Abstract
The outcome of hepatitis C virus (HCV) infection has been associated with antiviral CD4 T cell response, human leukocyte antigens (HLA) class II genotypes, and ethnicity. However, HLA class II molecules restrict the nature of CD4 T cell response, and HLA distributions differ between ethnic groups. In this study, we asked whether HLA class II genotypes associated with HCV clearance are shared between Caucasian and African Americans and whether they contribute to enhanced antiviral CD4 T cell response. In a cohort of 93 HCV-seropositive subjects from Northeast America with defined ethnicity, virological outcome, and HCV-specific CD4 T cell proliferation, we confirm the previously reported associations between HCV clearance and two HLA types (DQB1*03, DRB1*11) while identifying a new association with DRB3*02. Strikingly, these associations were identified only among Caucasian [DQB1*03: odds ratio (OR), 10.4; P = 0.031, DRB1*11: OR, 7.0, P = 0.019; DRB3*02: OR, 8.3, P = 0.005; DQB1*03-DRB3*02: OR, 13.5, P = 0.001) but not among African American patients. Furthermore, although HLA DQB1*03, DRB1*11, and DRB3*02 genotypes were associated with increased HCV-specific CD4 T cell response in univariate analyses, these associations were lost when controlling for virological outcomes.
Conclusion
We conclude that the immunogenetic basis for HCV clearance differs between ethnic groups and that the association between HLA class II and HCV clearance is not directly explained by antiviral CD4 T cell response.
Infection with the hepatitis C virus (HCV) presents with a variety of clinical manifestations. A minority of individuals will clear the virus whereas most become persistently viremic with subsequent course that ranges from minimal inflammation to progressive cirrhosis and, in some cases, hepatocellular carcinoma.1,2 Although the pathogenesis of HCV infection is not fully defined, a strong, sustained multispecific CD4 proliferative T cell response is associated with HCV clearance in HCV-infected patients.3–6 Furthermore, direct CD4 depletion resulted in chronic evolution in previously HCV-immune chimpanzees.7 These results suggest that CD4 T cells play a critical role in the outcome of HCV infection. Additional host factors may contribute to the outcome of HCV infection, including ethnicity and human leukocyte antigen (HLA) type. For example, chronic HCV infection is more prevalent among African Americans, who are also more resistant to antiviral therapy based on interferon-alpha (IFN-α),8–12 suggesting an underlying genetic mechanism of immune tolerance to HCV. Indeed, African Americans tend to display less severe biochemical and histological outcome of HCV infection compared with Caucasian Americans while maintaining HCV-specific CD4 T cell proliferative response.6,8,13 In addition to ethnicity, various immunogenetic factors have been associated with the outcome of HCV infection including HLA, natural killer inhibitory receptors, and single nucleotide polymorphisms for immune-associated genes.14–18 Given the relevance of CD4 T cells in HCV infection, a particular attention has been focused on HLA class II molecules, which bind exogenously processed antigenic peptides (or epitopes) and present them on cell surface for CD4 T cell recognition.19–21 Genotypic variants of class II molecules display distinctive pockets in the peptide-binding groove that interact with specific residues within the antigenic peptides, thus influencing the nature and specificity of antigen-specific CD4 T cell response.22 In HCV infection, the HLA class II genotypes DQB1*03 and DRB1*11 have been consistently linked with self-limited HCV infections23–28 and DQB1*02 with chronic evolution,29,30 mostly in European cohorts.
In this study, we examined whether HLA associations with virological outcomes differ between African Americans and Caucasian Americans and whether HLA associations with virological outcome are based on antiviral CD4 T cell response. We show that HLA DQB1*03, DRB1*11, and DRB3*02 correlate with HCV clearance in white but not black patients, demonstrating a novel ethnic disparity in immunogenetic associations relevant for HCV clearance. However, HLA class II genotypes associated with HCV clearance did not influence antiviral CD4 T cell response when controlling for virological status, suggesting that HLA class II associations with HCV clearance are not directly explained by antiviral CD4 T cell response and highlighting the need to validate immunogenetic associations with functional analyses.
Patients and Methods
Study Subjects
Ninety-three HCV-seropositive subjects including 70 chronically HCV-infected (“chronic” or C) and 23 HCV-recovered (“recovered” or R) subjects were selected from our existing cohort of 99 chronic and 31 recovered subjects that were also included in our previous studies examining HCV-specific CD4 T cell response6,31 based on availability of cells for HLA class II genotyping, ethnicity defined as either Caucasian American (“white”) or African American (“black”) and known HCV genotype or serotype. Patients with prior antiviral or immunosuppressive therapy, acute hepatitis C within 1 year, human immunodeficiency virus (HIV), or hepatitis B virus coinfection, autoimmune diseases, or conditions precluding phlebotomy were excluded. Study subjects were enrolled through the clinics at the Philadelphia Veterans Affairs Medical Center, Temple University, the Department of Transfusion Medicine at the National Institutes of Health, and the Clinical Research Center at the University of Pennsylvania. All subjects gave written informed consent to participate in this study, approved by the respective institutional review board. All subjects were assessed for demographic and clinical parameters as well as serum anti-HCV antibody by second-generation enzyme immunoassay and HCV RNA by quantitative or qualitative Roche COBAS reverse transcriptase polymerase chain reaction (Roche Diagnostics, Branchburg, NJ). HCV RNA-positive patients were tested for HCV genotype by INNOLIPA (Innogenetics, Ghent, Belgium). In HCV-recovered subjects, plasma HCV antibodies were serotyped using an HCV serotyping 1–6 assay (performed by David Parker and Lara Sandler, Murex Diagnostic, London, UK), as previously described.31
The clinical, demographic, and immunological profiles of the 93 selected subjects (Table 1) were consistent with our previous report with greater liver function abnormalities and suppressed HCV-specific CD4 T cell response in chronic patients compared with recovered subjects.6,31 Of note, HCV-associated risk factors were similar between the chronic and recovered subjects, including prior injection drug or cocaine use (77% C, 73% R) and transfusion (27% C, 17% R), as previously reported.31
Table 1.
