G) were as sensitive to geminin depletion as were cells derived from colon cancers.
Simply stated, cells derived from normal tissues, including those that were subsequently immortalized by a viral oncogene, appeared unaffected by geminin depletion, whereas cells derived from malignant cancers underwent DNA re-replication. Moreover, not all cells derived from malignant cancers were sensitive to siGem; cervix adenocarcinoma and skin melanoma cells were as insensitive to geminin depletion as were normal and immortalized cells. These results were obtained with three different siRNAs directed against different sites within the geminin structural gene (Fig. S7). Thus, DNA re-replication in these cells resulted from increased CDT1 activity as a consequence of reduced geminin activity.
DNA Re-Replication Suppressed Cell Proliferation by Inducing Apoptosis
To determine the effect of DNA re-replication on cell proliferation, HCT116 cells were transfected either with siGL2 or with siGem and then harvested at various times post-transfection. The population of cells transfected with siGL2 increased ~350-fold by 8 days with a doubling time of 18–20 hours, and geminin levels in these cells remained constant (Fig. 2A, B). In contrast, geminin levels were suppressed within 2 days of the first transfection with siGem (Fig. 2B), and they did not begin to proliferate (Fig. 2A) until their geminin levels were restored to normal about 6 days after the first transfection. Similarly, depletion of geminin suppressed cell proliferation in cells derived from colon adenocarcinoma (DLD-1), non-small lung carcinoma (H1299), and mammary gland adenocarcinoma (MCF7), but not in five normal cells (Fig. 4D).
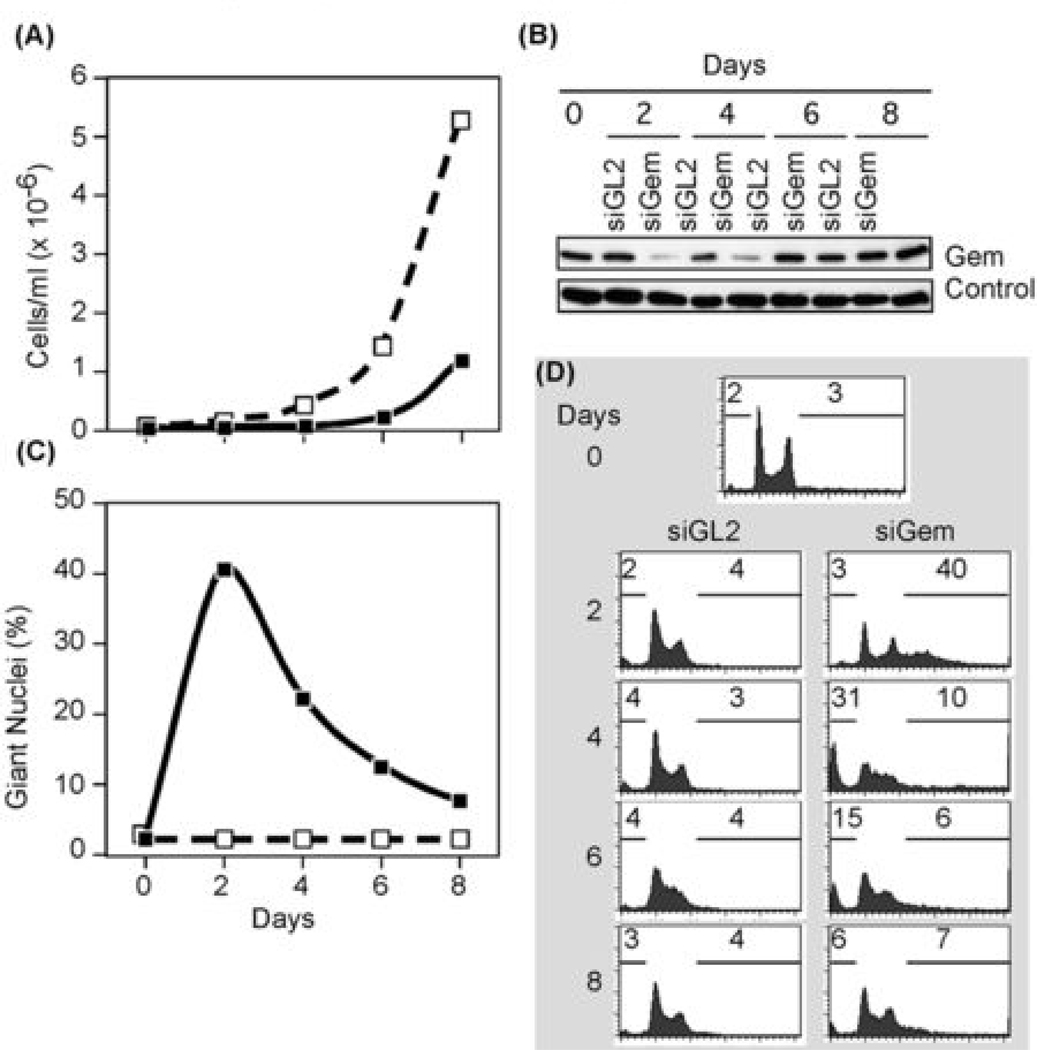
Figure 2.
