Abstract
Cripto is a developmental oncoprotein that signals via MAPK/ERK, PI3K/Akt and Smad2/3 pathways. However, the molecular basis for Cripto coupling to these pathways during embryogenesis and tumorigenesis is not fully understood. In this regard, we recently demonstrated that Cripto forms a cell surface complex with the HSP70 family member glucose-regulated protein-78 (GRP78). Here, we provide novel functional evidence demonstrating that cell surface GRP78 is a necessary mediator of Cripto signaling in human tumor, mammary epithelial, and embryonic stem cells. We show that targeted disruption of the cell surface Cripto/GRP78 complex using shRNAs or GRP78 immunoneutralization precludes Cripto activation of MAPK/PI3K pathways and modulation of activin-A, activin-B, Nodal, and TGF-β1 signaling. We further demonstrate that blockade of Cripto binding to cell surface GRP78 prevents Cripto from increasing cellular proliferation, downregulating E-Cadherin, decreasing cell adhesion and promoting pro-proliferative responses to activin-A and Nodal. Thus, disrupting the Cripto/GRP78 binding interface blocks oncogenic Cripto signaling and may have important therapeutic value in the treatment of cancer.
INTRODUCTION
Cripto (Cripto-1, TDGF1) is an extracellular, GPI-anchored signaling protein with important roles during embryonic development, stem cell function and cancer progression (Adewumi et al., 2007; Strizzi et al., 2005). While Cripto expression is generally low or absent in normal adult tissues, it is found at high levels in many human tumors and its overexpression promotes various tumorigenic attributes including cellular proliferation, migration and epithelial-to-mesenchymal transition (EMT) (Strizzi et al., 2005). Cripto transgenic mice were shown to develop mammary tumors (Strizzi et al., 2004; Wechselberger et al., 2005) and monoclonal antibodies targeting Cripto reduced the growth of tumor xenografts in nude mice (Adkins et al., 2003; Xing et al., 2004).
Cripto exerts its biological effects in part by modulating the signaling of TGF-β superfamily members that activate the Smad2/3 pathway. These ligands induce assembly of serine/threonine kinase transmembrane receptors (type I and type II) and trigger activation of the type I receptor kinase which phosphorylates cytoplasmic Smad2/3 proteins. Upon phosphorylation, Smads 2/3 translocate to the nucleus where they regulate transcription of target genes (Shi & Massague, 2003). Cripto has been shown to directly bind the type I receptors ALK4 (Yeo & Whitman, 2001) and ALK7 (Reissmann et al., 2001) and is an obligatory co-receptor for certain TGF-β ligands such as Nodal (Shen, 2007). This Cripto co-receptor function is essential during embryogenesis (Strizzi et al., 2005) and it has also been implicated in promoting tumor growth since Nodal plays a key role in promoting tumorigenicity of human melanoma and breast cancer cells (Postovit et al., 2008; Topczewska et al., 2006). In contrast to its role as a Nodal co-receptor, Cripto inhibits activin signaling (Adkins et al., 2003; Gray et al., 2003; Kelber et al., 2008) and cytostatic TGF-β1 effects (Gray et al., 2006; Shani et al., 2008; Shukla et al., 2008).
In addition to its role as a modulator of Smad2/3 signaling, soluble forms of Cripto also activate ras/raf/MAPK and PI3K/Akt pathways via c-Src leading to the designation of Cripto as a tumor growth factor (Bianco et al., 2003; Strizzi et al., 2005). The extracellular proteoglycan glypican-1 was shown to be required for this Cripto tumor growth factor activity (Bianco et al., 2003) but the receptor mechanism involved remains to be fully characterized. Interestingly, this pathway was shown to be independent of ALK4 and Nodal (Bianco et al., 2002), suggesting that Cripto regulates Smad2/3 and MAPK/PI3K pathways via separate, non-overlapping mechanisms. In an effort to further characterize Cripto signaling, we recently conducted a screen aimed at identifying novel Cripto binding proteins that led to the identification of Glucose Regulated Protein-78 (GRP78) (Shani et al., 2008). GRP78 is an ER chaperone in the heat shock protein 70 (HSP70) family that is highly expressed in tumors and that promotes tumor cell survival, chemoresistance and malignancy (Dong et al., 2008; Lee, 2007). Notably, GRP78 is localized to the plasma membrane of tumor cells where it has receptor-like functions associated with increased cellular proliferation, motility and survival (Misra et al., 2006; Misra et al., 2004; Philippova et al., 2008).
In the present study, we provide evidence indicating that Cripto binding to cell surface GRP78 is a necessary upstream event that mediates Cripto signaling via both MAPK/PI3K and Smad2/3 pathways. Importantly, blockade of this interaction precludes oncogenic Cripto effects, including increased cell proliferation, downregulation of E-Cadherin, decreased cell adhesion and promotion of pro-proliferative responses to activin-A and Nodal.
RESULTS
Cripto and GRP78 cooperatively regulate activin/Nodal/TGF-β signaling
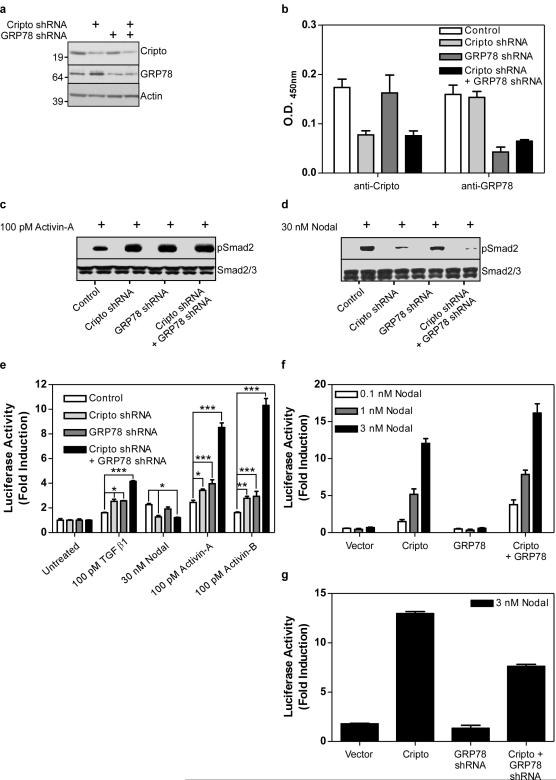
In order to test the function of the cell surface Cripto/GRP78 complex, we generated NCCIT cell populations stably expressing shRNAs targeting Cripto and/or GRP78. These shRNAs specifically reduced endogenous Cripto and GRP78 protein levels in total cell lysates (Figure 1a) and at the cell surface (Figure 1b). As shown in Figure 1c, activin-A-induced Smad2 phosphorylation was enhanced by Cripto knockdown consistent with our previous demonstration that Cripto overexpression inhibits activin-A signaling (Gray et al., 2003; Kelber et al., 2008). Interestingly, Activin-A-dependent Smad2 phosphorylation was similarly increased in GRP78 knockdown cells and also in cells with both Cripto and GRP78 knocked down (Figure 1c). By contrast to its effects on activin-A signaling, knockdown of Cripto reduced Nodal-dependent Smad2 phosphorylation, consistent with the requirement of Cripto as a Nodal co-receptor (Figure 1d). Knockdown of GRP78 alone appeared to cause a modest inhibition of Nodal signaling whereas knockdown of Cripto and GRP78 together inhibited Nodal signaling to a greater extent than knockdown of either Cripto or GRP78 alone (Figure 1d).
