Abstract
The vacuole/lysosome serves an essential role in allowing cellular components to be degraded and recycled under starvation conditions. Vacuolar hydrolases are key proteins in this process. In Saccharyomces cerevisiae, some resident vacuolar hydrolases are delivered by the cytoplasm to vacuole targeting (Cvt) pathway, which shares mechanistic features with autophagy. Autophagy is a degradative pathway that is used to degrade and recycle cellular components under starvation conditions. Both the Cvt pathway and autophagy employ double-membrane cytosolic vesicles to deliver cargo to the vacuole. As a result, these pathways share a common terminal step, the degradation of subvacuolar vesicles. We have identified a protein, Cvt17, which is essential for this membrane lytic event. Cvt17 is a membrane glycoprotein that contains a motif conserved in esterases and lipases. The active-site serine of this motif is required for subvacuolar vesicle lysis. This is the first characterization of a putative lipase implicated in vacuolar function in yeast.
One fundamental role of the yeast vacuole is in the recycling of biological macromolecules. The vacuole, like the lysosome in animal cells, is the primary site of degradation. Our understanding of the hydrolytic vacuolar enzymes that serve in protein turnover is well advanced. Progress has also been made in elucidating mechanisms that deliver the substrates of these vacuolar hydrolases. While research has focused on the biosynthesis and function of the vacuolar proteases, little is known about how lipids are recycled in this organelle, and a lipase that functions in membrane recycling has not been identified.
Nearly all vacuolar/lysosomal delivery pathways involve packaging of cargo within membrane-enclosed transport compartments. Because the vacuole/lysosome serves as the final destination for these numerous vesicle-mediated transport pathways, the issue of how membranes reaching the vacuole are recycled is an important one. Macroautophagy is the major degradative process in eukaryotes and is essential during starvation conditions (1). In yeast, autophagy overlaps with a biosynthetic process, the Cvt pathway, that delivers the hydrolase aminopeptidase I (API1; Ref. 2) from its site of synthesis in the cytoplasm to the vacuolar lumen. Cvt and autophagy employ many of the same molecular components and are mechanistically related (3-6). Both pathways involve the formation of double-membrane cytosolic vesicles, sequestering either precursor aminopeptidase I (prAPI) specifically, or in the case of autophagy, also enveloping bulk cytosol in a nonselective manner. Fusion of these vesicles with the vacuole results in the release of single-membrane subvacuolar vesicles within the lumen. These pathways require a mechanism for specific lysis of the internalized vesicles, so that vesicle cargo can be released into the vacuole lumen, and further require a mechanism for degradation of vesicle lipids.
To understand the molecular basis of these import and degradation pathways, we carried out a genetic screen for mutants defective in delivery of prAPI to the vacuole. We isolated a series of mutants, cvt, which accumulate the precursor form of API. The cvt mutants overlap with mutants defective in autophagy. The majority of these mutants are blocked at a stage involving formation of the sequestering vesicle (reviewed in Ref. 1). One mutant, cvt17, was found to be blocked in the breakdown of subvacuolar vesicles, suggesting that Cvt17 acts at a late stage of the import process (6). We report in this paper the cloning of the gene encoding CVT17. Immunological and biochemical studies demonstrate that Cvt17 is a glycosylated, integral membrane protein that transits through the secretory pathway. Cvt17 contains a domain that is conserved among lipases of the α/β-hydrolase-fold superfamily. Mutation of the putative lipase active site abolishes Cvt17 function in the Cvt and autophagy pathways, suggesting that lipase activity is critical to the role of this protein in these import processes.
EXPERIMENTAL PROCEDURES
Strains and Media
Wild type yeast strain SEY6210, was described previously (7), as were the mutant strains THY32 (cvt17-1; Ref. 3), MGY101 (apg5Δ::LEU2; Ref. 8), TVY1 (pep4Δ::LEU2; Ref. 9), WSY99 (ypt7Δ::HIS3; Ref. 10), and NNY20 (apg1Δ::LEU2; Ref. 11). Strain WPHYD7 SEY6210 aut7Δ::LEU2 will be described elsewhere.2 The cvt17Δ strain is described below. Yeast cells were grown as described previously (12).