Patient Characteristics
| Chronic Patients | Recovered Patients | Chronic vs. Recovered P-value |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| All Patients (n = 70) |
Blacks (n = 42) |
Whites (n = 28) |
B versus W P-value |
All Patients (n = 23) |
Blacks (n = 10) |
Whites (n = 13) |
B versus W P-value |
||
| A. Median demographic and clinical parametersa | |||||||||
| Age, years | 52 | 52 | 51 | 0.183 | 46 | 49 | 44 | 0.277 | 0.001 |
| ALT, IU/mL | 52 | 49 | 61 | 0.265 | 26 | 23 | 31 | 0.214 | <0.001 |
| Albumin, g/dL | 4.4 | 4.3 | 4.4 | 0.810 | 4.3 | 4.4 | 4.3 | 0.950 | 0.978 |
| Total bilirubin, mg/dL | 0.7 | 0.7 | 0.7 | 0.371 | 0.5 | 0.5 | 0.5 | 0.725 | 0.025 |
| Prothrombin time (INR) | 0.98 | 0.96 | 1.02 | 0.001 | 0.99 | 0.99 | 0.97 | 0.906 | 0.971 |
| Platelets, 103/mm3 | 230 | 242 | 206 | 0.006 | 252 | 260 | 250 | 0.390 | 0.069 |
| Sex (%M) | 97.1 | 97.6 | 96.4 | 1.000 | 82.6 | 90.0 | 76.9 | 0.604 | 0.031 |
| Virus type 1, % | 78.6 | 88.1 | 64.3 | 0.017 | 78.3 | 90 | 69.2 | 0.034 | |
| B. % HCV-specific CD4 T cell proliferation with SI > 3.0 among genotype 1-infected patients | |||||||||
| n = 55 | n = 37 | n = 18 | n = 55 | n = 18 | n = 9 | n = 9 | n = 18 | n = 73 | |
| Core | 7.3 | 10.8 | 0 | 0.291 | 44.4 | 22.2 | 66.7 | 0.153 | 0.001 |
| NS3-4 | 5.5 | 8.1 | 0 | 0.543 | 88.9 | 100.0 | 77.8 | 0.471 | <0.001 |
| NS5 | 9.1 | 13.5 | 0 | 0.160 | 72.2 | 88.9 | 55.6 | 0.294 | <0.001 |
| ≥1 Antigen | 16.4 | 24.3 | 0 | 0.023 | 94.4 | 100.0 | 88.9 | 1.000 | <0.001 |
A. Median demographic, clinical, and virological parameters for each ethnic subgroup of chronic and recovered patients are shown. Virus type was determined by molecular genotyping for chronic patients and serotyping for recovered patients. P-values for continuous variables were calculated using the nonparametric Mann-Whitney U test. B. HCV-specific CD4 proliferative T cell response was considered positive if the stimulation index was > 3.0. P-values for categoric variables were calculated using chi-square or Fisher’s exact test.
Abbreviations: ALT, alanine aminotransferase; INR, international normalized ratio.
Peripheral Blood Mononuclear Cells
Blood samples from each patient were layered onto Ficoll-Histopaque (Sigma Chemical Co, St Louis, MO), centrifuged to isolate the buffy coat layer, washed three times in Roswell Park Memorial Institute (Gibco BRL, Gaithersburg, MD), counted on the hemocytometer, and either used immediately in complete Roswell Park Memorial Institute with 10% AB serum or cryopreserved as previously described.6,31–33
Recombinant HCV Proteins
All recombinant HCV proteins and control superoxide dismutase proteins were provided by Dr. Michael Houghton (Chiron Corp., Emeryville, CA). These HCV genotype 1a–derived proteins coded for HCV core (c22, amino acids 2–120), NS3-4 (c200, amino acids 1192–1931), and NS5 (NS5, amino acids 2054–2995), as described previously.31
HLA Typing
Class II molecular typing was performed by polymerase chain reaction and sequence-specific probe hybridization at the HLA Laboratories at the National Institutes of Health and the University of Pennsylvania. The HLA typing was performed at the genotype level with two-digit intermediate/low resolution in 81 of 93 subjects and with allele level four-digit high resolution in 12 of 93 subjects. Four-digit allele specificities were collapsed into their respective two-digit categories to enhance statistical power. Because most HLA data involved the intermediate/low-level analysis, we used the terms HLA genotype or HLA types throughout the text to more accurately represent our data rather than using the term “allele,” appropriate for higher-resolution HLA typing.
HCV-Specific CD4 Proliferative T Cell Response
Freshly isolated peripheral blood mononuclear cells were stimulated in five replicates (0.2 million peripheral blood mononuclear cells/well) for 7 days with recombinant HCV and negative control superoxide dismutase proteins (10 µg/mL) as previously described.31 Results were expressed as a stimulation index (SI), calculated as the mean counts per minute in stimulated wells divided by that in control wells. To simplify analysis, an SI cutoff of 3.0 was used to define a positive response for each HCV antigens because an SI of 3.0 was always greater than 2 standard deviations above the average responses to all three HCV antigens in uninfected controls. For a positive control, peripheral blood mononuclear cells from all patients were stimulated with and without 2 µg/mL phytohemagglutinin in triplicates for 4 days.
Statistical Analysis
The clinical and immunological parameters of patient subgroups were compared using a nonparametric Mann-Whitney U test. HLA genotype frequencies were compared by the chi-squared test, or Fisher’s exact test when an expected cell value was less than 5. Three genotypes (DQB1*02, DQB1*03, and DRB1*11) were predicted, a priori, to be related to our outcome and response measures. For all other genotypes, a modified Bonferroni correction for multiple comparisons as recommended by Keppel et al.34 was used to determine the nominal level of significance. In the case of the DQB1 locus, the number of comparisons was 5 (α = 0.010). For the DRB loci, there were 13 comparisons in DRB1 (α = 0.004); three in DRB3 (α = 0.017); one in DRB4 (α = 0.050); and two in DRB5 (α = 0.025). Where necessary to avoid division by zero, odds ratios (ORs) were adjusted by the method of Woolf and Haldane. The Breslow-Day statistic was used to test the homogeneity of ORs across ethnic groups. Logistic regression was used to estimate the effects of HLA genotypes on CD4 T cell response controlling for virological outcome, employing a small delta adjustment (0.2) to stabilize the algorithm and prevent bias in the estimates. Statistical analysis was conducted using Stata version 9.0 (StataCorp, College Station, TX) and SPSS version 14.0 (Chicago, IL).