Figure 1. Cripto and GRP78 cooperatively regulate activin/Nodal/TGF-β signaling.
NCCIT cells stably expressing Cripto and/or GRP78 shRNAs were analyzed by Western blot (a) or intact cell surface ELISA (b) using the indicated antibodies. The same cells were treated with activin-A (c) or Nodal (d) and resulting levels of phospho-Smad2 (pSmad2) and Smad2 were measured by Western blot. NCCIT cells (e) and 293T cells (f, g) overexpressing the indicated proteins and/or shRNAs were transfected with a Smad2-responsive luciferase reporter and treated with the indicated doses of TGF-β ligands. Resulting luciferase activities were normalized and are presented as fold induction over untreated samples. *** p<0.001; ** p<0.005; * p<0.01.
Next, we tested whether Cripto and GRP78 cooperatively regulate activin-A, activin-B, Nodal and TGF-β1 induction of a Smad2-responsive luciferase reporter construct. As shown in Figure 1e, shRNA knockdown of either Cripto or GRP78 expression caused a modest but significant increase in activin-A, activin-B and TGF-β1 signaling. By contrast, shRNA knockdown of Cripto significantly decreased Nodal signaling (Figure 1e). Notably, knockdown of Cripto and GRP78 together caused activin-A, activin-B and TGF-β1 signaling to be enhanced to a greater extent than knockdown of either protein alone and reduced Nodal signaling to undetectable levels (Figure 1e). Since Nodal-dependent luciferase induction was relatively weak in NCCIT cells, we used the same Smad2-dependent reporter system in 293T cells which express cell surface GRP78 (Shani et al., 2008) but lack endogenous Cripto (Yan et al., 2002). In the presence of transfected Cripto, Nodal signaling was enhanced by GRP78 overexpression (Figure 1f) and was attenuated when endogenous GRP78 was knocked down (Figure 1g). Together, these results indicate that Cripto and GRP78 cooperatively regulate signaling via activin-A, activin-B, TGF-β1 and Nodal.
An antibody targeting the Cripto binding site on GRP78 inhibits Cripto modulation of activin/Nodal/TGF-β signaling
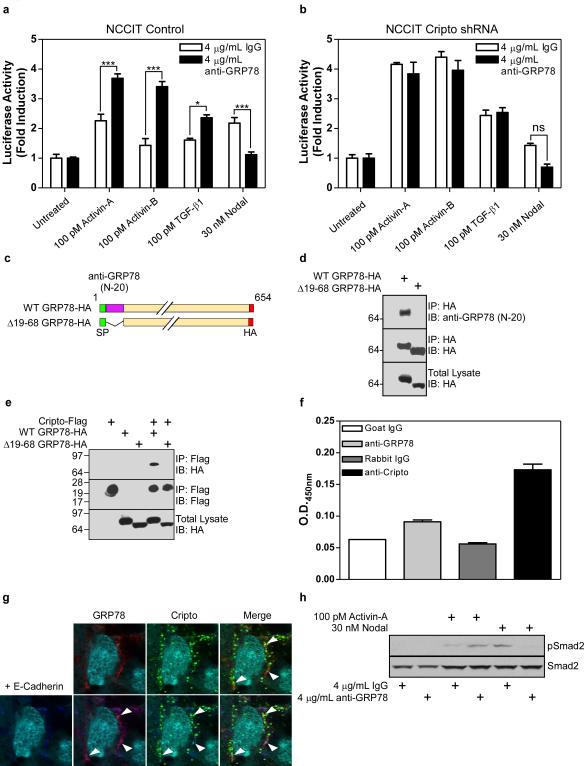
The antibody we have used to detect cell surface GRP78 (N-20, Santa Cruz) has been reported to block cell surface GRP78 receptor function (Davidson et al., 2005; Philippova et al., 2008) and we tested whether it blocks Cripto signaling via activin/Nodal/TGF-β ligands. As shown in Figure 2a, treatment of NCCIT control cells with this N-20 antibody increased activin-A, activin-B and TGF-β1 signaling and decreased Nodal signaling resembling the effect Cripto knockdown (Figure 1e). Significantly, N-20 antibody treatment did not affect activin-A, activin-B, Nodal or TGF-β1 signaling in Cripto knockdown cells (Figure 2b) indicating the antibody exerts its effects on these ligands by blocking Cripto function.
Figure 2. An antibody targeting the Cripto binding site on GRP78 inhibits Cripto modulation of activin/Nodal/TGF-β signaling.
NCCIT cells stably infected with empty vector (a) or Cripto shRNA (b) were transfected with a Smad2-responsive luciferase reporter and treated with the indicated doses of TGF-β ligands in the absence or presence of N-20 antibody as indicated. Resulting luciferase activities were normalized and are presented as fold induction over untreated samples. (c) Diagram illustrating wild type GRP78 and the Δ19-68 GRP78 construct lacking the N-20 epitope. (d, e) Lysates from 293T cells transfected with the indicated constructs were subjected to immunoprecipitation and Western blotting using anti-HA, anti-Flag and anti-GRP78 antibodies as indicated. H9 human ES cells were subjected to intact cell ELISA (f) or immunofluorescence (g) using the indicated antibodies. In (h), anti-Cripto staining is green and anti-GRP78 staining is red. (i) H9 ES cells were treated with activin-A or Nodal following anti-GRP78 treatment as indicated and resulting levels of phospho-Smad2 (pSmad2) and Smad2 were measured by Western blot using pSmad2 and Smad2 antibodies. *p<0.01; ***p<0.001.