Materials
We followed standard procedures to prepare antiserum to Cvt17, using synthetic peptides corresponding to amino acids 131–152 and 295–312 (Multiple Peptide Systems, San Diego, CA). Additional antisera were described previously (13-15). The copper-inducible promoter-based plasmid pCu426 was from Dr. Dennis J. Thiele (University of Michigan; Ref. 16). Other reagents are identical to those described previously (12, 17).
Cloning, Disruption, and Mutagenesis of CVT17
The CVT17 gene was cloned by transforming the cvt17-1 strain with a plasmid genomic library and screening for complementation of the starvation-sensitive phenotype on plates containing phloxine B (18). A secondary screen for prAPI accumulation was carried out as described previously (2). Sub-cloning of a complementing plasmid identified YCR068w as the complementing CVT17 ORF. The cvt17Δ strain (KTYD17) was made by inserting the LEU2 gene into the CVT17 ORF of strain SEY6210. At the time of this analysis, the Saccharomyces Genome Database (SGD) sequence had shown YCR068w as a 1.289-kilobase coding sequence. We amplified this region and subcloned it into pCu426 using PCR (all primer sequences available upon request). This construct (pCuCVT17ΔC(426)) failed to complement the prAPI defect of cvt17Δ. The construct was sequenced, and comparison to the SGD sequence revealed a single cytosine insertion in the PCR amplified gene, at nucleotide 956 of the ORF. Sequencing of the original genomic plasmid revealed an identical sequence to the PCR product, suggesting an error in the SGD sequence. A recent revision of the chromosome III sequence has corrected this error.
The rectified CVT17 sequence shifts the frame of the coding sequence, increasing the length of the encoded protein. The longer, full-length coding sequence was amplified by PCR and subcloned into pCu426 to generate pCuCVT17(426). For analysis of Cvt17 expression driven by the native promoter, CVT17 was amplified by PCR and cloned into pRS416 and pRS426, with inclusion of 0.5 kilobase before the start codon and 0.432 kilobase after the stop codon. A PCR-based overlap-extension strategy was used in site-directed mutagenesis of CVT17. Serine 332 was converted to alanine by changing the T at nucleotide 994 to G.
Other Procedures
Cell fractionation, immunoprecipitation of radiolabeled proteins, nitrogen starvation sensitivity, prAPI protease sensitivity, and reversal in SD-N were performed as described previously (8, 19). For endoglycosidase H (Endo H) treatment, cells were labeled for 10 min with no chase. Immunoprecipitated proteins were resuspended in 0.1% SDS, 50 mm sodium citrate, pH 5.5, 1 mm PMSF. After boiling for 7 min, an equal volume of 0.1% SDS and 0.1 m β-mercaptoethanol was added, then boiled for 7 min. Denatured proteins were incubated overnight at 37° C in the presence of 50 mm sodium citrate, pH 5.5, 16 mm β-mercaptoethanol, 0.03% SDS, 5 mm PMSF, and 2 units of Endo H. A second immunoprecipitation recovered the Cvt17 proteins.
Fluorescence Microscopy
Construction of the GFPAut7 fusion protein and FM 4-64 labeling of cells are described elsewhere.2 Cells were examined using a Nikon Eclipse E-800 fluorescence microscope. Images were captured by a Hamamatsu C4742–98 digital camera.
RESULTS
Cvt17 Is Required for Lysis of Subvacuolar Cvt Bodies and Autophagic Bodies
Previous analysis of cvt17-1 by electron microscopy suggested a defect in the breakdown of subvacuolar, prAPI-containing vesicles, called Cvt bodies (3, 6). We isolated the CVT17 gene and disrupted the chromosomal locus to generate a cvt17Δ strain (see “Experimental Procedures”). The null phenotype was accumulation of prAPI (Fig. 1A), as seen previously with the cvt17-1 mutant.
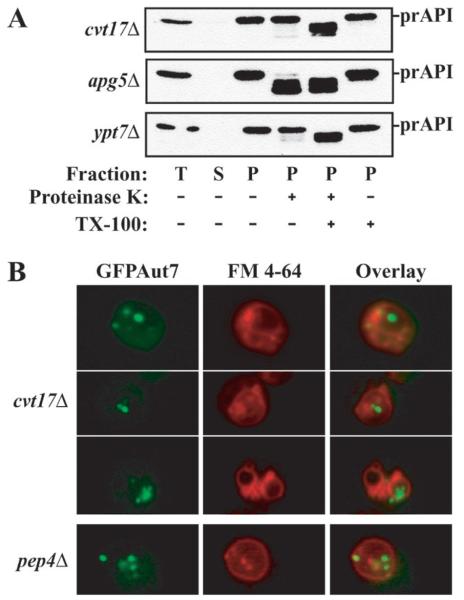
Fig. 1. The cvt17Δ strain accumulates Cvt bodies within the vacuole.
