SUMMARY
Accurate chromosome segregation during mitosis and meiosis depends on shugoshin proteins that prevent precocious dissociation of cohesin from centromeres. Shugoshins associate with PP2A, which is thought to de-phosphorylate cohesin and thereby prevent cleavage by separase during meiosis I. A crystal structure of a complex between a fragment of human Sgo1 and an AB’C PP2A holoenzyme reveals that Sgo1 forms a homodimeric parallel coiled-coil that docks simultaneously onto PP2A’s C and B’ subunits. Sgo1 homo-dimerization is a pre-requisite for PP2A binding. While hSgo1 interacts only with the AB’C holoenzymes, its relative Sgo2 interacts with all PP2A forms and may thus lead to dephosphorylation of distinct substrates. Mutant shugoshin proteins defective in the binding of PP2A cannot protect centromeric cohesin from separase during meiosis I or support the spindle assembly checkpoint in yeast. Finally, we provide evidence that PP2A’s recruitment to chromosomes may be sufficient to protect cohesin from separase in mammalian oocytes.
INTRODUCTION
The chromatids of bivalent chromosomes like their mitotic counterparts are held together by a multi-subunit complex called cohesin, whose α-kleisin (Rec8), Smc1, and Smc3 subunits form a tripartite ring within which sister DNAs are thought to be trapped (Haering et al., 2008; Nasmyth and Haering, 2005). The first meiotic division is triggered by destruction of sister chromatid cohesion by a thiol protease called separase, which opens the cohesin ring by cleaving it’s α-kleisin subunit (Buonomo et al., 2000). This splits the bivalent into a pair of dyad chromosomes that are segregated to opposite poles at the first meiotic division. Crucially, the two chromatids of each dyad remain associated with each other both during and after anaphase I because cohesin holding sister centromeres together is protected from cleavage by separase by a class of proteins called shugoshins (Goldstein, 1980; Kerrebrock et al., 1995; Kitajima et al., 2004; Rabitsch et al., 2004). The persistence of centromeric cohesion is essential to ensure that sister kinetochores and hence individual chromatids are pulled to opposite poles at the second meiotic division. A failure to protect centromeric cohesin from separase might contribute to the human aneuploidy caused by chromosome missegregation during meiosis I in oocytes, especially in older women (Vogt et al., 2008).
How do shugoshins protect sister chromatid cohesion at centromeres? Recent work indicate that shugoshins protect centromeric cohesin by interacting with protein phosphatise 2A (PP2A) (Kitajima et al., 2006; Riedel et al., 2006). PP2A holoenzymes are composed of a catalytic C subunit, a structural or scaffold A subunit, and a regulatory B subunit. The A and C subunits bind directly to each other forming a core enzyme. The B subunits in contrast are much more varied and mammals possess at least 18 types, which belong to four subfamilies: PR55/B, PR61/B’, PR72/B” and PR110/B”’ (Janssens and Goris, 2001; Lechward et al., 2001; Sontag, 2001; Yu, 2007). During meiosis in both fission and budding yeast, a shugoshin, namely Sgo1, is found stably associated with AB’C PP2A holo-enzyme (Riedel et al., 2006). This led to the suggestion that phosphorylation of cohesin’s Rec8 subunit may be required for its cleavage by separase (Brar et al., 2006; Riedel et al., 2006), as is at least partly the case for its Scc1 mitotic counterpart (Alexandru et al., 2001), and that by recruiting PP2A, Sgo1 prevents cleavage of centromeric Rec8 by inducing its de-phosphorylation. Consistent with this hypothesis, yeast mutants lacking PP2A’s B’ subunits fail to prevent Rec8’s removal from centromeres at the first meiotic division, which is accompanied by their precocious disjunction soon after the first meiotic division (Riedel et al., 2006).
However, inactivation of PP2A causes highly pleiotropic phenotypes because the enzyme has a wide variety of functions and substrates and the effect on chromosome segregation of mutating PP2A could conceivably be due to indirect effects. To address whether recruitment of PP2A really is a crucial part of the mechanism by which shugoshins protect centromeric cohesin, it is necessary to understand how PP2A actually binds to shugoshin and to use this information to investigate the phenotype of mutant proteins that are defective in their ability to bind the phosphatase. This sort of approach is equally important for disentangling the role of PP2A-shugoshin interactions during mitotic chromosome segregation in mammals, where hSgo1 has a crucial role in preventing dissociation of cohesin from centromeres (Kitajima et al., 2005: McGuinness et al., 2005; Salic et al., 2004) and it is unclear whether Sgo1’s role is to recruit PP2A to centromeres or the converse (Kitajima et al., 2006; Tang et al., 2006).