Geminin depletion suppressed cancer cell proliferation and induced apoptosis. (A) Colon carcinoma HCT116 cells were transfected with either siGem (■) or siGL2 (□),harvested at indicated times post-transfection, and the total number of cells recorded. (B) Cells treated as in (A) were subjected to Western immuno-blotting for geminin and actin. (C) The fraction of cells in (A) that remained attached to the dish with giant nuclei was determined by fluorescent microscopy. (D) Total cells (attached plus unattached) in (A) were analyzed by FACS. Percentage with less than 2N DNA content, and the percentage with greater than 4N DNA content are indicated.
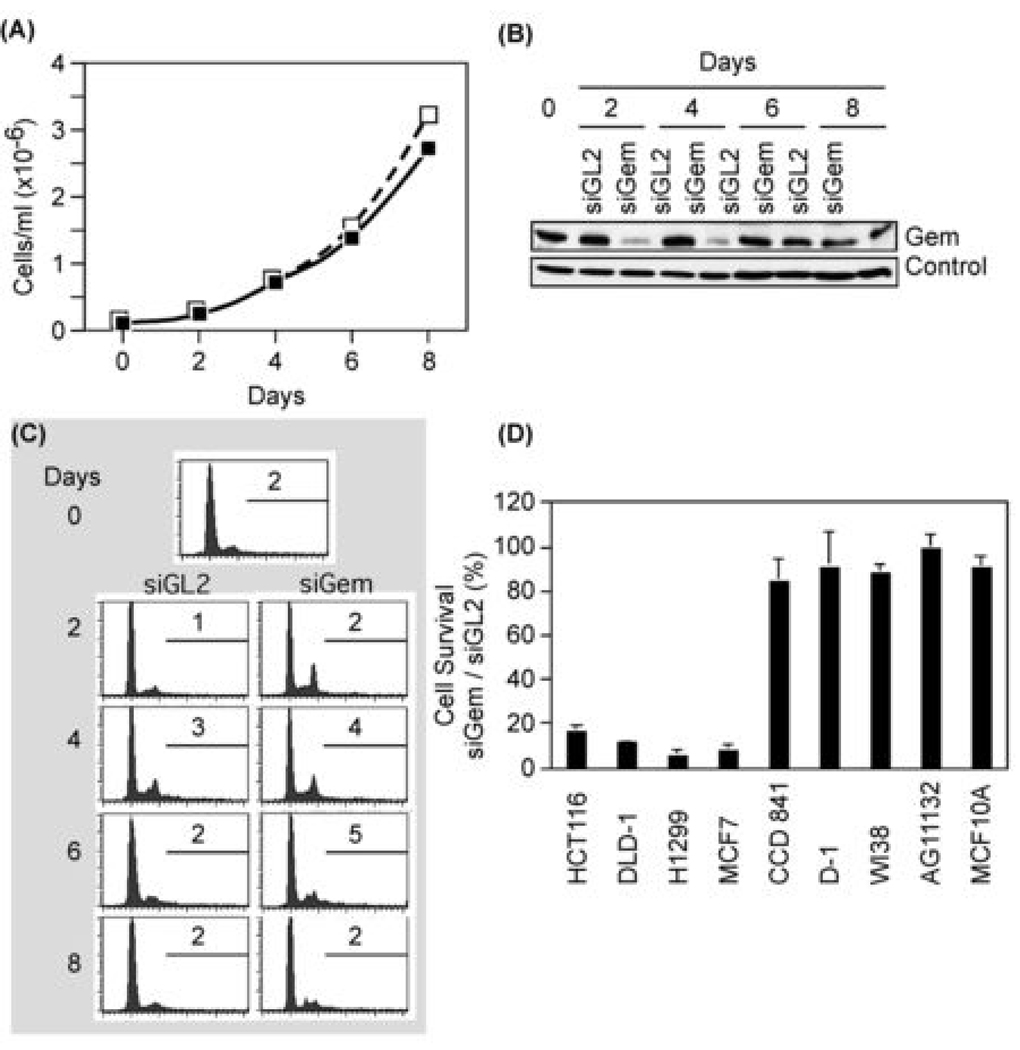
Figure 4.
Depletion of geminin did not suppress proliferation of normal cells and did not induce apoptosis. (A) The total number of normal skin D1 cells transfected with either siGL2 (□) or siGem (■) at the indicated times post-transfection. (B) Cells treated as in (A) were subjected to Western immuno-blotting for geminin and actin. ‘Control’ indicates an unidentified protein that cross-reacted with anti-geminin antibody. (C) Cells in (A) were analyzed by FACS, as in Fig. 1. Percentage of cells with >4N DNA is shown. (D) The indicated cells were transfected with either siGem or siGL2. Six days later the total number of cells was counted, and the ratio of cells surviving siGem transfection relative to those surviving siGL2 transfection was determined.