A, precursor API is enclosed within a membrane compartment in the cvt17Δ strain. Osmotically lysed spheroplasts (T) were separated into low speed supernatant (S) and pellet (P) fractions by a 5,000 × g centrifugation. P fractions were resuspended in lysis buffer alone or treated with proteinase K ± Triton X-100. B, the Cvt vesicle marker protein GFPAut7 accumulates in subvacuolar vesicles in cvt17Δ. pep4Δ and cvt17Δ cells harboring the pCuGFPAUT7(416) plasmid were grown to mid-log phase. GFPAut7 expression was induced with 10 μM CuSO4 for 3 h. Vacuoles were labeled with FM 4-64.
To determine the site of action of Cvt17, we analyzed the state of precursor API in cvt17Δ. Yeast cells were converted to spheroplasts and subjected to osmotic lysis. Addition of exogenous protease allowed us to determine whether the accumulated prAPI was blocked at a stage prior to Cvt vesicle formation/completion (protease-sensitive) or at a point following enclosure but prior to fusion with the vacuole or vesicle breakdown (protease-protected). As controls, we examined prAPI protease sensitivity in two strains that are defective in prAPI import. Apg5 is part of a novel protein conjugation complex that is required for both the Cvt and autophagy pathways (20). The apg5Δ strain is blocked in the completion of Cvt vesicles and accumulates membrane-associated, but protease-sensitive, prAPI (8). Ypt7 is a rab GTPase that is required for fusion of Cvt vesicles and autophagosomes with the vacuole (1). The ypt7Δ mutant accumulates prAPI in a protease-resistant state within completed cytosolic vesicles. Precursor API was found in a pelletable fraction in cvt17Δ, as it was in the apg5Δ and ypt7Δ mutant strains (Fig. 1A). In the apg5Δ strain, prAPI was sensitive to exogenous protease, even in the absence of detergent, consistent with a block in vesicle formation (8). In contrast, ypt7Δ and cvt17Δ cells contained prAPI that was not degraded to the mature form in the presence of exogenously added proteinase K unless detergent was also added, indicating that prAPI was within a membrane enclosed compartment (Fig. 1A; Ref. 21). These data, coupled with the vesicle accumulation phenotype of cvt17-1, suggest that prAPI was accumulating within subvacuolar Cvt vesicles in the cvt17Δ strain.
To assess whether the vesicles accumulating in cvt17Δ are indeed Cvt bodies, we examined whether a Cvt vesicle marker accumulated within subvacuolar vesicles during vegetative growth conditions. Aut7 is a component that is required for Cvt vesicle formation and is itself enclosed within Cvt vesicles (22). We have recently demonstrated the utility of a GFPAut7 construct as a marker for following the formation and movement of Cvt vesicles in vivo.2 To visualize the vacuole membrane, we used FM 4-64, a lipophilic red dye. In cvt17Δ, GFPAut7 is localized to Cvt bodies, seen as punctate structures accumulating within FM 4-64-labeled vacuoles (Fig. 1B, top panels). Some GFPAut7 punctate staining was also observed outside the vacuole. Mutants in the PEP4 gene, encoding vacuolar proteinase A, accumulate subvacuolar vesicles, including Cvt bodies by electron microscopy (5). Consistent with these previous observations, GFPAut7-labeled Cvt bodies also accumulated in the vacuoles of a pep4Δ strain (Fig. 1B, bottom panels) but did not accumulate in the vacuoles of wild type yeast (data not shown). The FM 4-64 vacuolar staining in cvt17Δ differed from that in other strains, including pep4Δ; we observed a relative increase in the number of FM 4-64-labeled compartments per cell in the cvt17Δ strain (Fig. 1B). While we do not know the molecular basis for the change in vacuolar morphology, the microscopy data provide direct evidence that cvt17Δ accumulates Cvt bodies within the vacuole.