The ability of the N-20 antibody to inhibit Cripto signaling, suggested that Cripto and the N-20 antibody compete for binding to GRP78. To test this, we generated a GRP78 mutant (Δ19-68 GRP78, Figure 2c) lacking the N-20 epitope and tested its ability to bind Cripto. As predicted, this GRP78 mutant did not bind the N-20 antibody (Figure 2d) and, importantly, it was also unable to bind Cripto (Figure 2e). These data indicate that the N-20 antibody and Cripto share a binding site on GRP78 and suggest that they may compete for GRP78 binding.
Cripto regulates proliferation, differentiation and pluripotency in hES cells (Minchiotti, 2005) and we tested whether GRP78 co-localizes with Cripto and regulates Cripto function in these cells. GRP78 and Cripto are both expressed at the surface of intact H9 hES cells (Figure 2f) where they co-localize as indicated by immunostaining using antibodies specifically targeting Cripto, GRP78 and the cell surface marker E-Cadherin (Figure 2g). This association has functional relevance since treatment of H9 hES cells with the N-20 antibody increased Smad2 phosphorylation in response to activin-A and decreased Smad2 phosphorylation in response to Nodal (Figure 2h). Together, these results in NCCIT cells and H9 hES cells demonstrate that Cripto and the N-20 antibody share the same binding site on GRP78 and that knockdown or immunoneutralization of cell surface GRP78 disrupts Cripto effects on Smad2/3 signaling.
Cripto signaling via cell surface GRP78 promotes MAPK/PI3K signaling and mitogenesis in NCCIT cells
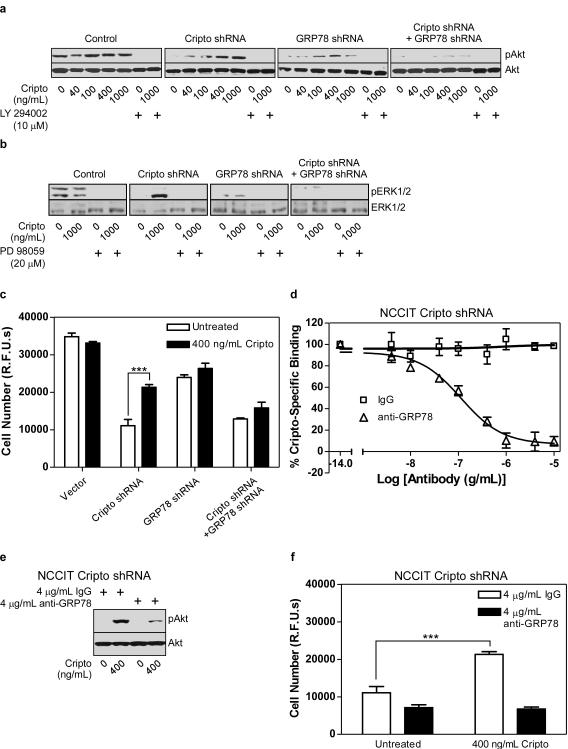
Next, we used NCCIT cells expressing Cripto and/or GRP78 shRNAs to test if GRP78 is required for Cripto-induced activation of MAPK and PI3K pathways. As shown in Figure 3a, control cells had high basal phospho-Akt levels that were unaffected by Cripto treatment. By contrast, basal phospho-Akt levels were drastically reduced in cells expressing Cripto and/or GRP78 shRNAs. Notably, Cripto treatment completely and dose-dependently rescued Akt phosphorylation in Cripto shRNA cells but had little if any effect on Akt phosphorylation when GRP78 was knocked down, either alone or in combination with Cripto. Finally, Cripto-induced phosphorylation of Akt was blocked by the PI3K inhibitor, LY294002, and therefore depends on PI3K activation (Figure 3a).
Figure 3. Cripto signaling via cell surface GRP78 promotes MAPK/PI3K signaling and mitogenesis in NCCIT cells.
NCCIT cells stably expressing the indicated shRNAs were serum starved and then treated with the indicated doses of soluble Cripto and either the PI3K inhibitor (LY2940002) (a) or the MEK1/2 inhibitor (PD98059) (b) as indicated. Cell lysates were subjected to Western blotting using anti-phospho-Akt (pAkt) and Akt (a) or phospho-ERK1/2 (pERK1/2) and ERK1/2 antibodies (b) as indicated. (c) The same NCCIT cells were treated with Cripto as indicated, grown for 8 days and then cell number was measured using the CyQuant proliferation assay kit. (d) NCCIT cells infected with Cripto shRNA were subjected to 125I-Cripto binding in the presence of a range of doses of anti-GRP78 or IgG control. Cripto specific binding represents the amount of 125I-Cripto binding that is blocked by an excess of unlabeled soluble Cripto. NCCIT cells infected with Cripto shRNA were (e) serum-starved and then treated with the indicated dose of soluble Cripto after pretreatment with the indicated dose of IgG or anti-GRP78 or (f) treated with soluble Cripto following pretreatment with IgG control or anti-GRP78 as indicated. Cells were grown for an additional 8 days and proliferation was measured using the CyQuant proliferation assay kit. *p<0.01; ***p<0.001.
We further tested the effects of Cripto and/or GRP78 knockdown on Cripto-induced phosphorylation of ERK1/2. As shown in Figure 3b, basal ERK1/2 phosphorylation was high in control NCCIT cells and unaffected by Cripto treatment suggesting that endogenous Cripto levels are sufficient for maximal Cripto-dependent activation of this pathway. By contrast, basal phospho-ERK1/2 was undetectable in Cripto knockdown cells and treatment of these cells with Cripto caused pronounced phosphorylation of ERK2 (p42). This observation is consistent with the previous demonstration that soluble Cripto triggers ERK2 phosphorylation in mammary epithelial cells (Kannan et al., 1997). Similar to Cripto knockdown cells, basal phospho-ERK levels were very low in cells expressing GRP78 shRNA. However, Cripto treatment of these cells was unable to stimulate ERK phosphorylation, suggesting that GRP78 is required for Cripto-dependent activation of the MAPK pathway (Figure 3b).