The failure of geminin depleted cancer cells to proliferate resulted from induction of apoptosis. Two characteristics of apoptosis are loss of cell adhesion and reduction in DNA content. Cells with giant nuclei appeared within two days of siGem transfection and then detached from the dish and disintegrated (Fig. 2C). Since cells were cultured in the absence of siGem, their place was taken by new cells that expressed geminin. FACS analysis confirmed that geminin depletion induced DNA re-replication in cancer cells within two days, but within four days, cells with >4N DNA content disappeared, and cells with <2N DNA content took their place (Fig. 2D). In contrast, cells transfected with siGL2 remained typical of proliferating cell populations (Fig. 2D) and did not accumulate cells with giant nuclei (Fig. 2C).
To confirm that siGem induced apoptosis in cancer cells, geminin depleted HCT116 cells were assayed both for nuclear DNA fragmentation and for a rounded cell morphology with blebbing, two additional characteristics of apoptosis (33). Nuclear DNA fragmentation was detected with the TUNEL assay in which terminal deoxyribonucleotidyl transferase is used to label free 3'-OH terminated DNA fragments with fluorescein-conjugated dUTP. HCT116 cells with giant nuclei were TUNEL positive, whereas HCT116 cells with normal nuclei were not (Fig. 3A). This was consistent with previous studies in which DNA strand breaks were detected in cells undergoing DNA re-replication (6). TUNEL positive nuclei were not detected in HCT116 cells treated with siGL2 (Fig. 3A). Cells with giant nuclei exhibited a rounded cell morphology with multiple blebs, whereas cells with normal nuclei did not (Fig. S6).
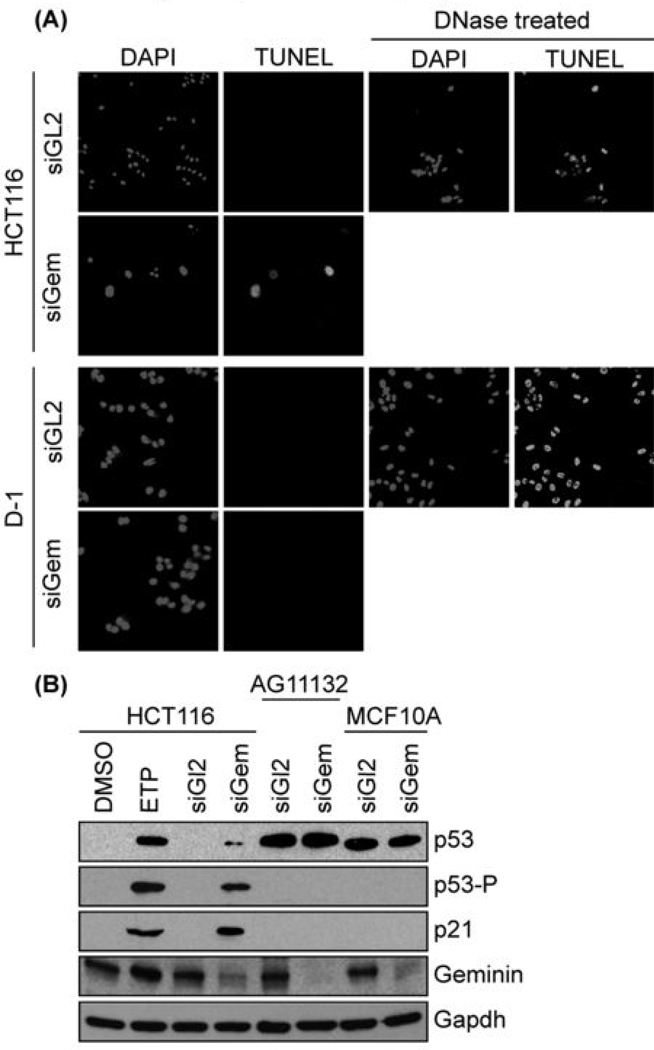
Figure 3.
Only cells that re-replicated their DNA experienced genotoxic stress and apoptosis. (A) Colon carcinoma HCT116 cells were transfected with either siGL2 or siGem. At 4 days post-transfection, cells not attached to the dish were isolated by centrifugation and subjected to the TUNEL assay. HCT116 cells treated with DNase I provided a TUNEL positive control. Normal skin D1 cells were treated likewise. (B) The indicated cells were treated with either siGL2 or siGem as in Fig. 1, and then subjected to Western immuno-blotting for the indicated protein (p53-P is p53 phosphorylated on serine 15). To demonstrate the effects of genotoxic stress, p53+/+ HCT116 cells were treated with 3 µM etoposide (ETP) for 16 hours in parallel with HCT116 cells treated with the same buffered solvent (DMSO). Longer exposures revealed p53 in HCT116 cells, and shorter exposures confirmed that p53 levels in AF11132 and MCF10A cells remained unchanged.