While many molecular components are shared between the Cvt and autophagy pathways, some cvt mutants are not defective in autophagy (3, 12). To test whether cvt17Δ has a defect in autophagy, we examined whether the strain can survive growth in nitrogen-depleted media. Because transport to the vacuole by autophagy is the primary mode for degradation of cytoplasmic constituents under starvation conditions, the process is essential for viability during nutrient limitation. Wild type cells can withstand nutrient deprivation for long periods of time, while autophagy mutants such as apg1Δ or protease-deficient strains such as pep4Δ show reduced viability following a shift from growth in nutrient-rich medium to nitrogen-deficient medium (Fig. 2A). Like apg1Δ and pep4Δ, the cvt17Δ strain was starvation-sensitive.
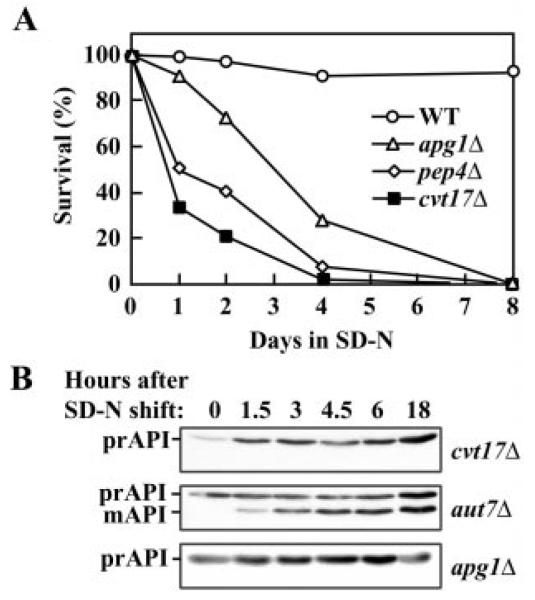
Fig. 2. The cvt17Δ strain is defective in autophagy.
A, log phase cultures grown in 1% yeast extract, 2% peptone, 2% glucose (YPD) were shifted to medium lacking nitrogen (SD-N). Viability was determined by plating aliquots on YPD plates. B, indicated strains were shifted to SD-N as in A, with aliquots taken at indicated times. Crude lysates were subjected to immunoblot analysis with anti-API antiserum.
Growing yeast cells transport prAPI to the vacuole via Cvt vesicles, while under nutrient deprivation, prAPI can be transported by autophagosomes (4, 5). If cvt17Δ is defective in lysis of autophagic bodies, as its starvation sensitivity suggests, we should see a block in prAPI maturation not only in rich media but also under starvation conditions. Indeed, both the Cvt and autophagic routes of delivery are blocked, as indicated by analysis of API following a shift to SD-N medium (Fig. 2B). Autophagy mutants display differential blocks in API import under nutrient-rich versus starvation conditions. For example, mutants such as aut7Δ that do not have a complete defect in autophagy accumulate prAPI in nutrient-rich media but are able to mature prAPI under starvation conditions (Ref. 19; Fig. 2B). In contrast, strains that have more severe autophagy defects, including apg1Δ, accumulate prAPI even after prolonged growth in SD-N. The cvt17Δ strain did not exhibit a rescue of the prAPI processing defect when cells were shifted to SD-N, indicating that it is defective in autophagy as well as the Cvt pathway (Fig. 2B). From these results we conclude that Cvt17 is involved in the lysis of autophagic bodies, as well as Cvt bodies.