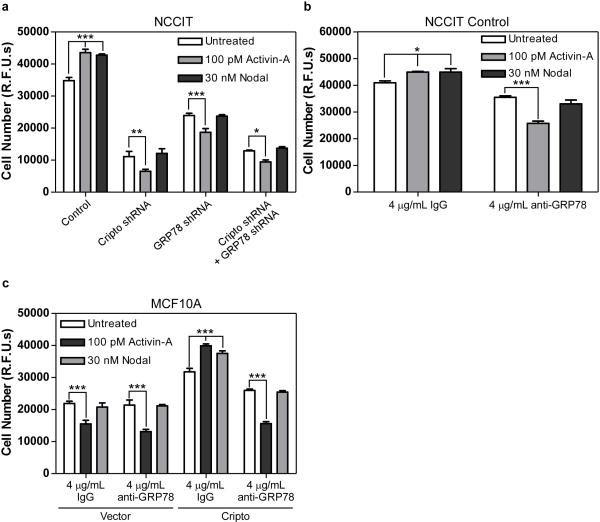
Next, we tested whether soluble Cripto promotes proliferation of NCCIT cells in a GRP78-dependent manner. As shown in Figure 3c, the basal proliferation rate of control NCCIT cells was unaffected by soluble Cripto treatment, consistent with our data showing that Cripto treatment has no effect on MAPK/PI3K signaling in these cells. By contrast, Cripto and/or GRP78 knockdown reduced basal NCCIT cell proliferation and, importantly, soluble Cripto treatment rescued proliferation of Cripto knockdown cells but had no effect on cells in which GRP78 was knocked down either alone or in combination with Cripto (Figure 3c). This result indicates that, similar to its effects on MAPK/PI3K pathways, the pro-proliferative effect of Cripto is GRP78-dependent.
Since the GRP78 antibody (N-20) and Cripto share overlapping binding sites on GRP78, we hypothesized that this antibody may also affect Cripto-induced MAPK/PI3K signaling and mitogenic effects. Initially, we tested the ability of this antibody to compete with soluble Cripto for binding to NCCIT cells stably expressing Cripto shRNA. As shown in Figure 3d, 125I-Cripto bound to these cells specifically and was displaced in a dose-dependent manner by the N-20 antibody but not by control IgG. Together with the results presented in Figure 2g, these data indicate that Cripto and the N-20 antibody directly compete for binding to the same N-terminal site on GRP78. Consistent with this, Cripto-dependent Akt phosphorylation in these cells was markedly attenuated by the N-20 antibody. Figure 3f further shows that the N-20 antibody completely blocked the pro-proliferative effect of Cripto treatment. Together, these data indicate that Cripto binding to cell surface GRP78 is required for both Cripto-dependent MAPK/PI3K signaling and mitogenic effects in NCCIT cells.
Cripto binding to cell surface GRP78 mediates oncogenic Cripto effects in human mammary epithelial cells
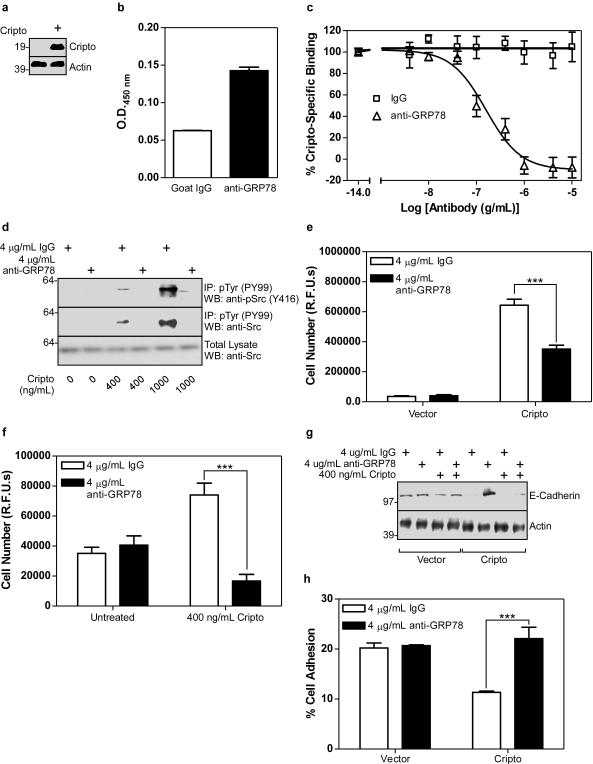
In order to test the role of cell surface GRP78 on Cripto signaling in human mammary epithelial cells, we generated MCF10A cell lines stably expressing an empty vector or a Cripto expression vector. As shown in Figure 4a, control cells had undetectable levels of Cripto expression while Cripto cells had high levels of Cripto expression. Importantly, GRP78 was surface-exposed in MCF10A control cells (Figure 4b) and treating them with the GRP78 N-20 antibody competitively blocked both 125I-Cripto binding (Figure 4c) and dose-dependent Cripto-induced phosphorylation of c-Src (Figure 4d). Cripto overexpression (Figure 4e) or Cripto treatment (Figure 4f) caused increased proliferation of MCF10A cells and this was also blocked by the N-20 antibody. Cripto causes migration and invasion of mammary epithelial cells and promotes downregulation of E-Cadherin expression and EMT (Strizzi et al., 2005; Strizzi et al., 2004). As shown in Figure 4g, either Cripto overexpression or Cripto treatment caused downregulation of E-Cadherin in MCF10A cells that was blocked by the N-20 antibody. Furthermore, this N-20 block was prevented by co-treatment with excess soluble Cripto, providing further evidence that Cripto and the N-20 antibody compete for GRP78 binding (Figure 4g). E-Cadherin promotes cell-cell adhesion and, as shown in Figure 4h, the amount of cell adhesion was reduced by ~50% in Cripto overexpressing cells relative to control cells. Importantly, while the N-20 antibody had no effect on adhesion of control cells, it abrogated Cripto-dependent loss of cell adhesion (Figure 4h) indicating Cripto binding to cell surface GRP78 is required for this effect. Altogether, these data demonstrate that antibody disruption of the cell surface Cripto/GRP78 complex blocks multiple oncogenic Cripto effects in MCF10A cells.
Figure 4. Cripto binding to cell surface GRP78 mediates oncogenic Cripto effects in human mammary epithelial cells.