Geminin depletion induces DNA re-replication in cancer cells, regardless of the presence or absence of the tumor suppressor protein p53, but p53+/+ cancer cells induce expression and phosphorylation of p53, as well as expression of the CDK-specific inhibitor p21 whose transcription is p53 dependent [(7); Fig. 3B]. These events are part of the DNA damage response in mammalian cells, as demonstrated by treatment of cells with etoposide, a specific inhibitor of topoisomerase II that induces DNA breaks (Fig. 3B). Thus, induction of DNA re-replication by siGem in cells from malignant cancers resulted in apoptosis within four to eight days post-transfection.
Geminin Depletion Did Not Affect Normal Cell Proliferation
Normal somatic cells typically proliferate slowly compared with cancer cells. For example, normal breast AG11132 cells doubled in 38–40 hours, whereas breast adenocarcinoma MCF7 cells doubled in 30–32 hours. Normal colon FHC cells doubled in 39 hours, whereas colon carcinoma HCT116 cells required only 18–20 hours. Since differences in the time required for cell division in mammalian cells reflect differences in the length of their G1-phase, the absence of DNA re-replication in normal cells treated with siGem may result simply from fewer S, G2 and M phase cells, the only cells expressing geminin protein. In that case, normal cells may simply require more time to respond to geminin depletion. However, this was not the case.
The rate of proliferation for normal primary skin D1 fibroblasts was not affected significantly by siGem relative to siGL2 (Fig. 4A), despite the fact that geminin levels were reduced markedly in cells transfected with siGem, but not in cells transfected with siGL2 (Fig. 4B). Furthermore, recovery of geminin expression in D1 cells transfected with siGem (Fig. 4B) followed the same time course as observed with HCT116 cells (Fig. 2B). FACS profiles for both siGL2 and siGem treated D1 cells were characteristic of proliferating cell populations, and they exhibited no significant change in the fraction of cells containing >4N DNA (Fig. 4C).
To determine whether or not normal cells accumulated DNA damage in response to geminin depletion, each cell type was treated with either siGL2 or siGem and then assayed for nuclear DNA fragmentation. Under conditions that readily detected TUNEL positive cells in cancer cells treated with siGem, no TUNEL positive cells were detected in normal cells such as D1 (Fig. 3A), and other noncarcinogenic cells such as colon CCD841 CoN, lung WI-38, breast AG11132 and MCF10A (Fig. 4D; data not shown). Moreover, the amount of p53 in normal cells was not increased by geminin depletion and p53 was not phosphorylated. Moreover, siGem did not induce expression of p21 (Fig. 3B). Therefore, siGem did not induce genotoxic stress in normal cells under conditions where it did so in cancer cells.
Sensitivity To siGem Did Not Correlate With Cellular Levels Of Either Geminin Or CDT1
The simplest explanation for why geminin depletion induces DNA re-replication in cancer cells but not in normal cells is that the ratio of CDT1 to geminin is increased to a greater extent in cancer cells than in normal cells. To assess this possibility, geminin and CDT1 protein levels were assayed before and after geminin depletion in normal skin D1 cells and in colon carcinoma HCT116 cells (Fig. 5). Geminin depleted D1 cells experienced a 3 to 4-fold decrease in CDT1, although the ratio of CDT1 to geminin actually increased 5-fold. Similarly, geminin depleted HCT116 cells experienced at least a 10-fold decrease in CDT1 [as previously reported (7)], although with little, if any, change in the CDT1/geminin ratio. Therefore, the ability of geminin depletion to induce DNA re-replication in HCT116 cells, but not in D1 cells, did not correlate with increased CDT1 protein levels.
Figure 5.