Putative Serine Nucleophile in a Consensus Lipase Active Site Is Required for Cvt17 Function
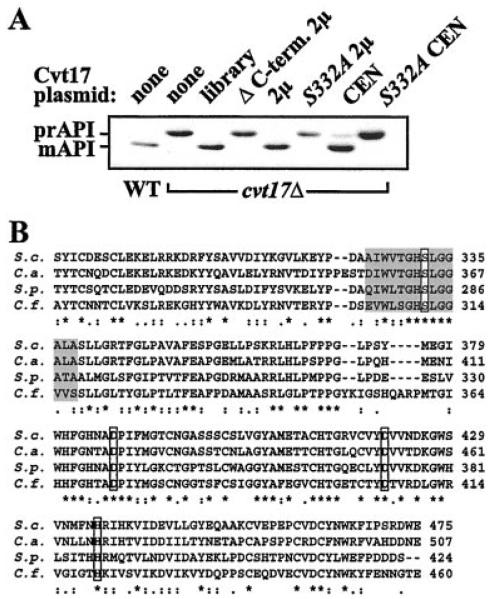
Our original analysis of CVT17 revealed an SGD error in the coding region of this ORF (see “Experimental Procedures”). We discovered that the predicted coding sequence that was given in the data base at that time did not complement the prAPI accumulation phenotype in cvt17Δ (Fig. 3A, ΔC-term. 2 μ). Sequencing revealed the presence of an extra cytosine base in the gene, at nucleotide 956 of the ORF. This insertion shifts the predicted reading frame, resulting in a protein with a predicted molecular mass of 58.5 kDa, rather than the 49.9 kDa predicted in the original data base. The full-length gene is able to complement the cvt17Δ strain, as shown by the accumulation of mAPI (Fig. 3A, 2μ and CEN). Importantly, the frameshift occurred just 5′ to a consensus site that is conserved in lipases and esterases (EC 3.1.1.3). The presence of this motif was revealed by a BLOCK search using the full-length, correct Cvt17 sequence. The sequence from amino acid 324 to 338 matches the consensus pattern of serine-active lipases (PROSITE reference PS00120; [LIV]-X-[LIVFY]-[LIVMST]-G-[HYWV]-S-X-G-[GSTAC]; Fig. 3B). A BLAST search identified related proteins in several distinct fungi, including Candida albicans (GenBank™ accession number AL033391; 43% identity), as well as the fission yeast Schizosaccharomyces pombe (GenBank™ accession number Z99753; 48% identity) and the tomato pathogen Cladosporium fulvum (GenBank™ accession number Y14554; 38% identity (23)). Alignments of the partial sequences over an area of significant homology are shown in Fig. 3B. Cvt17 is most similar to the subfamily of fungal lipases within the larger α/β-hydrolase-fold superfamily (24, 25). Triglcyeride lipases are lipolytic enzymes that hydrolyze the ester bond of triglycerides. In addition to the conserved serine within the active site consensus motif, a histidine residue and an aspartic acid residue act in hydrolysis in a charge-relay system (26). We found two well conserved aspartic acid residues in Cvt17 and other related fungal genes as well as a conserved histidine (Fig. 3B).
Fig. 3. The nucleophilic serine of a putative lipase domain is required for Cvt17 function.
A, wild type yeast or strain cvt17Δ with the indicated CVT17-containing plasmids were grown to mid-log stage. Western blots with anti-API antiserum are shown. Plasmids: library, complementing library plasmid; ΔC-term. 2μ, truncated CVT17 gene with the last 87 residues deleted cloned in pCu426; 2μ and CEN, full-length CVT17 gene in pRS426 and pRS416, respectively; S332A 2μ and S332A CEN, Cvt17 with Ser to Ala mutation in the putative catalytic site cloned in pRS426 and pRS416, respectively. B, sequence alignments of Cvt17 and related fungal proteins (see “Results” for details). Gaps in the alignment (−), fully conserved residues (*), and residues with strong (:) and weaker (.) similarity are indicated. Putative catalytic triad residues (S-D-H) are indicated by outlined boxes; only one of the conserved Asp residues is expected to be active. The gray box indicates the sequence that matches the consensus lipase pattern. ClustalX software was used to align sequences.
To determine whether the serine of the putative lipase domain is required for the function of Cvt17, we mutated the serine at position 332 to an alanine residue. Western blot analysis of API in the cvt17Δ strain that contained the mutated gene (cvt17S332A) showed accumulation of the precursor form of API, even when the mutant gene was expressed from a high copy plasmid (Fig. 3A). In contrast, the wild type gene allowed for prAPI maturation on both high and low copy plasmids. Thus, Cvt17 cannot function to break down subvacuolar vesicles and allow prAPI to be released into the vacuole without this serine within the G-X-S-X-G lipase motif.