Results
HCV clearance Is Associated with HLA Class II Genotypes DQB1*03, DRB1*11, and DRB3*02 in White but Not Black Patients
The relationship between HLA class II and the outcome of HCV infection was examined in 93 HCV-seropositive subjects including 70 chronically HCV-infected (“chronic”) and 23 HCV-recovered (“recovered”) subjects with either Caucasian American (“white”) or African American (“black”) ethnicity, selected based on available HCV-specific CD4 T cell proliferation assay results, ethnicity, and genotype from previous studies,6,31 as well as HLA class II typing as described in Patients and Methods. Thus, the clinical, demographic, and immunological profiles of these subjects (Table 1) were consistent with our previous reports.6,31
As shown in Fig. 1 (top panel), there was no significant difference in the prevalence of 5 DQB1 and 19 DRB1-5 HLA class II genotypes between all chronic and recovered subjects. However, when the comparison was restricted to white patients (Fig. 1, middle panel), two HLA types, DQB1*03 and DRB1*11, previously associated with HCV clearance23–28 were also more prevalent among recovered than chronically HCV-infected subjects in our study: (DQB1*03: 92.3% R versus 53.6% C, OR = 10.4, 95% CI = 1.2–91.2, P = 0.031; DRB1*11: 53.8% R versus 14.3% C, OR = 7.0, 95% CI = 1.5–32.0, P = 0.019). Although DQB1*02 was previously associated with HCV persistence,29,30 this association did not reach a statistical significance in our study (All: 30% R versus 46% C, OR = 0.5, 95% CI = 0.2–1.4, P = 0.198; white patients: 23% R versus 54% C, OR = 0.3, 95% CI = 0.06–1.2, P = 0.067; black patients: 40% R versus 41% C, OR = 1.0, 95% CI = 0.2–4.0, P = 0.978). A significant positive association with HCV clearance was also identified among white patients for DRB3*02 (69.2% R versus 21.4% C, OR = 8.3, 95% CI = 1.9–36.4, P = 0.005, Pc = 0.015), a genotype without a previously known association with HCV clearance. Importantly, none of the HLA types was associated with virological outcome among black patients, suggesting an ethnic difference in immunogenetic contribution to HCV clearance (Fig. 1, bottom panel).
Fig. 1.
HLA class II genotype prevalence in patients with chronic or spontaneously resolved HCV infection by ethnic group. The bar graphs show the relative prevalence of HLA class II genotypes within the DQB1, DRB1, DRB3, DRB4, and DRB5 loci in HCV-recovered (white bars) and chronic (black bars) patients. Top panel refers to all patients (n = 93), middle panel to white patients (n = 41), and bottom panel to black patients (n = 52). Red asterisks in the middle panel refer to HLA types that were significantly associated with HCV clearance: DQB1*03 (P = 0.031), DRB1*11 (P = 0.019), and DRB3*02 (P = 0.005). P-values were calculated using chi-squared or Fisher’s exact test. Because DRB3*02 was not previously associated with HCV clearance, a Bonferroni-type correction was applied for DRB3*02 with continued statistical significance (Pc = 0.015).
The association between DRB3*02 and HCV clearance was not by chance because DRB3*02 is known to be linked to DRB1*11,35 and they define a known haplotype with DQB1*03 and DRB1*11.36 As shown in Table 2, white DRB3*02+ patients were more likely to be DQB1*03+ (87% versus 54%, P = 0.033), DRB1*11+ (73% versus 0%, P < 0.001), or double-positive for both DQB1*03+ and DRB1*11+ (73% versus 0%, P < 0.001), compared with DRB3*02– patients. However, the association between DRB3*02 and DQB1*03 was lost among blacks. Examined in combination (Table 3), DQB1*03-DRB3*02 haplotype was clearly associated with viral clearance (56.5% R versus 24.3% C, OR = 4.1, 95% CI = 1.5–10.9, P = 0.004) in all patients. However, this association was attributable to the white patients in whom both DQB1*03-DRB3*02 (69.2% R versus 14.3% C, OR = 13.5, 95% CI = 2.8–65.8, P = 0.001) and DQB1*03-DRB1*11-DRB3*02 (53.8% R versus 14.3% C, OR = 7.0, 95% CI = 1.5–32.0, P = 0.019) haplotypes correlated with HCV clearance, whereas no such associations were observed among black patients. These findings show that HLA DRB3*02 is associated with HCV clearance as part of a haplotype that also includes HLA DQB1*03 and DRB1*11. Furthermore, we show that these HLA-associations were limited to white patients.
Table 2.
HLA DQB1*03+ or DRB1*11 Genotype Distribution Relative to HLA DRB3*02 Among HCV-Seropositive Persons
| All Patients | White Patients | Black Patients | |||||||
|---|---|---|---|---|---|---|---|---|---|
| DRB3*02 | DRB3*02 | DRB3*02 | |||||||
| + (n = 45) | − (n = 48) | P-Value | +(n = 15) | − (n = 26) | P-Value | + (n = 30) | − (n = 22) | P-Value | |
| %DQB1*03+ | 67% | 48% | 0.094 | 87% | 54% | 0.033 | 30% | 27% | 0.830 |
| %DRB1*11+ | 60% | 2% | <0.001 | 73% | 0% | <0.001 | 53% | 5% | <0.001 |
| %DQB1*03+ DRB1*11+ | 51% | 2% | <0.001 | 73% | 0% | <0.001 | 40% | 5% | 0.004 |
P-values were calculated using chi-square or Fisher’s exact test. Significant P-values (≤0.05) are shown in bold.
Table 3.
Distribution of DQB1*03, DRB1*11, and DRB3*02 in HCV-Seropositive Persons With and Without Spontaneous Viral Clearance
| HLA Genotype | % All Patients | % White Patientsa | % Black Patients | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype Frequency | R (n = 23) |
C (n = 70) |
OR (95% CI, P-value) | R (n = 13) |
C (n = 28) |
OR (95% CI, P-value) | R (n = 10) |
C (n = 42) |
OR (95% CI, P-value) |
| DQB1*03 | 73.9 | 51.4 | 2.7 (0.9–7.6, 0.059) | 92.3 | 53.6 | 10.4 (1.2–91.2, 0.031) | 50.0 | 50.0 | 1.0 (0.3–4.0, 1.000) |
| DRB1*11 | 39.1 | 27.1 | 1.7 (0.6–4.6, 0.277) | 53.8 | 14.3 | 7.0 (1.5–32.0, 0.019) | 20.0 | 35.7 | 0.5 (0.1–2.4, 0.467) |
| DRB3*02 | 60.9 | 44.3 | 2.0 (0.7–5.1, 0.167) | 69.2 | 21.4 | 8.3 (1.9–36.4, 0.005) | 50.0 | 59.5 | 0.7 (0.2–2.7, 0.725) |
| DQB1*03-DRB3*02 | 58.5 | 24.3 | 4.1 (1.5–10.9, 0.004) | 69.2 | 14.3 | 13.5 (2.8–65.8, 0.001) | 40.0 | 31.0 | 1.5 (0.4–6.2, 0.711) |
| DRB1*11-DRB3*02 | 39.1 | 25.7 | 1.9 (0.7–5.0, 0.219) | 53.8 | 14.3 | 7.0 (1.5–32.0, 0.019) | 20.0 | 33.3 | 0.5 (0.1–2.7, 0.705) |
| DQB1*03-DRB1*11 | 39.1 | 21.4 | 2.4 (0.9–6.5, 0.092) | 53.8 | 14.3 | 7.0 (1.5–32.0, 0.019) | 20.0 | 26.2 | 0.7 (0.1–3.8, 1.000) |
| DQB1*03-DRB1*11-DRB3*02 | 39.1 | 20.0 | 2.6 (0.9–7.1, 0.065) | 53.8 | 14.3 | 7.0 (1.5–32.0, 0.019) | 20.0 | 23.8 | 0.8 (0.1–4.4, 1.000) |
Percent of individuals positive for the HLA types (either alone or in combination) is shown among all, black, and white patients with spontaneous HCV clearance (R) or chronic infection (C) with respective odds ratios. 95% confidence intervals, and P-values calculated by chi-squared and Fisher’s exact test. Because significant associations for HLA DQB1*03 and DRB1*11 were anticipated, the P-values were not further corrected. For DRB3*02, the number of comparisons was 3 (adjusted α = 0.07), resulting in corrected P-value of 0.015, which remained significant.