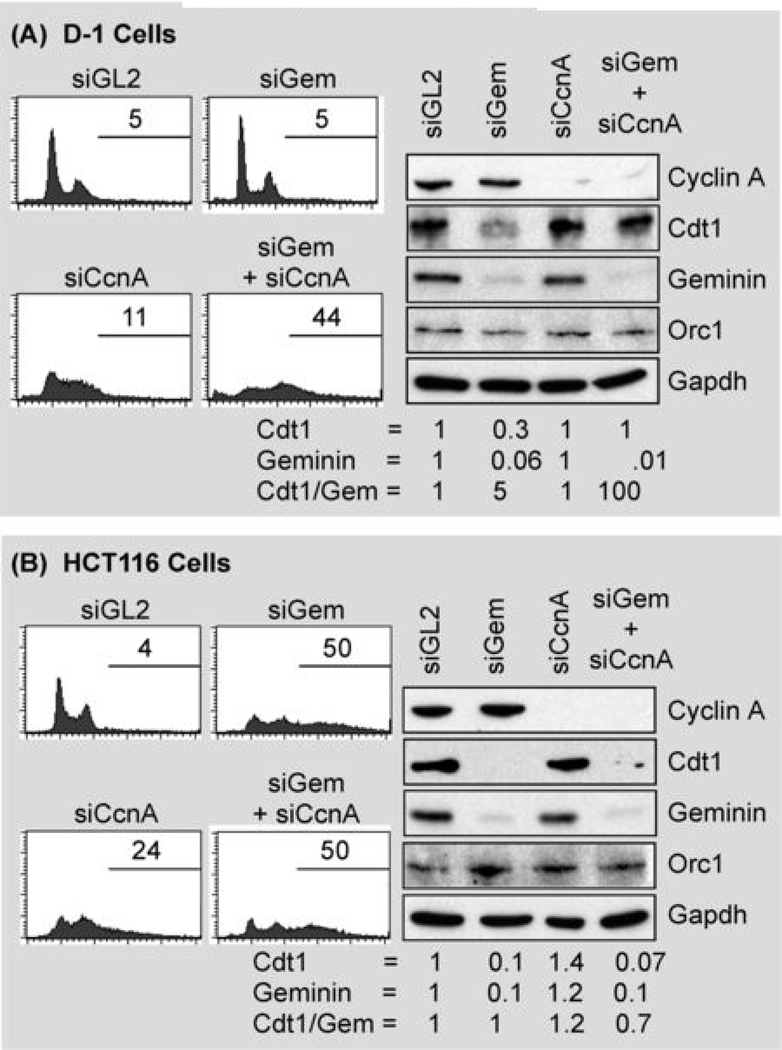
Normal cells prevent DNA re-replication through multiple convergent pathways that are dependent on both geminin and cyclin A. Normal skin D1 cells (A) and colon carcinoma HCT116 cells (B) were transfected either with siGL2, with siGem, with siCcnA, or with both siGem and siCcnA. Cells were harvested at two days post-transfection and subjected to FACS analysis to determine the fraction of cells with greater than 4N DNA content. The same cells were subjected to Western immuno-blotting to determine the relative levels of cyclin A, CDT1, geminin, and Orc1. Protein levels were quantified using Multi Gauge software (Fuji Film). Cdt1 and geminin protein levels were normalized relative to Gapdh, and then to either Cdt1 or geminin in siGl2 treated cells.
The levels of geminin and CDT1 protein were compared in other cells as well. As expected, the levels of both geminin and CDT1 were about 5-fold greater in colon cancer cells (HCT116, DLD-1, SW480, COLO320DM) than in normal colon cells (CCD841CoN, FHC), and the ratios of geminin to CDT1 in each of these cells was essentially unchanged (Fig. S8A). As expected, cell populations with high geminin levels contained a greater proportion of cells in S and G2/M phases than cell populations with lower geminin levels (Fig. 8SB). Normal skin D1 cells were similar to normal colon cells. Therefore, one would not expect depletion of geminin to be more injurious to cancer cells than to normal cells.
On the other hand, normal breast cells (AG11132, AG11134, MCF10A) exhibited higher geminin levels but lower CDT1 levels than breast adenocarcinoma MCF7 cells (Fig. 8SA). Again, these results were consistent with the relative fraction of cells in S and G2/M phases (Fig. 8SB). Thus, a simple correlation between geminin levels, geminin to CDT1 ratios, and sensitivity to geminin depletion was not apparent. Normal colon cells contained low levels of geminin, because they consisted predominantly of G1-phase cells, and normal breast cells contained high levels of geminin, because they contained a higher proportion of proliferating cells, but neither cell type underwent DNA re-replication when transfected with siGem.
Induction Of DNA Re-Replication In Normal Cells Required Suppression Of Multiple Regulatory Pathways
Several different but convergent pathways have been described that contribute to restricting genome duplication to once each time a cell divides (3, 4, 13). At least three of them involve CDK-dependent phosphorylation of preRC proteins, and in metazoa, cyclin A is the principle activator of CDK activities that suppress DNA re-replication. To determine whether or not suppression of multiple pathways induces DNA re-replication in normal cells, D1 cells were treated with both siGem and siRNA targeted against cyclin A (siCcnA). Remarkably, this combination induced DNA re-replication in normal cells to an extent similar to that observed in geminin depleted HCT116 cells (Fig. 5). The response to depletion of cyclin A alone in D1 cells was detectable but modest in comparison to depletion of both cyclin A and geminin. Similar results were obtained with normal breast (AG11132, MCF10A) and colon (CCD841 CoN) cells (Table I). In contrast, the response to depletion of both cyclin A and geminin in HCT116 cells was essentially the same as with depletion of geminin alone. Therefore, DNA re-replication is prevented primarily by the ratio of geminin to CDT1 in HCT116 cells, whereas DNA re-replication in D1 cells is prevented by cyclin A-dependent pathways as well as by geminin.
This conclusion was confirmed by changes in the levels of CDT1 and geminin in these experiments (Fig. 5). Depletion of cyclin A in D1 cells did not affect the levels of either CDT1 or geminin, whereas depletion of both cyclin A and geminin prevented loss of CDT1 (Fig. 5), consistent with a critical role for CDT1 in DNA re-replication. However, while depletion of cyclin A did not affect CDT1 and geminin levels in HCT116 cells, neither did it prevent loss of CDT1 in geminin depleted cells. Since neither siGem nor siGem plus siCcnA increased the ratio of CDT1 to geminin in HCT116 cells, elevation of CDT1 activity does not account for DNA re-replication under these conditions. Both CDK and geminin-dependent pathways prevent DNA re-replication in normal cells, whereas cancer cells rely primarily, if not exclusively, on geminin.