PP2A binds to a human Sgo1 fragment containing its N-terminal 176 amino acids and the interaction is abolished by mutation of a highly conserved asparagine (N61I) within a region predicted to form a coiled-coil (Tang et al., 1998; Tang et al., 2006). The N61I mutation abolishes persistence of sister centromere cohesion during meiosis in Drosophila (Kerrebrock et al., 1995), but it might have rather pleiotropic consequences as it supposedly also affects the ability of hSgo1/ MEI-S332 to bind chromosomes (Tang et al., 1998; Tang et al., 2006) and possibly disrupts Sgo1’s putative coiled-coil.
A crystal structure of a complex formed between a fragment of human Sgo1 and an AαB56γCα PP2A holoenzyme (from now on referred to as AB’C PP2A or PP2A) reveals that Sgo1 forms a parallel coiled-coil whose N- and C-terminal ends bind to C and B’ PP2A subunits respectively. Analysis of phenotypes of shugoshin mutants defective in PP2A binding demonstrate that recruitment of PP2A by Sgo1 is essential for the protection of sister chromatid cohesion at centromeres at meiosis I and for the SAC during mitosis in yeast and that recruitment of PP2A to chromosome arms may be sufficient to block the resolution of chiasmata in mouse oocytes. Another important implication of our findings is that PP2A’s specificity is not determined solely by its regulatory B subunits.
RESULTS
Shugoshin’s putative coiled-coil is responsible for PP2A binding
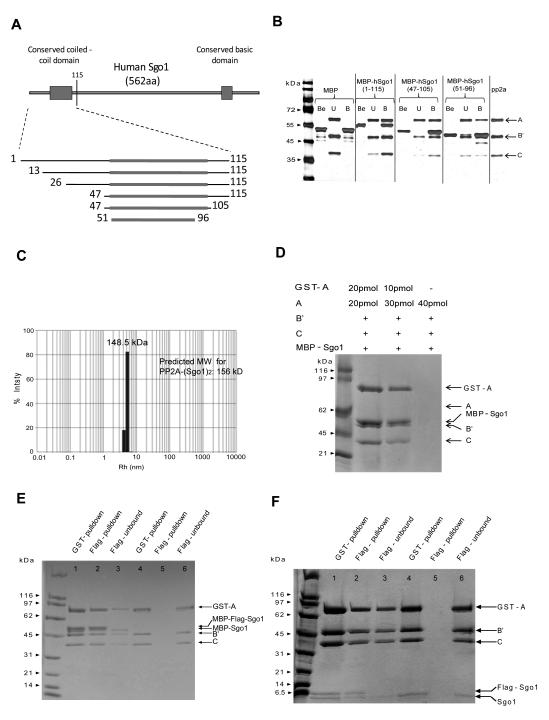
Recombinant human Sgo1 fragments containing hSgo1’s N-terminal 176 residues (Figure 1A) aggregate but MBP-Sgo1 fusion proteins are soluble and interact with PP2A. Binding studies with a variety of MBP-Sgo1 fusion proteins reveal that the putative coiled-coil (residues 47-105) confers interaction with AB’C PP2A holoenzyme (Figure 1B). A slightly shorter fragment (51-96) also confers binding but with much lower affinity (Figures 1B and S1).
Figure 1. One AB’C PP2A holoenzyme binds with two Sgo1 molecules in the solution.
A. Domain structure of Sgo1. MBP fusions of the indicated fragments were tested positive for binding to AB’C PP2A (data not shown except in B.).
B. The coiled-coil domain of Sgo1 is sufficient for binding to AB’C PP2A holoenzyme. Purified AB’C PP2A holoenzyme was pulled down by corresponding MBP-Sgo1 fragments. The SDS-PAGE gel was silver stained. Be, U and B represent the MBP-Sgo1 sample before PP2A binding, unbound PP2A and total proteins bound to the amyloid beads, respectively.
C. Size distribution of the PP2A-Sgo1(51-105) complex measured by dynamic light scattering.
D. There is one PP2A holoenzyme in each PP2A-Sgo1 complex. Indicated amounts of GST-A and A subunits were incubated with MBP-Sgo1(51-105), as well as the B’ and C subunits of PP2A prior to GST affinity pull down. Bead bound proteins were eluted in a buffer containing glutathione and resolved on a SDS-PAGE gel.