All HLA DRB1*11+ white patients (n = 11) also carried DQB1*03 and DRB3*02.
HLA DQB1*03, DRB1*11 and DRB3*02 Genotypes Are Positively Associated with HCV-Specific CD4 T Cell Responsiveness in White but Not Black Patients
We next looked for associations between the protective HLA class II types (DQB1*03, DRB1*11, and DRB3*02) and HCV-specific CD4 T cell response, restricting the analysis to 73 subjects with evidence of current or prior HCV genotype 1 infection (55 chronic HCV genotype 1 infection and 18 serotype 1–positive HCV-recovered subjects), because genotype 1–derived HCV antigens were used in our immunological analyses. Among all subjects, DQB1*03 was positively associated with CD4 response to HCV core (OR 4.5, 95% CI = 0.9–22.4, P = 0.048), and DRB3*02 was positively associated with CD4 T cell response to HCV NS5 (OR 3.0, 95% CI = 1.0–9.2, P = 0.049), whereas DQB1*03-DRB3*02 haplotype was associated with CD4 T cell responses to all three HCV antigens (Core: OR 5.6, 95% CI = 1.5–22.2, P = 0.015; NS3-4: OR 3.2, 95% CI = 1.1–9.4, P = 0.033; NS5: OR 3.7, 95% CI = 1.2–11.1, P = 0.018) (Table 4). Among white subjects, DRB3*02 and DRB1*11 (either alone or in combination) were associated with significant CD4 T cell responses to two or more HCV antigens despite a low sample size. Of note, because these HLA types were tightly linked, an independent assessment of their effect on HCV-specific CD4 T cell response is difficult. For example, because DRB3*02 always occurred with DQB1*03 among CA patients, DRB3*02 and DRB3*02-DQB1*03 haplotype showed identical associations with HCV-specific CD4 T cell response. Also, because all seven DRB1*11+ patients also expressed DQB1*03 and DRB3*02, the associations in DRB1*11-containing haplotypes were identical. However, there was no association between the three HLA types and HCV-specific CD4 T cell responsiveness among black patients. The ethnic differences in HLA associations with CD4 T cell response was significant for the nonstructural HCV antigens by the Breslow-Day statistic testing for homogeneity of odds ratios: DRB3*02 with NS3-4 (PBD = 0.011) and NS5 (PBD = 0.051); DRB1*11 with NS3-4 (PBD = 0.049) and NS5 (PBD = 0.018). Thus, the ethnic disparity in HLA associations with the outcome of HCV infection extended to virus-specific CD4 T cell responsiveness, an important determinant of HCV clearance.
Table 4.
Odds Ratios for CD4 Proliferative Response to HCV Core, NS3-4, and NS5 Proteins in Patients Positive for HLA DQB1*03, DRB1*1, or DRB3*02 Relative to Ethnicity
| All Patients (n = 73) | White Patients (n = 27) | Black Patients (n = 46) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Core | NS3-4 OR (P-value) |
NS5 | N | Core | NS3-4 OR (P-value) |
NS5 | N | Core | NS3-4 OR (P-value) |
NS5 | |
| DQB1*03 | 42 | 4.5 (0.048) | 1.9 (0.264) | 1.7 (0.336) | 18 | 10.0 (0.071)b | 4.0 (0.363) | 2.3 (0.636) | 24 | 2.0 (0.67) | 1.4 (0.62) | 1.7 (0.43) |
| DRB1*11 | 21 | 3.1 (0.191) | 1.7 (0.366) | 2.6 (0.090) | 7 | 4.3 (0.290) | 7.6 (0.050) | 25.3 (0.009) | 14 | 2.6 (0.35) | 0.7 (0.73) | 1.0 (1.00) |
| DRB3*02 | 34 | 2.7 (0.127) | 1.9 (0.250) | 3.0 (0.049) | 8 | 8.5 (0.044) | 14.2 (0.011) | 18.0 (0.017) | 26 | 1.6 (0.68) | 0.7 (0.60) | 1.3 (0.67) |
| DQB1*03-DRB3*02 | 24 | 5.6 (0.015) | 3.2 (0.033) | 3.7 (0.018) | 8 | 8.5 (0.044) | 14.2 (0.011) | 18.0 (0.017) | 16 | 4.7 (0.16) | 1.5 (0.73) | 2.0 (0.33) |
| DRB1*11-DRB3*02 | 20 | 3.4 (0.077) | 1.8 (0.283) | 2.9 (0.075) | 7a | 4.3 (0.290) | 7.6 (0.050) | 25.3 (0.009) | 13 | 3.0 (0.33) | 0.8 (1.00) | 1.2 (1.00) |
| DQB1*03-DRB1*11 | 19 | 3.7 (0.067) | 2.0 (0.235) | 3.2 (0.062) | 7a | 4.3 (0.290) | 7.6 (0.050) | 25.3 (0.009) | 12 | 3.4 (0.17) | 0.9 (1.00) | 1.4 (0.72) |
| DQB1*03-DRB1*11- DRB3*02 |
18 | 4.1 (0.060) | 2.3 (0.215) | 3.6 (0.055) | 7a | 4.3 (0.290) | 7.6 (0.050) | 25.3 (0.009) | 11 | 4.0 (0.14) | 1.1 (1.00) | 1.7 (0.70) |
Odds ratios (OR) with respective P-values in parentheses were calculated by chi-squared or Fisher’s exact test. Only patients with genotype 1 or serotype 1 infection are included. HCV-specific CD4 T cell response with stimulation index > 3.0 was considered positive.