(a) Cell lysates from control or Cripto-infected MCF10A cells were analyzed by Western blot using Cripto and actin antibodies as indicated. Human mammary epithelial MCF10A cells infected with empty vector were subjected to intact cell ELISA using IgG or anti-GRP78 (b) or to 125I-Cripto binding in the presence of a range of doses of anti-GRP78 or control IgG antibody as indicated (c). (d) MCF10A cells infected with empty vector were serum starved and then treated with the indicated doses of soluble Cripto, anti-GRP78 and/or IgG as indicated. Resulting cell lysates were subjected to immunoprecipitation with anti-phospho-Tyr (pTyr) antibody and Western blotting with anti-phospho-Src (pSrc, Y416) or anti-Src antibodies as indicated. MCF10A cells infected with either empty vector or Cripto (e) or with empty vector (f) were treated with soluble Cripto, IgG and/or anti-GRP78 as indicated. Cells were grown for 8 days and proliferation was measured using the CyQuant proliferation assay kit. (g) MCF10A cells infected with empty vector or Cripto were treated with soluble Cripto after pretreatment with IgG or anti-GRP78 as indicated. Cell lysates were analyzed by Western blot using E-Cadherin and actin antibodies. (h) MCF10A cells infected with empty vector or Cripto were pretreated with IgG or anti-GRP78, plated and allowed to adhere. Resulting cell adhesion was quantified using the CyQuant adhesion assay. ***p<0.001.
The cell surface interaction between Cripto and GRP78 facilitates pro-proliferative effects of activin-A and Nodal
We have shown that GRP78 mediates Cripto regulation of activin- and Nodal-induced Smad2 signaling (Figures 1 and 2) and here we tested how cell surface Cripto/GRP78 complexes impact activin-A- and Nodal-induced effects on cellular proliferation. As shown in Figure 5a, activin-A and Nodal both increased proliferation of control NCCIT cells whereas activin-A had cytostatic effects and Nodal had no effect on NCCIT cells when Cripto and/or GRP78 were knocked down. As shown in Figure 5b, the N-20 antibody had an effect similar to that of Cripto and/or GRP78 knockdown since it blocked the pro-proliferative effects of activin-A and Nodal and caused activin-A to become cytostatic in nature. We conducted parallel proliferation studies in MCF10A cells and, as shown in Figure 5c, activin-A substantially inhibited proliferation of MCF10A control cells while Nodal had no effect, consistent with the lack of detectable Cripto expression in these cells (Figure 4a). By contrast, MCF10A cells overexpressing Cripto had an increased rate of proliferation relative to control cells and were no longer growth-inhibited by activin-A. Rather, activin-A and Nodal each increased proliferation of these cells (Figure 5c). Thus, adding exogenous Cripto to MCF10A cells caused them to resemble NCCIT cells. Consistent with this, treatment of Cripto-expressing MCF10A cells with the N-20 antibody blocked the pro-proliferative effects of activin-A and Nodal and caused activin-A to revert to its antiproliferative status (Figure 5c). Together, these data demonstrate that the cell surface interaction between Cripto and GRP78 facilitates pro-proliferative responses to both activin-A and Nodal whereas disruption of this complex blocks Nodal effects and causes activin-A to become an antiproliferative cytokine.
Figure 5. The cell surface interaction between Cripto and GRP78 facilitates pro-proliferative effects of activin and Nodal.
NCCIT cells infected with empty vector, Cripto and/or GRP78 shRNAs (a, b) and MCF10A cells infected with empty vector or Cripto (c) were left untreated or treated with activin-A or Nodal in the absence or presence of IgG or anti-GRP78 as indicated. Cells were grown for an additional 8 days and proliferation was measured using the CyQuant proliferation assay kit. *** p<0.001; ** p<0.005; * p<0.01.
Cripto binding to cell surface GRP78 promotes the tumorigenic phenotype
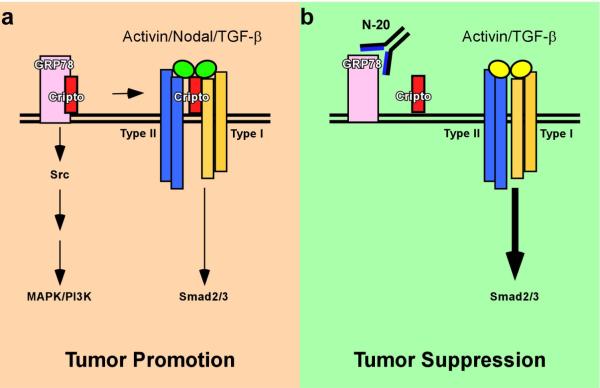
Our data support a model in which the cell surface Cripto/GRP78 complex acts as a signaling node that promotes the tumor phenotype (Figure 6a). According to this model, Cripto and GRP78 cooperatively inhibit cytostatic Smad2/3 signaling in response to activin and TGF-β and cause activin, Nodal and TGF-β to adopt pro-proliferative effects. Concurrently, GRP78 mediates Cripto-dependent activation of c-Src, ERK and Akt, increased cellular proliferation, decreased E-Cadherin expression and decreased cell adhesion. Importantly, we have shown that an antibody targeting GRP78 disrupts the cell surface Cripto/GRP78 complex and inhibits oncogenic Cripto signaling via c-Src/MAPK/PI3K and Smad2/3 pathways (Figure 6b).
Figure 6. Cripto binding to cell surface GRP78 promotes the tumorigenic phenotype.
(a) The cell surface Cripto/GRP78 complex has a pro-tumorigenic effect and a dual signaling role. Cripto binding to cell surface GRP78 leads to MAPK and PI3K signaling. Cripto binding to cell surface GRP78 also facilitates Cripto effects on signaling by activin/Nodal/TGF-β ligands leading to intermediate levels of Smad2/3 activation. The signaling effects mediated by the cell surface Cripto/GRP78 complex may increase tumor growth and metastasis. (b) Immunoneutralization of cell surface GRP78 blocks Cripto binding to surface exposed GRP78 in tumor and mammary epithelial cells and inhibits Cripto-dependent signaling via MAPK, PI3K and Smad2/3 pathways. Under these conditions, activin and TGF-β can elicit high levels of Smad2/3 signaling resulting in cytostatic responses and tumor suppression.
DISCUSSION
Independent lines of evidence have established Cripto and GRP78 as critical survival/pro-proliferation factors during normal developmental processes and cancer progression (Lee, 2007; Strizzi et al., 2005). For example, both Cripto (Strizzi et al., 2005) and GRP78 (Dong et al., 2008; Lee, 2007) increase malignancy and provide a competitive growth advantage to tumor cells by increasing tumor cell survival, proliferation and angiogenesis. Here we provide a novel functional link between these proteins and show that targeted disruption of the cell surface Cripto/GRP78 complex bocks oncogenic Cripto signaling via MAPK/PI3K and Smad2/3 pathways. We further demonstrate that blocking GRP78 inhibits the ability of Cripto to promote cellular proliferation and to decrease E-Cadherin expression and cell adhesion. We draw this evidence from human embryonal carcinoma, embryonic stem and mammary epithelial cells suggesting a broad range of biological functions for the Cripto/GRP78 complex.