DISCUSSION
Studies on cells from flies, frogs, plants, and mammals suggest that CDT1 activity is the rate limiting step in preRC assembly in metazoa, and that geminin is the critical regulator of CDT1 activity [reviewed in (3, 18, 29, 30, 34–38)]. Nevertheless, exceptions have been reported. Whereas geminin depletion induced DNA re-replication in some cancer cells (5–7), it did not do so in HeLa (20, 39, 40), MCF10A (40), and 293T (41) cells. Moreover, over-expression of both CDT1 and Cdc6 induced DNA re-replication in some, but not all, lung carcinoma cells, and it did not induce DNA re-replication in primary human and rodent cells, 293T cells, HeLa cells and other cervical carcinoma cell lines (19, 42). These results suggested to us that CDT1 activity may be the rate limiting factor for preRC assembly in rapidly proliferating cells such cleavage stage embryos, preimplantation embryos, and cancer cells, but not in normal cells.
To test this hypothesis, siRNA targeted against the human geminin gene was transfected into 23 different cell lines derived from eight different human tissues (Table I and Table SI). In six of these tissues, cells were derived from both malignant and normal biopsies. Remarkably, all of the cells that re-replicated their DNA had been derived from malignant cancers; none were from normal tissues. Moreover, cells derived from normal tissues that were later immortalized with SV40 T-antigen (293T, hFOB 1.19) continued to proliferate normally even when their geminin levels were reduced more than 10-fold. Similarly, three cell lines derived from malignant cancers (HeLa, WM-266-4, A375) also were insensitive to suppression of their geminin levels. We attribute these effects to suppression of geminin synthesis, because all three of the siGem oligoribonucleotides tested suppressed geminin protein in both cancer and non-cancer cells, and either over-expression of a non-degradable form of geminin (28), or co-suppression of both CDT1 and geminin has been shown to prevent induction of DNA re-replication by two of these same molecules (5, 7). These studies, carried out under the same conditions using the same siRNAs, demonstrate that whereas some malignant cancer cells are sensitive to geminin depletion, most primary and secondary mammalian cells are insensitive. Therefore, drugs targeted specifically against geminin activity should be excellent candidates to selectively induce DNA re-replication in some forms of malignant cancer without interfering either with the function or proliferation of non-cancer cells.
Previous studies reported that inhibition either of the ATM and ATR protein kinases, or the CHK1 protein kinase two days after siGem treatment of HCT116 cells rapidly induced apoptosis in cells that had undergone DNA re-replication (7). Therefore, DNA re-replication induced by geminin depletion activates DNA damage response pathways that prevent cells from entering mitosis until the damage is repaired, because suppression of this pathway resulted in apoptosis. When these pathways are inhibited, induction of apoptosis is immediate (6, 7). Here we show that induction of apoptosis occurs spontaneously within a few days of siGem transfection without addition of protein kinase inhibitors. Since siGem did not induce DNA re-replication, DNA damage, or DNA damage response genes in either normal or immortalized cells, it did not induce apoptosis. Normal cells continued to proliferate. Therefore, inhibition of geminin activity should, in principle, selectively kill cancer cells in vivo.
Induction of DNA re-replication by geminin depletion implies that the level of Cdt1 activity is rate limiting for origin licensing, and that a reduction in geminin results in an increase in Cdt1 activity. However, suppression of geminin in either cancer or non-cancer cells resulted in a concomitant decrease of Cdt1 protein, the result of ubiquitin-mediated degradation of Cdt1 when it is not bound to geminin (43). Suppression of both geminin and cyclin A restored Cdt1 protein levels to normal in non-cancer cells and induced DNA re-replication. Similar results have been reported with Drosophila cells (27), and confirms the existence multiple, convergent pathways that restrict genome duplication to once per cell division. For example, CDT1 activity in humans is down-regulated during S-phase in three ways (3, 44, 45). It is phosphorylated by cyclin A:CDK2 and then ubiquitinated by SCFskp2. It is also ubiquitinated by CDRDdb1 in the presence of PCNA, a protein that stimulates DNA polymerase activity at replication forks and that also binds to CDT1. Both reactions result in degradation of CDT1 by the 26S proteasome, although only SCF requires prior phosphorylation of its substrate. Finally, geminin binds specifically to CDT1, thereby inhibiting its activity while simultaneously protecting CDT1 from ubiquitination and degradation. In addition to inactivation of CDT1, cyclin A dependent phosphorylation of Orc1 prevents it binding to chromatin during mitosis, and cyclin A dependent phosphorylation of Cdc6 affects its nuclear localization (3, 40). All of these mechanisms could contribute to restricting genome duplication to once per cell division in normal cells, but the fact that suppression of geminin alone can induce DNA re-replication in some cancer cells means that one or more of these pathways is inactive in those cells.