All of the white patients who carried the DRB1*11 allele (n = 11) also carried DQB1*03 and DRB3*02.
OR was Haldane-Woolf adjusted.
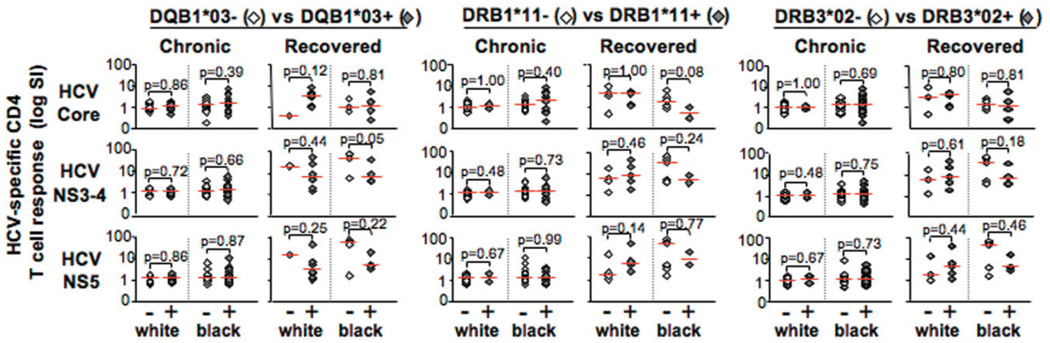
Lack of Positive Association Between HLA Types and HCV-Specific CD4 T Cell Response Among Viro-logically Distinct Groups
The foregoing results implied that the HLA genotypes could influence HCV-specific CD4 T cell response (thus contributing to HCV clearance or persistence), as previously suggested for DQB1*03.27,37 However, the prolonged viremia experienced by chronic patients can induce a functional suppression in antigen-specific T cells via immune exhaustion,38,39 whereas virus-specific memory T cell response may be vigorously maintained after viral clearance.40 In an effort to avoid the confounding effect of viremia on antiviral T cell response, the effect of HLA genotypes on HCV-specific CD4 T cell response was examined separately among the HCV-recovered subjects and the chronic patients. Although the number was small, the presence of DQB1*03, DRB1*11, and DRB3*02 genotypes did not associate with increased HCV-specific CD4 T cell responsiveness among HCV-recovered subjects regardless of ethnicity (P > 0.05 for all comparisons) (Fig. 2). Conversely, HCV-specific CD4 T cell responses were mostly weak among the chronic patients regardless of HLA status (although greater CD4 T cell responsiveness was observed for black patients, as previously reported).6 Accordingly, in multivariate analysis controlling for virological status, there were no significant associations between any of the HLA types and HCV-specific CD4 T cell response among white subjects. Further analysis of 54 subjects for interferon gamma (IFN-γ) response to the same HCV antigens failed to show significant associations between HCV-specific CD4 IFN-γ responses and the three HLA class II genotypes among white or black patients (data not shown). Taken together, we show a novel ethnic disparity in HLA class II genotypes associated with HCV clearance. However, these HLA associations are not directly associated with an enhanced antiviral CD4 T cell response.
Fig. 2.
Presence of HLA DQB1*03, DRB1*11 and DRB3*02 is not associated with increased antigen-specific CD4 T cell proliferation among subjects with similar virological outcomes. Presence of DQB1*03, DRB1* 11, or DRB3*02 had no significant effect on CD4 proliferative 1 cell response to HCV core, NS3-4, and NS5 among persons with similar virological status (chronic versus recovered) and race (black or white). CD4 T cell proliferation is shown as SI in a logarithmic scale. Patients positive for a HLA genotype are represented by shaded diamonds; patients negative for a genotype are represented by unfilled diamonds (also indicated as − or +, respectively). Only subjects with HCV genotype 1 infection by Innolipa or serotyping were included (N = 73). Comparison of median stimulation indexes between persons with and without specific HLA type in each ethnic/virological group using the nonparametric Mann-Whitney U test showed no statistically significant associations (P > 0.05).
Discussion
Gene products of the HLA class II region have been implicated in many immune-mediated diseases, such as DQB1*0302 with celiac disease,41 DRB1*03/04 with autoimmune hepatitis,42 DRB1, DQA1, and DQB1 with type 1 diabetes,43 although the underlying pathways are not fully understood. In concert with other biomarkers, HLA type is an important factor in clinical diagnosis and management of various diseases, and in defining vaccine strategies.44 In HCV infection, a number of HLA class II types has been associated with the outcome of HCV infection, most prominently DQB1*02, DQB1*03, and DRB1*11.16,45,46 In this study, we report that DRB3*02, in addition to DQB1*03 and DRB1*11, is associated with HCV clearance among Caucasian but not African American subjects from North America, suggesting that immunogenetic basis for HCV clearance may differ between these ethnic groups. Although these HLA class II types were initially associated with increased HCV-specific CD4 T cell response among all subjects (both chronic and recovered), further analysis controlling for virological outcome showed that the presence of these HLA genotypes does not correlate with increased HCV-specific CD4 T cell response, suggesting an alternative mechanism whereby these HLA genotypes may contribute to HCV clearance.
Race or ethnicity can contribute to variable associations in population-based studies of inherited traits.47 For example, different HLA associations with HCV clearance have been found among Irish,48,49 Hispanic,50 European,23–25,51 and North American cohorts.28 In particular, HCV clearance was repeatedly associated with HLA DQB1*03 or DRB1*11 in studies examining white patients of European origin.23–26,51 Conversely, Thio and coworkers28 examined both injection drug user and hemophilia cohorts in North America and found only a weak association between DQB1*03 and HCV clearance among black (P = 0.054) but not white patients. In our study, also based on Northeast America, DQB1*03, DRB1*11, and DRB3*02 were significantly associated with HCV clearance among white patients whereas none of the 24 HLA class II genotypes correlated with HCV clearance (or persistence) among black patients. Thus, our findings are more in keeping with the European studies while differing from the Thio et al. study. We speculate that the differences between the Thio et al. study and ours derive from differences in the comorbid factors and route of HCV transmission. For example, we examined patients without HIV coinfection, whereas the study by Thio et al. included a substantial proportion of patients coinfected with HIV (39%–4l%) or hepatitis B virus (5%–17%). Furthermore, injection drug use was the primary HCV risk factor for most of our subjects, whereas the Thio et al. study included two hemophilia cohorts with recurrent high-dose HCV exposure (for example, by blood products and clotting factors). Because mode of HCV transmission also may contribute to HCV outcome and HLA associations,14 DQB1*03, DRB1*11, and DRB3*02 could be protective among white (but not black) persons in the context of low-dose HCV exposure (for example, by injection drug use associated with most of our subjects) without HIV or hepatitis B virus coinfection. The ethnic difference in HLA associations with HCV clearances suggest that these “protective” HLA types do not contribute to HCV clearance among black patients or that additional antiviral pathways exist in black patients that may overcome any effect of these HLA types.