Although generally restricted to the lumen of the ER in normal tissues, GRP78 is localized to the cell surface of tumor cells and has been identified as a tumor-specific antigen in primary human breast and prostate cancer samples (Arap et al., 2004; Jakobsen et al., 2007). In addition, GRP78 antibodies were found in the serum of prostate cancer patients and shown to serve as a biomarker of increased cancer aggressiveness (Arap et al., 2004). Cell surface GRP78 has also been validated as a tumor-selective target since chimeric peptides composed of GRP78 binding motifs fused to an apoptosis-inducing sequence inhibited tumor growth in mouse models of prostate and breast cancer (Arap et al., 2004; Liu et al., 2007). Importantly, cell surface GRP78 appears to have an oncogenic function since it mediates activation of MAPK and PI3K pathways after binding α2-macroglobulin in 1-LN prostate carcinoma cells resulting in pro-proliferative and anti-apoptotic behavior (Misra et al., 2006). T-Cadherin binding to cell surface GRP78 was also shown to have an essential role in mediating survival signal transduction via Akt in endothelial cells (Philippova et al., 2008). In addition, tumorigenesis and Akt activation resulting from Pten loss in prostate epithelium is blocked by prostate-specific disruption of GRP78 expression (Fu et al., 2008), consistent with the ability of cell surface GRP78 to initiate Akt signaling.
Our discovery that GRP78 mediates Cripto modulation of multiple TGF-β superfamily members suggests that surface-exposed GRP78 may serve as an anchor or scaffold that allows Cripto to adopt a conformation or an orientation necessary for its functional interaction with these ligands and their receptors. By contrast, our demonstration that GRP78 mediates Cripto-dependent activation of c-Src and MAPK/PI3K pathways resembles previous findings suggesting GRP78 can operate as a cell surface signaling receptor (Misra et al., 2006; Philippova et al., 2008). Alternatively, cell surface GRP78 may indirectly couple Cripto to c-Src, MAPK and PI3K activation as a critical component of a larger protein complex, possibly involving other proteins that have been implicated in Cripto signaling such as glypican-1 (Bianco et al., 2003), ErbB4 (Bianco et al., 1999) and/or Tomoregulin-1 (Harms & Chang, 2003). We previously reported that the CFC domain of Cripto binds GRP78 (Shani et al., 2008) and we have shown here that the extreme N-terminus of GRP78 is required for Cripto binding. Interestingly, T-Cadherin (Philippova et al., 2008) and α2-macroglobulin (Gonzalez-Gronow et al., 2006) also bind cell surface-exposed GRP78 near its N-terminus suggesting these proteins and Cripto may share a common mode of activating GRP78 receptor function. Importantly, while the exact mechanism of GRP78-mediated Cripto signaling remains to be fully elucidated, our data clearly demonstrate that Cripto binding to cell surface GRP78 is required for oncogenic Cripto signaling.
Our results show that shRNA knockdown of Cripto and/or GRP78 or antibody blockade of GRP78 in NCCIT cells enhances activin-A, activin-B and TGF-β1 signaling while inhibiting Nodal signaling. These data represent the first demonstration that endogenously expressed Cripto inhibits activin signaling and confirm our previous demonstration that endogenous Cripto blocks signaling by TGF-β1 (Gray et al., 2006). Knockdown of Cripto and GRP78 together affected the signaling of these TGF-β ligands to a greater extent than knockdown of either protein alone indicating Cripto and GRP78 function cooperatively. In support of this, a GRP78 blocking antibody that disrupts Cripto/GRP78 binding abolished Cripto-dependent effects on signaling by activin/Nodal/TGF-β ligands. Since Cripto-dependent Nodal signaling has essential functions during early embryogenesis (Shen, 2007), our data point to a novel and critical role for cell surface GRP78 during development. In support of this, we have also made the exciting observation that GRP78 is present at the surface of human ES cells where it co-localizes with Cripto. Furthermore, we have shown that an immunoneutralizing antibody that targets the Cripto binding site on GRP78 has opposing effects on Nodal and activin-A signaling similar to those observed following Cripto knockdown in NCCIT cells. Aberrantly activated developmental programs frequently give rise to tumor growth and metastasis in the adult, and our data suggest that the cell surface Cripto/GRP78 complex has a physiological role during development that is dysreglated during tumorigenesis.
Our data indicate that GRP78 is a necessary mediator of Cripto tumor growth factor activity since knockdown or antibody blockade of cell surface GRP78 prevented Cripto-dependent activation of PI3K/Akt and MAPK/ERK pathways. Cripto has been reported to activate these pathways via c-Src (Bianco et al., 2003) and we have shown here that Cripto-dependent c-Src activation in human mammary epithelial MCF10A cells is blocked by GRP78 immunoneutralization. Furthermore, our results show that shRNA knockdown or antibody blockade of cell surface GRP78 prevents Cripto from increasing cellular proliferation of NCCIT and MCF10A cells. E-Cadherin mediates calcium-dependent cell-cell adhesion and its loss is associated with invasive, metastatic cancer (Peinado et al., 2007). Importantly, we have shown that blocking cell surface GRP78 prevents Cripto-induced loss of E-Cadherin expression and cellular adhesion. Thus, our data support a necessary role for GRP78 in mediating Cripto’s effects on proliferation, migration, invasion and EMT.
TGF-β ligands that activate Smad2/3 signaling can inhibit or promote tumorigenesis depending on the cellular context and on whether tumor cells have become refractory to the antiproliferative effects of the Smad2/3 pathway (Rahimi & Leof, 2007). We have shown here that activin-A has opposing effects on cellular proliferation depending on the presence or absence of cell surface Cripto/GRP78 complexes. Activin-A had pro-proliferative effects on NCCIT cells and MCF10A cells in which cell surface Cripto/GRP78 complexes were intact but had antiproliferative effects when these complexes were disrupted by knockdown or immunoneutralization. Notably, this finding resembles our previous demonstration that co-expression of Cripto and GRP78 caused TGF-β1 to switch from having cytostatic effects to having pro-proliferative effects on PC3 prostate carcinoma cells (Shani et al., 2008). Nodal also increased proliferation in cells that expressed intact Cripto/GRP78 complexes but, unlike activin-A and TGF-β1, Nodal had no effect on the proliferation of cells in which Cripto/GRP78 complexes were disrupted. This difference between activin-A and TGF-β1 as opposed to Nodal likely reflects the fact that Cripto is required for Nodal signaling but not for activin-A or TGF-β1 signaling. Thus, our results indicate that Cripto and GRP78 can cooperate to promote tumor growth in part by facilitating mitogenic effects of Nodal and causing activin-A and TGF-β1 to switch from being cytostatic to pro-proliferative in nature.