Since geminin protects CDT1 from ubiquitin-dependent degradation (43), suppression of geminin also reduced CDT1 protein levels in both normal and cancer cells. The fact that cancer cells could still re-replicate their DNA under these conditions can be attributed to the approximately 10-fold excess of CDT1 (Fig. S8) and other preRC proteins [(46); unpublished results of S. Ghosh & M.L. DePamphilis] in cancer cells compared with normal cells. The fact that suppression of both geminin and cyclin A restored CDT1 levels in normal cells but not in cancer cells (Fig. 5) suggests that normal cells use the SCF pathway to degrade CDT1 whereas some cancer cells use the CDR pathway. The fact that both cyclin A and geminin prevent DNA re-replication in normal cells but only geminin prevents DNA re-replication in some cancer cells could be explained if both SCF and geminin maintained CDT1 activity rate limiting in normal cells, but only geminin maintained CDT1 activity rate limiting in some cancer cells. However, this does not appear to be the case, because over-expression of CDT1 fails to induce DNA re-replication in mammalian cells (19, 31). Therefore, either cyclin A dependent phosphorylation of ORC1 or CDC6 plays a role, or there is as yet an undiscovered regulatory pathway in mammalian cells.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the intramural program of the National Institute of Child Health and Human Development. Antiserum against geminin was kindly provided by Dr. Anindya Dutta.
REFERENCES
- 1.Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. Embo J. 2006;25:5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly TJ, Stillman B. Duplication of DNA in Eukaryotic Cells. In: DePamphilis ML, editor. DNA Replication and Human Disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 1–30. [Google Scholar]
- 3.DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Blow JJ, Gillespie PJ. Replication licensing and cancer--a fatal entanglement? Nat Rev Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melixetian M, Ballabeni A, Masiero L, et al. Loss of Geminin induces rereplication in the presence of functional p53. J Cell Biol. 2004;165:473–482. doi: 10.1083/jcb.200403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W, Dutta A. An ATR- and BRCA1-mediated Fanconi anemia pathway is required for activating the G2/M checkpoint and DNA damage repair upon rereplication. Mol Cell Biol. 2006;26:4601–4611. doi: 10.1128/MCB.02141-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montanari M, Boninsegna A, Faraglia B, et al. Increased expression of geminin stimulates the growth of mammary epithelial cells and is a frequent event in human tumors. J Cell Physiol. 2005;202:215–222. doi: 10.1002/jcp.20120. [DOI] [PubMed] [Google Scholar]
- 9.Wohlschlegel JA, Kutok JL, Weng AP, Dutta A. Expression of geminin as a marker of cell proliferation in normal tissues and malignancies. The American journal of pathology. 2002;161:267–273. doi: 10.1016/S0002-9440(10)64178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shreeram S, Sparks A, Lane DP, Blow JJ. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng D, Tu Z, Wu W, Liang C. Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res. 2003;63:7356–7364. [PubMed] [Google Scholar]
- 12.Machida YJ, Teer JK, Dutta A. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J Biol Chem. 2005;280:27624–27630. doi: 10.1074/jbc.M502615200. [DOI] [PubMed] [Google Scholar]
- 13.Schwob E, Labib K. Regulating Initiation Events in Yeasts. In: DePamphilis ML, editor. DNA Replication and Human Disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 295–312. [Google Scholar]
- 14.Baldinger T, Gossen M. Binding of Drosophila ORC proteins to anaphase chromosomes requires cessation of mitotic cyclin-dependent kinase activity. Mol Cell Biol. 2009;29:140–149. doi: 10.1128/MCB.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu JR, Gilbert DM. The replication origin decision point is a mitogen-independent, 2-aminopurine-sensitive, G1-phase event that precedes restriction point control. Mol Cell Biol. 1997;17:4312–4321. doi: 10.1128/mcb.17.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JR, Gilbert DM. Lovastatin arrests CHO cells between the origin decision point and the restriction point. FEBS letters. 2000;484:108–112. doi: 10.1016/s0014-5793(00)02135-9. [DOI] [PubMed] [Google Scholar]
- 17.Wu JR, Keezer SM, Gilbert DM. Transformation abrogates an early G1-phase arrest point required for specification of the Chinese hamster DHFR replication origin. Embo J. 1998;17:1810–1818. doi: 10.1093/emboj/17.6.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivaprasad U, Dutta A, Bell SP. Assembly of Pre-replication Complexes. In: DePamphilis ML, editor. DNA Replication and Human Disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 63–88. [Google Scholar]
- 19.Vaziri C, Saxena S, Jeon Y, et al. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 20.Nishitani H, Lygerou Z, Nishimoto T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J Biol Chem. 2004;279:30807–30816. doi: 10.1074/jbc.M312644200. [DOI] [PubMed] [Google Scholar]