The three protective HLA genotypes also correlated with HCV-specific CD4 T cell responsiveness among white patients in our initial analysis. An association between CD4 T helper response and DQB1*03 was reported previously,27 suggesting a causal link between the HLA type and enhancement of HCV-specific CD4 T cell response that results in HCV clearance. However, HCV-recovered patients without the protective genotypes still maintained a vigorous HCV-specific T cell responsiveness similar to the response detected in HCV-recovered subjects with the protective HLA types. Conversely, HCV-specific CD4 T cell response in chronically HCV-infected patients with the protective HLA genotypes was no better than the response in chronic patients without the HLA genotypes. Indeed, the associations between the HLA types and CD4 T cell response were lost when comparing the antiviral T cell response relative with HLA type among patients with similar virological outcome and in a multivariate analysis controlling for virological status, although subtle differences may have been lost because of small sample size. Thus, there was no direct association between the HLA class II genotypes and HCV-specific CD4 T cell responsiveness, contrary to our initial expectations. Similarly, no significant independent association was identified between class II HLA alleles, haplotypes, and antiviral CD4 T cell IFN-γ responses in the large Virahep-C cohort with established chronic HCV infection in a recent study.52 Although this study did not examine HLA DRB3*02 or patients with HCV clearance, their results further support the lack of direct association between class II HLA types and HCV-specific CD4 T cell responses in patients with HCV clearance and persistence.
One interpretation for our finding is that these HLA types can promote more robust antiviral CD4 T cell response during acute infection but that such effect may be lost over time. Nevertheless, it is notable that the strength of antiviral memory T cell response is generally believed to reflect its initial “burst size” during primary infection.40 Our analysis was also largely focused on HCV-specific CD4 T cell proliferation, a key determinant of the virological outcome in HCV infection.3–5,53 However, antigen-specific proliferation is only one aspect of CD4 T cell function. For example, CD4 T cells also play a critical immune regulatory role, provide help to induce and maintain efficient antiviral memory CD8 T cell response, and secrete important antiviral cytokines (for example, IFN-γ).54,55 However, we found no associations between HCV-specific CD4 T cell IFN-γ response and the three HLA class II genotypes among white or black patients (data not shown). The associations between HLA genotypes and HCV-specific T cell response (both CD4 and CD8) would be best examined prospectively in patients with acute evolving hepatitis C, although this is a challenging task because of the difficulty in identifying patients with acute hepatitis C5 and a multicenter approach needed to ensure adequate representation of specific ethnicities, HLA types, and virological outcomes. In addition, it would be important to define T cell epitopes restricted by the relevant HLA types to more precisely monitor the antiviral immune responses. At a more basic level, it would be interesting to examine the expression of these HLA molecules on antigen-presenting cells at the protein level and to define their capacity for antigen processing and presentation as well as T cell stimulation56 in future studies.
We are mindful of further limitations of this study. One concern is our modest sample size (n = 93), which can result in a type II error in which faint but important associations may have been missed. Conversely, we believe that there is low likelihood of detecting a false positive (type I error) because our findings conform to published literature as well as the pattern of linkage disequilibrium. It is also important to acknowledge that direct causality is difficult to ascertain in most human studies, particularly for immunogenetic studies because various genes may operate as proxies or surrogates through linkage disequilibrium, for the effects of genes at other loci. Hence, among whites, the protective genotypes DQB1*03, DRB1*11, and DRB3*02, though forming a well-known haplotype, may nonetheless be serving as a “stand-in” for another pathway to viral clearance. Contrary to our initial expectation that association between HLA class II types and HCV clearance can be explained by their capacity to induce a vigorous antiviral CD4 T cell response, our findings suggest that this proposition may have been too simplistic, albeit with a limited sample size. They also raise a more basic question regarding HLA associations and highlight the need to validate immunogenetic associations with further functional analysis.
In summary, we demonstrate significant HLA class II types that are associated with the outcome of HCV infection with a distinct ethnic disparity. However, we also show that these HLA class II types do not directly enhance antiviral CD4 T cell response. Future studies are needed to define the underlying mechanisms whereby HLA class II types participate in the outcome of HCV infection, and how these pathways diverge with ethnicity.
Acknowledgment
We thank Mary Valiga, RN, and Marcia Johnson, NP, for patient recruitment; Jason Stadanlick and Gina Suh for technical assistance; Colleen Brensinger for statistical support; Dr. Harvey J. Alter for patient samples; Dr. Toni Simonis, Julia Hackett, and Sharon Adams at the HLA Laboratory at the Warren G. Magnuson Clinical Center for HLA class II typing; and Dr. Michael Houghton for the HCV recombinant proteins. We also gratefully acknowledge all of our study subjects.
Supported by the National Institutes of Health (NIH) grants AI47519 and AA12849; the Philadelphia VA Medical Research; NIH/National Institute of Diabetes and Digestive and Kidney Diseases Center of Molecular Studies in Digestive and Liver Diseases P30DK50306 and its Molecular Biology and Cell Culture Core Facilities; the NIH Public Health Service Research Grant M01-RR00040; R.H. was also supported by University of Pennsylvania Summer Undergraduate Scholars Program and Howard Hughes Interdisciplinary Scholar Award at Haverford College.