In summary, we have shown that Cripto binding to cell surface GRP78 is required for Cripto signaling in human tumor, ES and mammary epithelial cells. Cripto and GRP78 have each been independently identified as cell surface tumor-selective targets in vivo and we have shown here that they also co-localize at the surface of hESCs. Our findings indicate that the cell surface Cripto/GRP78 complex has important functions during development and tumorigenesis. Furthermore, we demonstrate that targeting this complex represents a novel approach for abrogating tumor growth and metastasis.
EXPERIMENTAL PROCEDURES
Cell Lines, Transfection and Infection
HEK 293T cells and NCCIT cells were cultured as recommended by American Type Culture Collection (ATCC). MCF10A cells were cultured as previously described (Debnath et al., 2003). H9 (WiCell Inc., Madison, Wisconsin, USA) human embryonic stem cells were maintained in defined mTeSR-1 media (StemCell Technologies, Vancouver, BC, Canada) on growth-factor-depleted matrigel (BD Biosciences, San Jose, CA) coated plates. NCCIT cells and 293T cells were transfected with Lipofectamine 2000 (Invitrogen) and Perfectin (Gene Therapy Systems), respectively, according to manufacturer’s instructions. Lentivirus was produced as previously described (Miyoshi et al., 1998). Cells were infected as previously described (Gray et al., 2006).
Detection of Cell Surface Proteins
Protein detection via intact cell surface ELISA was performed as previously described (Gray et al., 2000).
Immunoprecipitation and Western blotting
Phosphoprotein analysis was essentially carried out as previously described (Kelber et al., 2008). Cells were grown to confluence in 24-well plates, rinsed with serum-free media and serum-starved for 4 hours. Appropriate inhibitors or blocking antibodies were added as indicated for 1 hour. Following growth factor treatment, cells were harvested and lysates were analyzed by Western blot. Co-immunoprecipitation studies were carried out as previously described (Kelber et al., 2008).
Luciferase Assays
Luciferase assays were carried out in NCCIT cells using the A3-luciferase reporter as previously described (Gray et al., 2003).
Confocal Microscopy
H9 cells were cultured on matrigel-coated coverslips. Staining and fixing was carried out to minimize cell lysis and maximize cell surface staining (see supplemental materials and methods for details). Sequential scanning of each excited wavelength was utilized to prevent bleed-through between fluorochromes in analysis.
Cell Proliferation Assays
The Invitrogen CyQUANT Cell Proliferation kit was used according to the manufacturer’s protocol. Cells were plated on 96-well plates at a density of 500 (NCCIT) or 200 (MCF10A) cells/well. 24 hours later, cells were either treated with indicated combinations of blocking agents (goat IgG or anti-GRP78 (N-20)) and growth factors or left untreated in quadruplicate. Cell number was measured eight days later.
Cripto Binding Assays
125I-Cripto binding to the cell surface of NCCIT and MCF10A cells was carried out essentially as previously described (Harrison et al., 2004).
E-Cadherin expression and Cell Adhesion
MCF10A cells were plated at 4×105 cells/well in 6-well plates. 24 hours later cells were pretreated with goat IgG or anti-GRP78 (N-20) for 1 hour and then treated with 400 ng/ml soluble Cripto or left untreated. 48 hours after treatment, cell lysates were analyzed by Western blot, as described above. Cells were also analyzed for their adhesive properties using the Invitrogen CyQUANT Prolifertion kit, as recommended by the manufacturer.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NCI, National Institutes of Health, Grant R01CA107420, the Foundation for Medical research, Inc., and the Robert J. Jr. and Helen Kleberg Foundaton. A portion of this work was also supported by an NIH training grant T32 CA009370 (to ADP) and the G. Harold and Leila Y. Mathers Charitable Foundation (to JCIB). Wylie W. Vale is a senior investigator of the Foundation for Medical Research, Inc. The authors thank Dr. Travis Berggren and Margaret Lutz of The Salk Institute Stem Cell Core for valuable technical assistance.
REFERENCES
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo AB, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K, Fleck RA, Ford A, Gertow K, Gertsenstein M, Gokhale PJ, Hamilton RS, Hampl A, Healy LE, Hovatta O, Hyllner J, Imreh MP, Itskovitz-Eldor J, Jackson J, Johnson JL, Jones M, Kee K, King BL, Knowles BB, Lako M, Lebrin F, Mallon BS, Manning D, Mayshar Y, McKay RD, Michalska AE, Mikkola M, Mileikovsky M, Minger SL, Moore HD, Mummery CL, Nagy A, Nakatsuji N, O’Brien CM, Oh SK, Olsson C, Otonkoski T, Park KY, Passier R, Patel H, Patel M, Pedersen R, Pera MF, Piekarczyk MS, Pera RA, Reubinoff BE, Robins AJ, Rossant J, Rugg-Gunn P, Schulz TC, Semb H, Sherrer ES, Siemen H, Stacey GN, Stojkovic M, Suemori H, Szatkiewicz J, Turetsky T, Tuuri T, van den Brink S, Vintersten K, Vuoristo S, Ward D, Weaver TA, Young LA, Zhang W. Nat Biotechnol. 2007;25:803–16. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Adkins HB, Bianco C, Schiffer SG, Rayhorn P, Zafari M, Cheung AE, Orozco O, Olson D, De Luca A, Chen LL, Miatkowski K, Benjamin C, Normanno N, Williams KP, Jarpe M, LePage D, Salomon D, Sanicola M. J Clin Invest. 2003;112:575–87. doi: 10.1172/JCI17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arap MA, Lahdenranta J, Mintz PJ, Hajitou A, Sarkis AS, Arap W, Pasqualini R. Cancer Cell. 2004;6:275–84. doi: 10.1016/j.ccr.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Bianco C, Adkins HB, Wechselberger C, Seno M, Normanno N, De Luca A, Sun Y, Khan N, Kenney N, Ebert A, Williams KP, Sanicola M, Salomon DS. Mol Cell Biol. 2002;22:2586–97. doi: 10.1128/MCB.22.8.2586-2597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Kannan S, De Santis M, Seno M, Tang CK, Martinez-Lacaci I, Kim N, Wallace-Jones B, Lippman ME, Ebert AD, Wechselberger C, Salomon DS. J Biol Chem. 1999;274:8624–9. doi: 10.1074/jbc.274.13.8624. [DOI] [PubMed] [Google Scholar]