Cvt17 Is a Glycosylated, Integral Membrane Protein
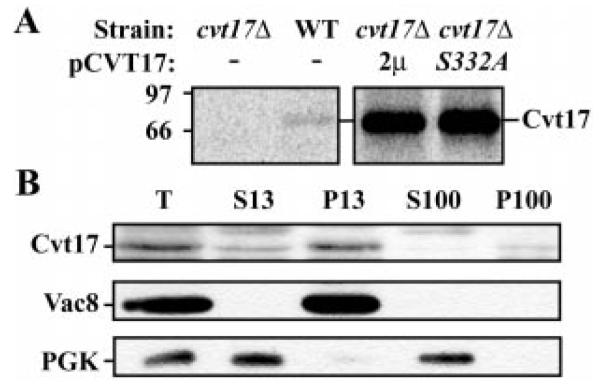
The potential assignment of Cvt17 as a lipase required for the degradation of subvacuolar vesicles led us to examine its bio-synthesis. To this end, we generated polyclonal antiserum against the protein. Immunoprecipitation revealed a single predominant band of ~70 kDa. This band was not detected in the cvt17Δ strain, and expression of CVT17 from a high copy plasmid greatly increased the abundance of this protein (Fig. 4A, 2μ lanes), suggesting that the serum was specific for Cvt17. We also immunoprecipitated Cvt17 from the strain harboring the mutant protein, which lacked the putative catalytic serine (cvt17S332A 2μ). The altered protein was recovered in equivalent amounts to the wild type protein, indicating that the single point mutation did not affect the stability of the protein.
Fig. 4. Cvt17-specific antiserum recognizes a 70-kDa membrane protein.
A, cells from cvt17Δ, wild type and cvt17Δ cells harboring 2μ wild type CVT17 and S332A mutant plasmids were radiolabeled for 5 min followed by a 10-min chase. Crude extracts were immunoprecipitated with antiserum to Cvt17. B, spheroplasts of wild type cells were osmotically lysed, and the resultant lysate (T) was centrifuged at 13,000 × g to yield supernatant (S13) and pellet (P13) fractions. S13 was further separated into high speed S100 and P100 fractions by centrifugation at 100,000 × g. Samples were subjected to immunoblot analysis with antisera to Cvt17, the vacuolar membrane-associated protein Vac8, and the cytosolic protein phosphoglycerate kinase (PGK).
Analysis of the Cvt17 primary sequence indicates a stretch of hydrophobic amino acids (residues 13–35) that might serve as a membrane anchor for the protein. Following osmotic lysis and subcellular fractionation, Cvt17 was predominately localized to a 13,000 × g pelletable fraction (Fig. 4B). We investigated whether this localization was due to direct interaction with a membrane by subjecting the P13 fraction to different wash conditions. Alkali extraction efficiently removed the peripheral membrane protein Vma2 from the membrane but had no effect on Cvt17 or the integral membrane protein Dpm1 (data not shown). Treatment with the detergent Triton X-100 resulted in the solubilization of all three proteins. These results suggest that Cvt17 is an integral membrane protein.
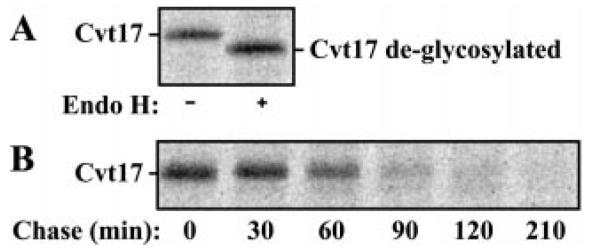
The predicted molecular mass of Cvt17 is 58.5 kDa; however, the protein migrated with a molecular mass of 70 kDa following SDS-polyacrylamide gel electrophoresis (Fig. 4A). The Cvt17 sequence has three predicted N-linked glycosylation sites. To determine whether the aberrant migration was due to glycosylation, we treated immunoprecipitated Cvt17 with Endo H. Following treatment with the deglycosylating enzyme, the molecular mass of Cvt17 shifted to ~61 kDa (Fig. 5A). This change in molecular mass correlates well with the predicted removal of three N-linked carbohydrate side chains. This post-translational modification reveals that Cvt17 must reside at least temporarily in the secretory pathway. Furthermore, the observation that Cvt17 is glycosylated allows us to infer the membrane topology of the protein. All three putative glycosylation sites are C-terminal to the transmembrane domain, suggesting that Cvt17 is a type II integral membrane protein. Upon integration into the ER membrane, the N-terminal 13 amino acids would remain exposed to the cytosol, with the majority of the protein located in the lumen of the ER.