E and F. There are at least two Sgo1 molecules in each PP2A-Sgo1 complex. MBP-Sgo1(51-105) was incubated either with equal amount of (lanes 1, 2 and 3) or without (lanes 4, 5 and 6) MBP-FLAG-Sgo1(51-105) and with PP2A GST-A, B’ and C subunits prior to GST affinity pull down. Bead bound proteins were eluted in a buffer containing glutathione. Eluted proteins were incubated with FLAG affinity beads before elution in a buffer containing FLAG peptide. Eluates and unbound fractions were resolved on a SDS-PAGE gel (E). This experiment was repeated using Sgo1(51-105) and FLAG-Sgo1(51-105) peptides, instead of MBP-fusions of these peptides (F).
Two shugoshin molecules interact with a single PP2A holoenzyme in solution
Because the coiled-coil region is predicted to form a homodimer, we determined the molecular stoichiometry of the PP2A-Sgo1 complex. Both size exclusion chromatography and dynamic light scattering indicate that the complex formed between PP2A and MBP-Sgo1(51-105) has a molecular weight ∼250 kD, while that formed with Sgo1(51-105) after MBP had been removed by cleavage has a molecular weight of ∼148-160 kD (Figures 1C and S1). Because the molecular weights of the AB’C PP2A holoenzyme, Sgo1(51-105) and MBP are 146 kD, 6 kD and 45 kD, respectively, the molecular stoichiometry of the PP2A-Sgo1 complex in solution is likely 1:2. This molar ratio is consistent with MBP-Sgo1 bands having twice the intensity of PP2A subunits with an equivalent molecular weight in coomassie blue-stained SDS gels of purified PP2A-Sgo1 complexes purified either using the GST-tag on the PP2A A subunit or the MBP moiety of MBP-Sgo1 and/or by gel-filtration (Figures 1B, 1D, 1E and S1A). To show that there is only a single PP2A holoenzyme in the complex, we incubated GST-tagged PP2A A subunit (GST-A) with excessive untagged-A, B, C subunits and MBP-Sgo1. If there were two or more PP2A holoenzymes, then GST-A should pull down untagged PP2A A subunits, which we do not observe (Figure 1D). The complexes contain at least two Sgo1 molecules, because after mixing Sgo1 fragments with different lengths and tags (one was fused to MBP, and the other to MBP and a FLAG epitope), addition of PP2A holoenzyme enabled FLAG affinity beads to pull down both MBP-Sgo1 proteins (Figure 1E). Dimerization of Sgo1 fragments within PP2A complexes is not caused by their fusion to MBP because co-purification occurs even when the latter is removed (Figure 1F, Supplementary data).
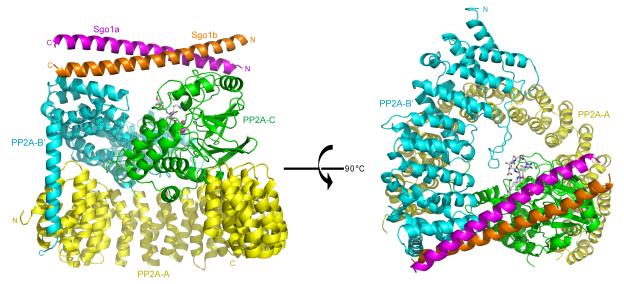
Crystal structure of the PP2A-Sgo1 complex: overall architecture
We failed to obtain useful crystals with complexes containing Sgo1(51-105) or longer fragments but eventually obtained a crystal structure with Sgo1(51-96), which lacks a complete coiled-coil region (Figures S1B and S1C). Despite not forming stable homodimers in solution (data not shown), MBP-Sgo1(51-96) does pull down PP2A holoenzyme (Figure 1B), though the complex dissociates partially during gel filtration producing a peak fraction containing a mixture of the PP2A-Sgo1 complex and unbound PP2A holoemzyme (Figure S1B and S1C). It is this mixture that forms useful crystals, presumably because “free” PP2A holoenzymes shield the hydrophobic surface of Sgo1’s coiled-coil not covered by the binding of the first PP2A holoenzyme (see below).