- Bianco C, Strizzi L, Rehman A, Normanno N, Wechselberger C, Sun Y, Khan N, Hirota M, Adkins H, Williams K, Margolis RU, Sanicola M, Salomon DS. Cancer Res. 2003;63:1192–7. [PubMed] [Google Scholar]
- Davidson DJ, Haskell C, Majest S, Kherzai A, Egan DA, Walter KA, Schneider A, Gubbins EF, Solomon L, Chen Z, Lesniewski R, Henkin J. Cancer Res. 2005;65:4663–72. doi: 10.1158/0008-5472.CAN-04-3426. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Dong D, Ni M, Li J, Xiong S, Ye W, Virrey JJ, Mao C, Ye R, Wang M, Pen L, Dubeau L, Groshen S, Hofman FM, Lee AS. Cancer Res. 2008;68:498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- Fu Y, Wey S, Wang M, Ye R, Liao CP, Roy-Burman P, Lee AS. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0807691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gronow M, Cuchacovich M, Llanos C, Urzua C, Gawdi G, Pizzo SV. Cancer Res. 2006;66:11424–31. doi: 10.1158/0008-5472.CAN-06-1721. [DOI] [PubMed] [Google Scholar]
- Gray PC, Greenwald J, Blount AL, Kunitake KS, Donaldson CJ, Choe S, Vale W. J. Biol. Chem. 2000;275:3206–12. doi: 10.1074/jbc.275.5.3206. [DOI] [PubMed] [Google Scholar]
- Gray PC, Harrison CA, Vale W. Proc Natl Acad Sci U S A. 2003;100:5193–8. doi: 10.1073/pnas.0531290100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PC, Shani G, Aung K, Kelber J, Vale W. Mol Cell Biol. 2006;26:9268–78. doi: 10.1128/MCB.01168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms PW, Chang C. Genes Dev. 2003;17:2624–9. doi: 10.1101/gad.1127703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CA, Gray PC, Fischer WH, Donaldson C, Choe S, Vale W. J Biol Chem. 2004;279:28036–44. doi: 10.1074/jbc.M402782200. [DOI] [PubMed] [Google Scholar]
- Jakobsen CG, Rasmussen N, Laenkholm AV, Ditzel HJ. Cancer Res. 2007;67:9507–17. doi: 10.1158/0008-5472.CAN-06-4686. [DOI] [PubMed] [Google Scholar]
- Kannan S, De Santis M, Lohmeyer M, Riese DJ, 2nd, Smith GH, Hynes N, Seno M, Brandt R, Bianco C, Persico G, Kenney N, Normanno N, Martinez-Lacaci I, Ciardiello F, Stern DF, Gullick WJ, Salomon DS. J Biol Chem. 1997;272:3330–5. doi: 10.1074/jbc.272.6.3330. [DOI] [PubMed] [Google Scholar]
- Kelber JA, Shani G, Booker EC, Vale WW, Gray PC. J Biol Chem. 2008;283:4490–500. doi: 10.1074/jbc.M704960200. [DOI] [PubMed] [Google Scholar]
- Lee AS. Cancer Res. 2007;67:3496–9. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- Liu Y, Steiniger SC, Kim Y, Kaufmann GF, Felding-Habermann B, Janda KD. Mol Pharm. 2007 doi: 10.1021/mp060122j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchiotti G. Oncogene. 2005;24:5668–75. doi: 10.1038/sj.onc.1208917. [DOI] [PubMed] [Google Scholar]
- Misra UK, Deedwania R, Pizzo SV. J Biol Chem. 2006;281:13694–707. doi: 10.1074/jbc.M511694200. [DOI] [PubMed] [Google Scholar]
- Misra UK, Gonzalez-Gronow M, Gawdi G, Wang F, Pizzo SV. Cell Signal. 2004;16:929–38. doi: 10.1016/j.cellsig.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. J Virol. 1998;72:8150–7. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Philippova M, Ivanov D, Joshi MB, Kyriakakis E, Rupp K, Afonyushkin T, Bochkov V, Erne P, Resink TJ. Mol Cell Biol. 2008 doi: 10.1128/MCB.00157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postovit LM, Margaryan NV, Seftor EA, Kirschmann DA, Lipavsky A, Wheaton WW, Abbott DE, Seftor RE, Hendrix MJ. Proc Natl Acad Sci U S A. 2008;105:4329–34. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi RA, Leof EB. J Cell Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico MG, Ibanez CF, Brivanlou AH. Genes Dev. 2001;15:2010–22. doi: 10.1101/gad.201801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani G, Fischer WH, Justice NJ, Kelber JA, Vale W, Gray PC. Mol Cell Biol. 2008;28:666–77. doi: 10.1128/MCB.01716-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM. Development. 2007;134:1023–34. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shukla A, Ho Y, Liu X, Ryscavage A, Glick AB. Mol Cancer Res. 2008;6:509–16. doi: 10.1158/1541-7786.MCR-07-0396. [DOI] [PubMed] [Google Scholar]
- Strizzi L, Bianco C, Normanno N, Salomon D. Oncogene. 2005;24:5731–41. doi: 10.1038/sj.onc.1208918. [DOI] [PubMed] [Google Scholar]
- Strizzi L, Bianco C, Normanno N, Seno M, Wechselberger C, Wallace-Jones B, Khan NI, Hirota M, Sun Y, Sanicola M, Salomon DS. J Cell Physiol. 2004;201:266–76. doi: 10.1002/jcp.20062. [DOI] [PubMed] [Google Scholar]
- Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ. Nat Med. 2006;12:925–32. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- Wechselberger C, Strizzi L, Kenney N, Hirota M, Sun Y, Ebert A, Orozco O, Bianco C, Khan NI, Wallace-Jones B, Normanno N, Adkins H, Sanicola M, Salomon DS. Oncogene. 2005 doi: 10.1038/sj.onc.1208417. [DOI] [PubMed] [Google Scholar]
- Xing PX, Hu XF, Pietersz GA, Hosick HL, McKenzie IF. Cancer Res. 2004;64:4018–23. doi: 10.1158/0008-5472.CAN-03-3888. [DOI] [PubMed] [Google Scholar]
- Yan YT, Liu JJ, Luo Y, E C, Haltiwanger RS, Abate-Shen C, Shen MM. Mol Cell Biol. 2002;22:4439–49. doi: 10.1128/MCB.22.13.4439-4449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C, Whitman M. Mol Cell. 2001;7:949–57. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.