Abbreviations
- C
chronically hepatitis C virus–infected
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- HLA
human leukocyte antigen
- IFN-γ
interferon gamma
- OR
odds ratio
- R
hepatitis C virus–recovered
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Hoofnagle JH. Hepatitis C: the clinical spectrum of disease. HEPATOLOGY. 1997;26 Suppl:15S–20S. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 2.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 3.Diepolder HM, Zachoval R, Hoffmann RM, Wierenga EA, Santantonio T, Jung MC, et al. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet. 1995;346:1006–1007. doi: 10.1016/s0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 4.Missale G, Bertoni R, Lamonaca V, Valli A, Massari M, Mori C, et al. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Invest. 1996;98:706–714. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan DE, Sugimoto K, Newton K, Valiga ME, Ikeda F, Aytaman A, et al. Discordant role of CD4 T-cell response relative to neutralizing antibody and CD8 T-cell responses in acute hepatitis C. Gastroenterology. 2007;132:654–666. doi: 10.1053/j.gastro.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 6.Sugimoto K, Stadanlick J, Ikeda F, Brensinger C, Furth EE, Alter HJ, et al. Influence of ethnicity in the outcome of hepatitis C virus infection and cellular immune response. Hepatology. 2003;37:590–599. doi: 10.1053/jhep.2003.50103. [DOI] [PubMed] [Google Scholar]
- 7.Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 8.Howell C, Jeffers L, Hoofnagle JH. Hepatitis C in African Americans: summary of a workshop. Gastroenterology. 2000;119:1385–1396. doi: 10.1053/gast.2000.19582. [DOI] [PubMed] [Google Scholar]
- 9.Reddy KR, Hoofnagle JH, Tong MJ, Lee WM, Pockros P, Heathcote EJ, et al. Racial differences in responses to therapy with interferon in chronic hepatitis C. Consensus Interferon Study Group. Hepatology. 1999;30:787–793. doi: 10.1002/hep.510300319. [DOI] [PubMed] [Google Scholar]
- 10.Fleckenstein J. Chronic hepatitis C in African Americans and other minority groups. Curr Gastroenterol Rep. 2004;6:66–70. doi: 10.1007/s11894-004-0028-z. [DOI] [PubMed] [Google Scholar]
- 11.He XS, Ji X, Hale MB, Cheung R, Ahmed A, Guo Y, et al. Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology. 2006;44:352–359. doi: 10.1002/hep.21267. [DOI] [PubMed] [Google Scholar]
- 12.Conjeevaram HS, Fried MW, Jeffers LJ, Terrault NA, Wiley-Lucas TE, Afdhal N, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Wiley TE, Brown J, Chan J. Hepatitis C infection in African Americans: its natural history and histological progression. Am J Gastroenterol. 2002;97:700–706. doi: 10.1111/j.1572-0241.2002.05555.x. [DOI] [PubMed] [Google Scholar]
- 14.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 15.Rauch A, Laird R, McKinnon E, Telenti A, Furrer H, Weber R, et al. Influence of inhibitory killer immunoglobulin-like receptors and their HLA-C ligands on resolving hepatitis C virus infection. Tissue Antigens. 2007;69 Suppl 1:237–240. doi: 10.1111/j.1399-0039.2006.773_4.x. [DOI] [PubMed] [Google Scholar]
- 16.Yee LJ. Host genetic determinants in hepatitis C virus infection. Genes Immun. 2004;5:237–245. doi: 10.1038/sj.gene.6364090. [DOI] [PubMed] [Google Scholar]
- 17.Oleksyk TK, Thio CL, Truelove AL, Goedert JJ, Donfield SM, Kirk GD, et al. Single nucleotide polymorphisms and haplotypes in the IL10 region associated with HCV clearance. Genes Immun. 2005;6:347–357. doi: 10.1038/sj.gene.6364188. [DOI] [PubMed] [Google Scholar]
- 18.Kimura T, Saito T, Yoshimura M, Yixuan S, Baba M, Ji G, et al. Association of transforming growth factor-beta 1 functional polymorphisms with natural clearance of hepatitis C virus. J Infect Dis. 2006;193:1371–1374. doi: 10.1086/503436. [DOI] [PubMed] [Google Scholar]
- 19.Fremont DH, Hendrickson WA, Marrack P, Kappler J. Structures of an MHC class II molecule with covalently bound single peptides. Science. 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 20.Fleury S, Thibodeau J, Croteau G, Labrecque N, Aronson HE, Cantin C, et al. HLA-DR polymorphism affects the interaction with CD4. J Exp Med. 1995;182:733–741. doi: 10.1084/jem.182.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villadangos JA. Presentation of antigens by MHC class II molecules: getting the most out of them. Mol Immunol. 2001;38:329–346. doi: 10.1016/s0161-5890(01)00069-4. [DOI] [PubMed] [Google Scholar]
- 22.Wang JH, Reinherz EL. Structural basis of T cell recognition of peptides bound to MHC molecules. Mol Immunol. 2002;38:1039–1049. doi: 10.1016/s0161-5890(02)00033-0. [DOI] [PubMed] [Google Scholar]
- 23.Alric L, Fort M, Izopet J, Vinel JP, Charlet JP, Selves J, et al. Genes of the major histocompatibility complex class II influence the outcome of hepatitis C virus infection. Gastroenterology. 1997;113:1675–1681. doi: 10.1053/gast.1997.v113.pm9352872. [DOI] [PubMed] [Google Scholar]
- 24.Minton EJ, Smillie D, Neal KR, Irving WL, Underwood JC, James V. Association between MHC class II alleles and clearance of circulating hepatitis C virus: members of the Trent Hepatitis C Virus Study Group. J Infect Dis. 1998;178:39–44. doi: 10.1086/515599. [DOI] [PubMed] [Google Scholar]
- 25.Thursz M, Yallop R, Goldin R, Trepo C, Thomas HC. Influence of MHC class II genotype on outcome of infection with hepatitis C virus. The HENCORE group. Hepatitis C European Network for Cooperative Research. Lancet. 1999;354:2119–2124. doi: 10.1016/s0140-6736(99)91443-5. [DOI] [PubMed] [Google Scholar]
- 26.Mangia A, Gentile R, Cascavilla I, Margaglione M, Villani MR, Stella F, et al. HLA class II favors clearance of HCV infection and progression of the chronic liver damage. J Hepatol. 1999;30:984–989. doi: 10.1016/s0168-8278(99)80250-5. [DOI] [PubMed] [Google Scholar]
- 27.Harcourt G, Hellier S, Bunce M, Satsangi J, Collier J, Chapman R, et al. Effect of HLA class II genotype on T helper lymphocyte responses and viral control in hepatitis C virus infection. J Viral Hepatol. 2001;8:174–179. doi: 10.1046/j.1365-2893.2001.00289.x. [DOI] [PubMed] [Google Scholar]
- 28.Thio CL, Thomas DL, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, et al. Racial differences in HLA class II associations with hepatitis C virus outcomes. J Infect Dis. 2001;184:16–21. doi: 10.1086/321005. [DOI] [PubMed] [Google Scholar]