Fig. 5. Cvt17 is a short-lived glycoprotein.
A, cells were labeled for 10 min, followed by immunoprecipitation with antiserum to Cvt17. Immunoprecipitated proteins were mock treated or treated with Endo H. B, wild type cells were labeled for 10 min. Crude extracts from samples collected at the indicated chase times were immunoprecipitated with Cvt17-specific antiserum.
The phenotype of the cvt17 mutant, accumulation of subvacuolar vesicles, coupled with its characterization as a secretory pathway protein suggests that Cvt17 might function within the vacuole lumen. Accordingly, we attempted to localize the Cvt17 protein. Subcellular fractionation based on velocity sedimentation indicated that Cvt17 was predominantly localized within a 13,000 × g fraction (Fig. 4B). The P13 fraction contains various subcellular compartments, including the vacuole. To directly determine whether Cvt17 was located within the vacuole, we purified vacuoles from yeast spheroplasts. However, we were unable to detect Cvt17 in the vacuolar fraction using this technique (data not shown). As an alternative approach, we fractionated cell lysates on an OptiPrep density gradient. Using this technique, in the presence of substantial amounts of protease inhibitors, we were able to detect small amounts of Cvt17 in fractions that contained the vacuolar protein marker alkaline phosphatase (data not shown). However, the majority of the Cvt17 was found in fractions that were distinct from the vacuole fractions. Thus, Cvt17 is found predominately in a membrane fraction residing outside the vacuole under steady-state conditions.
To examine the localization of Cvt17 in vivo, we engineered GFP fusions with the protein at both the N and C termini. Both constructs only partially complemented the Cvt trafficking defect of the cvt17Δ strain, as assessed by API Western blotting (data not shown). Furthermore, we were unable to visualize strong fluorescent signals anywhere in cells containing these GFP fusion constructs. An explanation for this lack of signal might be an inherent instability of Cvt17. To examine stability of Cvt17 directly, we carried out a pulse-chase analysis. Yeast cells were labeled with [35S]methionine/cysteine and subjected to a nonradioactive chase. At each time point, samples were removed and immunoprecipitated. We found that Cvt17 was unstable in wild type cells; the protein was degraded with a half-life of ~45 min (Fig. 5B). The inherent instability of this protein made its localization problematic as much of it was degraded during fractionation procedures.
DISCUSSION
We have identified a protein that is required for the turnover of membrane vesicles in the vacuole of yeast. Sequence analysis of this protein reveals the presence of a domain found in lipases and esterases, and we have confirmed the importance of this domain by using site-directed mutagenesis of a putative active site serine residue within the lipase motif. Phenotypic analysis of yeast expressing the altered Cvt17 protein shows that the protein is inactive in the Cvt pathway without this residue. The cvt17Δ mutant is unable to degrade both Cvt bodies and autophagic bodies and is extremely sensitive to starvation (Fig. 2A). The ability to degrade membranes that are released into the vacuole lumen is required for starvation survival. In the absence of this degradative capacity, the cell is unable to break down lipid and, as a result, is unable to access cargo contained within autophagic bodies. The occurrence of genes with similarity to CVT17 in other fungi suggests that this may be a conserved protein in the kingdom.
The maintenance of membrane-bound vesicles in the cvt17Δ strain suggests a role for the protein in degrading a phospholipid bilayer. Sequence comparisons, however, place the protein in a class of hydrolytic enzymes that act primarily on triglycerides, not phospholipids. It will be necessary to perform biochemical studies with the purified enzyme to investigate its substrate specificity. To function properly, lipases must be targeted to the appropriate substrates. The membrane source of Cvt vesicles and autophagosomes is not known. If Cvt17 is incorporated into Cvt vesicles and autophagosomes, its topology would place the Cvt17 active site on the outer surface of the Cvt and autophagic bodies. To prevent membrane degradation during transit, it is possible that Cvt17 becomes active only in the vacuole. If this is the case, the lytic activity of this protein must be suppressed during transit. Some aspect of the vacuolar milieu may then activate Cvt17 as is thought to be the case for proteinase A. If Cvt17 acts directly to degrade subvacuolar vesicles, it should reside within the vacuole. Subcellular fractionation did reveal a small population of vacuolar Cvt17 that may play a primary role in directly degrading Cvt and autophagic bodies. Most of the protein, however, did not cofractionate with this organelle. In agreement with these results, the rapid Cvt17 degradation that we observed (Fig. 5B) was independent of the major vacuole proteases (data not shown).