The crystal structure was determined at 2.7 Å resolution (Table 1). Each asymmetric unit within the crystal lattice contains one human PP2A AB’C holoenzyme, one Sgo1(51-96) peptide and one microcystin PP2A inhibitor molecule. The Sgo1(51-96) peptide forms a single long helix. Two Sgo1 peptides, related by a 2-fold crystallographic symmetry, form a parallel coiled-coil homodimer and interact with both B and C subunits of two PP2A holoenzymes (Figure S2). In this way, two symmetry-related PP2A holoenzymes interact with symmetrical surfaces of the Sgo1 coiled-coil dimer. Because only a single PP2A holoenzyme interacts with the Sgo1 dimer in solution, the two surfaces of the Sgo1 coiled-coil homodimer must be slightly different, though this is not apparent in the crystal structure. We imagine that the two surfaces of Sgo1’s coiled-coil have different affinities for PP2A, with the lower affinity surface only binding PP2A under crystallization conditions, when protein concentrations are much higher. Structure-based mutagenesis (see below) confirmed that the PP2A-Sgo1 interface observed in our crystal structure is physiologically relevant. For the rest of this work, we will discuss the interaction of one PP2A AB’C complex with two Sgo1(51-96) peptides, and for simplicity, refer to them as Sgo1a and Sgo1b, respectively (Figure 2).
Table 1.
Statistics of crystallographic analysis of the PP2A-Sgo1 complex.
| Data collection | |
| Space group | C2221 |
| Cell dimensions:a, b, c (Å) | 104.94, 145.86, 294.15 |
| Resolution (Å) | 50.0-2.7 (2.8-2.7) |
| aRsym | 0.135 (0.494) |
| I / σI | 12.2 (1.6) |
| Completeness (%) | 92.0 (51.3) |
| Redundancy | 6.2 (2.8) |
| Refinement | |
| Resolution (Å) | 50.0-2.7 |
| No. reflections | 54842 |
| bRwork / Rfree | 22.6% / 27.7% |
| No. atoms | |
| proteins | 10802 |
| Ions | 2 |
| B-factor | |
| Protein | 47.5 |
| Ions | 37.2 |
| R.m.s. deviation | |
| Bond lengths (Å) | 0.011 |
| Bond angles (1) | 1.52 |
| cRamachandran plot (core, disallowed, %) | 91.5 %, 0 % |
Values in the parentheses refer to the outer-shell bin.
Rsym = Σij|(Ii(j) — <I(j)>|/Σij Ii(j), where Ii(j) is the intensity of the i-th observation of reflection j. < I(j) >is the weighted mean of all measurements of j.
R = Σj∥Fobs(j)| — |Fcalc(j)∥/Σj|Fobs(j)|. Rwork and Rfree were calculated with the working and test reflection sets, respectively.
As defined in PROCHECK.
Figure 2. Overall structure of PP2A-Sgo1 complex.
A, B’, C subunits of PP2A, Sgo1a and Sgo1b are shown and labeled in yellow, cyan, green, orange and purple, respectively. The PP2A inhibitor microcysteine is represented by stick mode and two manganese atoms binding with PP2A are showed in pink and sphere mode. The orthogonal view of PP2A-Sgo1 complex is shown on the right. All structural images were prepared using the PyMOL Molecular Graphics System (DeLano Scientific).
The structure of PP2A holoenzyme in the complex is similar to the lower resolution structures containing PP2A alone (Cho and Xu, 2007; Xu et al., 2006). Briefly, the 15 HEAT-repeats of the A subunit form a horseshoe shaped scaffold that holds B and C subunits using the ridge formed by intra-repeat loops. The only significant structural difference concerns the C-terminal region of the B’ subunit’s conserved domain, which was mostly invisible in previous PP2A structures. It forms a long-helix and interacts with the A subunit’s most N-terminal HEAT repeat. This suggests that the divergent C-terminal domain of B’ subunits reside “below” or inside the horseshoe scaffold and have a role in PP2A localization (Figure S3).