- 21.Thomer M, May NR, Aggarwal BD, Kwok G, Calvi BR. Drosophila double-parked is sufficient to induce re-replication during development and is regulated by cyclin E/CDK2. Development. 2004;131:4807–4818. doi: 10.1242/dev.01348. [DOI] [PubMed] [Google Scholar]
- 22.Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005;19:114–126. doi: 10.1101/gad.1255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiorano D, Krasinska L, Lutzmann M, Mechali M. Recombinant Cdt1 induces rereplication of G2 nuclei in Xenopus egg extracts. Curr Biol. 2005;15:146–153. doi: 10.1016/j.cub.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida K, Takisawa H, Kubota Y. Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells. 2005;10:63–73. doi: 10.1111/j.1365-2443.2005.00815.x. [DOI] [PubMed] [Google Scholar]
- 25.Li A, Blow JJ. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. Embo J. 2005;24:395–404. doi: 10.1038/sj.emboj.7600520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellano MM, Boniotti MB, Caro E, Schnittger A, Gutierrez C. DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell. 2004;16:2380–2393. doi: 10.1105/tpc.104.022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mihaylov IS, Kondo T, Jones L, et al. Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol Cell Biol. 2002;22:1868–1880. doi: 10.1128/MCB.22.6.1868-1880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tachibana KE, Gonzalez MA, Guarguaglini G, Nigg EA, Laskey RA. Depletion of licensing inhibitor geminin causes centrosome overduplication and mitotic defects. EMBO Rep. 2005;6:1052–1057. doi: 10.1038/sj.embor.7400527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez MA, Tachibana KE, Adams DJ, et al. Geminin is essential to prevent endoreduplication and to form pluripotent cells during mammalian development. Genes Dev. 2006;20:1880–1884. doi: 10.1101/gad.379706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hara K, Nakayama KI, Nakayama K. Geminin is essential for the development of preimplantation mouse embryos. Genes Cells. 2006;11:1281–1293. doi: 10.1111/j.1365-2443.2006.01019.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu E, Lee AY, Chiba T, Olson E, Sun P, Wu X. The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J Cell Biol. 2007;179:643–657. doi: 10.1083/jcb.200704138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan W, Shao R. Transduction of a mesenchyme-specific gene periostin into 293T cells induces cell invasive activity through epithelial-mesenchymal transformation. J Biol Chem. 2006;281:19700–19708. doi: 10.1074/jbc.M601856200. [DOI] [PubMed] [Google Scholar]
- 33.Otsuki Y, Li Z, Shibata MA. Apoptotic detection methods--from morphology to gene. Prog Histochem Cytochem. 2003;38:275–339. doi: 10.1016/s0079-6336(03)80002-5. [DOI] [PubMed] [Google Scholar]
- 34.Kerns SL, Torke SJ, Benjamin JM, McGarry TJ. Geminin prevents rereplication during xenopus development. J Biol Chem. 2007;282:5514–5521. doi: 10.1074/jbc.M609289200. [DOI] [PubMed] [Google Scholar]
- 35.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nature reviews. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tada S. Cdt1 and geminin: role during cell cycle progression and DNA damage in higher eukaryotes. Front Biosci. 2007;12:1629–1641. doi: 10.2741/2175. [DOI] [PubMed] [Google Scholar]
- 37.DePamphilis ML, Blow JJ. Regulating Initiation Events in Metazoa. In: DePamphilis ML, editor. DNA Replication and Human Disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 313–334. [Google Scholar]
- 38.Saxena S, Dutta A. Geminin-Cdt1 balance is critical for genetic stability. Mutation research. 2005;569:111–121. doi: 10.1016/j.mrfmmm.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 39.Kulartz M, Knippers R. The replicative regulator protein geminin on chromatin in the HeLa cell cycle. J Biol Chem. 2004;279:41686–41694. doi: 10.1074/jbc.M405798200. [DOI] [PubMed] [Google Scholar]
- 40.Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teer JK, Dutta A. Human Cdt1 lacking the evolutionarily conserved region that interacts with MCM2-7 is capable of inducing re-replication. J Biol Chem. 2008;283:6817–6825. doi: 10.1074/jbc.M708767200. [DOI] [PubMed] [Google Scholar]
- 42.Tatsumi Y, Sugimoto N, Yugawa T, Narisawa-Saito M, Kiyono T, Fujita M. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. Journal of cell science. 2006;119:3128–3140. doi: 10.1242/jcs.03031. [DOI] [PubMed] [Google Scholar]
- 43.Ballabeni A, Melixetian M, Zamponi R, Masiero L, Marinoni F, Helin K. Human Geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. Embo J. 2004;23:3122–3132. doi: 10.1038/sj.emboj.7600314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujita M. Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell division. 2006;1:22. doi: 10.1186/1747-1028-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNairn AJ, Gilbert DM. Overexpression of ORC subunits and increased ORC-chromatin association in transformed mammalian cells. Journal of cellular biochemistry. 2005;96:879–887. doi: 10.1002/jcb.20609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.