The reasons for maintaining a sizable population of Cvt17 outside the vacuole are as yet obscure. It is possible that Cvt17 performs some additional unknown function outside the vacuole. Another possibility is that Cvt17 acts exclusively outside the vacuole and only indirectly influences the lytic competence of subvacuolar vesicles; Cvt17 might act as a lipase to specifically alter the lipid composition of Cvt vesicles and autophagosomes as they form. Specific lipid composition might be a requirement for the subsequent lysis of Cvt bodies and autophagic bodies. Lipids could even provide the molecular basis for recognition within the vacuole lumen, allowing an alternate vacuolar lipase to distinguish between subvacuolar vesicles destined for degradation and the bounding vacuolar membrane, which must remain intact.
Cvt17 function, and specifically the function of a lipase active site domain, is required for the lysis of subvacuolar Cvt and autophagic bodies. Characterization of this protein is an important first step in understanding vacuolar function in the turnover of lipids and in the terminal steps of the Cvt and autophagic pathways. Continued analysis of Cvt17 will provide important information about membrane recycling in the vacuole.
Acknowledgments
We thank Jacob Teter for technical assistance, Bob Fuller for use of the fluorescence microscope, and E. J. Brace for help with microscopy. We thank Maria Hutchins for comments on the manuscript and for technical assistance.
Footnotes
This work was supported by Public Health Service Grant GM53396 from the National Institutes of Health (to D. J. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: API, aminopeptidase I; prAPI, precursor API; Cvt, cytoplasm to vacuole targeting; Endo H, endoglycosidase H; ER, endoplasmic reticulum; GFP, green fluorescent protein; PMSF, phenylmethylsulfonyl fluoride; SD-N, synthetic minimal medium lacking nitrogen; ORF, open reading frame; SGD, Saccharomyces Genome Database; PCR, polymerase chain reaction.
Kim, J., Huang, W.-P., and Klionsky, D. J. (2001) J. Cell Biol. 152, in press
REFERENCES
- 1.Kim J, Klionsky DJ. Annu. Rev. Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 2.Harding TM, Morano KA, Scott SV, Klionsky DJ. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 4.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott SV, Baba M, Ohsumi Y, Klionsky DJ. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Mol. Biol. Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. J. Biol. Chem. 1998;19:15818–15829. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]
- 10.Wurmser A, Emr S. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Gene. Amst. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 12.Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 13.Klionsky DJ, Cueva R, Yaver DS. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomashek JJ, Sonnenburg JL, Artimovich JM, Klionsky DJ. J. Biol. Chem. 1996;271:10397–10404. doi: 10.1074/jbc.271.17.10397. [DOI] [PubMed] [Google Scholar]
- 15.Wang YX, Catlett NL, Weisman LS. J. Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labbé S, Thiele DJ. Methods Enzymol. 1997;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 17.Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. J. Cell Biol. 2000;148:465–479. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukada M, Ohsumi Y. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 19.Abeliovich H, Dunn WA, Jr., Kim J, Klionsky DJ. J. Cell Biol. 2000;151:1025–1033. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W-P, Scott SV, Kim J, Klionsky DJ. J. Cell Biol. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 23.Coleman M, Henricot B, Arnau J, Oliver RP. Mol. Plant Microbe Interact. 1997;10:1106–1109. doi: 10.1094/MPMI.1997.10.9.1106. [DOI] [PubMed] [Google Scholar]
- 24.Cousin X, Hotelier T, Giles K, Toutant JP, Chatonnet A. Nucleic Acids Res. 1998;26:226–228. doi: 10.1093/nar/26.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derewenda ZS, Sharp AM. Trends Biochem. Sci. 1993;18:20–25. doi: 10.1016/0968-0004(93)90082-x. [DOI] [PubMed] [Google Scholar]
- 26.Blow D. Nature. 1990;343:694–695. doi: 10.1038/343694a0. [DOI] [PubMed] [Google Scholar]