- 29.Zavaglia C, Martinetti M, Silini E, Bottelli R, Daielli C, Asti M, et al. Association between HLA class II alleles and protection from or susceptibility to chronic hepatitis C. J Hepatol. 1998;28:1–7. doi: 10.1016/s0168-8278(98)80195-5. [DOI] [PubMed] [Google Scholar]
- 30.McKiernan SM, Hagan R, Curry M, McDonald GS, Nolan N, Crowley J, et al. The MHC is a major determinant of viral status, but not fibrotic stage, in individuals infected with hepatitis C. Gastroenterology. 2000;118:1124–1130. doi: 10.1016/s0016-5085(00)70365-9. [DOI] [PubMed] [Google Scholar]
- 31.Sugimoto K, Kaplan DE, Ikeda F, Ding J, Schwartz J, Nunes FA, et al. Strain-specific T-cell suppression and protective immunity in patients with chronic hepatitis C virus infection. J Virol. 2005;79:6976–6983. doi: 10.1128/JVI.79.11.6976-6983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugimoto K, Ikeda F, Stadanlick J, Nunes FA, Alter HJ, Chang KM. Suppression of HCV-specific T cells without differential hierarchy demonstrated ex-vivo in persistent HCV infection. Hepatology. 2003;38:1437–1448. doi: 10.1016/j.hep.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Chang KM, Rehermann B, McHutchison JG, Pasquinelli C, Southwood S, Sette A, et al. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Invest. 1997;100:2376–2385. doi: 10.1172/JCI119778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keppel G, Suafley W, Jr, Tokunuga H. Introduction to Design and Analysis: A Student’s Handbook. 2nd ed. New York, NY: W.H. Freeman; 1992. [Google Scholar]
- 35.Tang TF, Huang AY, Pappas A, Slack R, Ng J, Hartzman RJ, et al. Relative frequencies of DRB1*11 alleles and their DRB3 associations in five major population groups in a United States bone marrow registry. Hum Immunol. 2000;61:820–827. doi: 10.1016/s0198-8859(00)00145-2. [DOI] [PubMed] [Google Scholar]
- 36.Hristova-Dimceva A, Verduijn W, Schipper RF, Schreuder GM. HLA-DRB and -DQB1 polymorphism in the Macedonian population. Tissue Antigens. 2000;55:53–56. doi: 10.1034/j.1399-0039.2000.550109.x. [DOI] [PubMed] [Google Scholar]
- 37.Cramp ME, Rossol S, Carucci P, Williams R, Naoumov NV, Donaldson PT. The influence of the HLA class II allele DQB1*0301 on HCV specific T helper cell. J Hepatol. 1998;28:94. [Google Scholar]
- 38.Wherry EJ, Ahmed R. Memory CD8 T-cell differentiation during viral infection. J Virol. 2004;78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klenerman P, Hill A. T cells and viral persistence: lessons from diverse infections. Nat Immunol. 2005;6:873–879. doi: 10.1038/ni1241. [DOI] [PubMed] [Google Scholar]
- 40.Hou S, Hyland L, Ryan KW, Portner A, Doherty PC. Virus-specific CD8+ T-cell memory determined by clonal burst size. Nature. 1994;369:652–654. doi: 10.1038/369652a0. [DOI] [PubMed] [Google Scholar]
- 41.Henderson KN, Tye-Din JA, Reid HH, Chen Z, Borg NA, Beissbarth T, et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity. 2007;27:23–34. doi: 10.1016/j.immuni.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Strettell MD, Donaldson PT, Thomson LJ, Santrach PJ, Moore SB, Czaja AJ, et al. Allelic basis for HLA-encoded susceptibility to type 1 autoimmune hepatitis. Gastroenterology. 1997;112:2028–2035. doi: 10.1053/gast.1997.v112.pm9178696. [DOI] [PubMed] [Google Scholar]
- 43.Lie BA, Thorsby E. Several genes in the extended human MHC contribute to predisposition to autoimmune diseases. Curr Opin Immunol. 2005;17:526–531. doi: 10.1016/j.coi.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Provenzano M, Panelli MC, Mocellin S, Bracci L, Sais G, Stroncek DF, et al. MHC-peptide specificity and T-cell epitope mapping: where immunotherapy starts. Trends Mol Med. 2006;12:465–472. doi: 10.1016/j.molmed.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Hong X, Yu RB, Sun NX, Wang B, Xu YC, Wu GL. Human leukocyte antigen class II DQB1*0301, DRB1*1101 alleles and spontaneous clearance of hepatitis C virus infection: a meta-analysis. World J Gastroenterol. 2005;11:7302–7307. doi: 10.3748/wjg.v11.i46.7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh R, Kaul R, Kaul A, Khan K. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J Gastroenterol. 2007;13:1770–1787. doi: 10.3748/wjg.v13.i12.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 48.McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, et al. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 49.Fanning LJ, Levis J, Kenny-Walsh E, Wynne F, Whelton M, Shanahan F. Viral clearance in hepatitis C (1b) infection: relationship with human leukocyte antigen class II in a homogeneous population. Hepatology. 2000;31:1334–1337. doi: 10.1053/jhep.2000.7437. [DOI] [PubMed] [Google Scholar]
- 50.Azocar J, Clavijo OP, Yunis EJ. MHC class II genes in HCV viral clearance of hepatitis C infected Hispanic patients. Hum Immunol. 2003;64:99–102. doi: 10.1016/s0198-8859(02)00722-x. [DOI] [PubMed] [Google Scholar]
- 51.Cramp ME, Carucci P, Underhill J, Naoumov NV, Williams R, Donaldson PT. Association between HLA class II genotype and spontaneous clearance of hepatitis C viraemia. J Hepatol. 1998;29:207–213. doi: 10.1016/s0168-8278(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 52.Rosen HR, Weston SJ, Im K, Yang H, Burton JR, Jr, Erlich H, et al. Selective decrease in hepatitis C virus-specific immunity among African Americans and outcome of antiviral therapy. Hepatology. 2007;46:350–358. doi: 10.1002/hep.21714. [DOI] [PubMed] [Google Scholar]
- 53.Ulsenheimer A, Lucas M, Seth NP, Tilman Gerlach J, Gruener NH, Loughry A, et al. Transient immunological control during acute hepatitis C virus infection: ex vivo analysis of helper T-cell responses. J Viral Hepatol. 2006;13:708–714. doi: 10.1111/j.1365-2893.2006.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masopust D, Kaech SM, Wherry EJ, Ahmed R. The role of programming in memory T-cell development. Curr Opin Immunol. 2004;16:217–225. doi: 10.1016/j.coi.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hershberg RM, Cho DH, Youakim A, Bradley MB, Lee JS, Framson PE, et al. Highly polarized HLA class II antigen processing and presentation by human intestinal epithelial cells. J Clin Invest. 1998;102:792–803. doi: 10.1172/JCI3201. [DOI] [PMC free article] [PubMed] [Google Scholar]