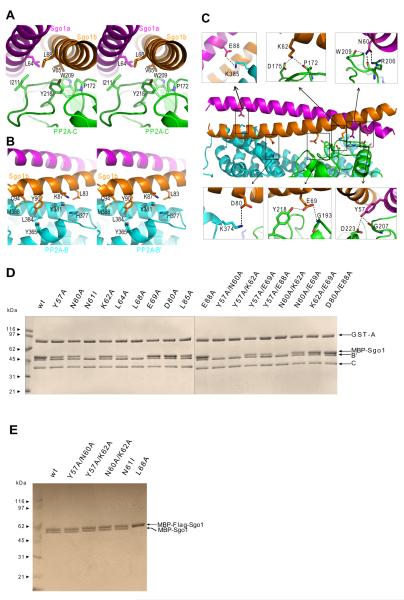
Both Sgo1a and Sgo1b interact with both B’ and C subunits of PP2A. The N- and C-terminal ends of Sgo1’s coiled-coil form discrete interfaces with C and B’ subunits respectively. Each binding interface contains a hydrophobic core as well as peripheral hydrogen bonds and salt bridges. The contact area between the C subunit and the N-terminal half of the Sgo1 homodimer (1328 Å2) is larger than that between the B’ subunit and its C-terminal half (1046 Å2). Several highly conserved hydrophobic Sgo1 residues (L64, L68, V75, I81, I82, L85 and L92), as well as two other residues (L53 and L54) found in Sgo1 but not Sgo2, appear to stabilize the Sgo1a-Sgo1b dimerization (Figure S4).
The PP2A-Sgo1 interface: hydrophobic interactions buttressed by hydrophilic interactions
The N-terminal region of Sgo1’s coiled-coil interacts directly with residues in two loops (between α9 and α10 and between β6 and α8) of the PP2A C subunit. Hydrophobic interactions involve Sgo1a’s L64 and Sgo1b’s L68, which stack on each other and are surrounded by the aliphatic parts of P172, W209, I211 and Y218 of the C subunit (Figure 3A). Hydrogen bonds involve Y57 and N60 from Sgo1a and E69 and K62 from Sgo1b (Figures 3A, 3C and S5). The hydroxyl group of the conserved Sgo1a Y57 sidechain interacts with the carbonyl of G207 on the longer of the two loops and D223 on α10. The side chain of Sgo1a’s conserved N60 is nestled in a surface groove and forms multiple hydrogen bonds with C subunit residues 206-209. Oδfrom N60 forms a hydrogen bond with Nε and Nη of R206 while Nδof N60 interacts with the carbonyls of R206 and W209. On Sgo1b, Nζ of K62 forms a hydrogen bond with the carbonyl of P172 and a salt bridge with D175, residues situated on the shorter loop. In addition, Sgo1b E69 docks into a surface groove and thereby interacts with the main chain amide nitrogen of G193 and with Oη of Y218, which is also on the longer loop. Surprisingly, Sgo1 residue N61, previously found to be critical for interaction with PP2A, does not participate directly in the PP2A-Sgo1 interface. N61 residues from Sgo1a and Sgo1b instead form a pair of strong hydrogen bonds (2.48 Å) in the middle of the coiled-coil interface (Figure S4).
Figure 3. The specific interactions between PP2A and Sgo1 and the effect of Sgo1 mutations on the interaction.
A. Stereo views of hydrophobic interactions between PP2A-C and Sgo1. The residues involved in the hydrophobic core are indicated and represented in the stick mode.
B. Stereo views of hydropholic interactions between PP2A-B’ and Sgo1.
C. Hydrophilic interactions between PP2A and Sgo1. The residues (Y57, N60, K62, E69, D80 and E88 as indicated) from both Sgo1 molecules form hydrogen bonds or salt bridges with PP2A B’ and C subunits, as indicated by dashed lines.
D. The binding assay of PP2A with various Sgo1 mutants. Wild type (wt), various single or double mutant Sgo1s, as indicated, were tested to bind with PP2A.
E. Dimerization assay of several selected mutant Sgo1. The mutant Sgo1s which cannot bind with PP2A (in the panel A) were selected to test the dimerization. The mutant MBP-Sgo1 was pull downed by the FLAG-tagged same mutant MBP-Sgo1 and followed elution by 100 ng/ml FLAG peptide solution and detected by SDS-PAGE analysis.
The PP2A holoenzyme contains one of four types of B subunit (PR55/B, PR61/B’, PR72/B” and PR110/B”’). Importantly, only PR61/B’ was found associated with Sgo1 from meiotic yeast cells (Riedel et al., 2006) or mitotic mammalian cells (Kitajima et al., 2006; Riedel et al., 2006). The crystal structure reveals that the C-terminal end of Sgo1’s coiled-coil region (residues 80-94) interacts with the last pseudo HEAT repeat of the conserved domain of PP2A’s B’ subunit (Figures 3B, 3C and S5). The aliphatic parts ofthe sidechains from Sgo1b’s L83, K87, Y90 and C94 interact with Y365, H377, Y381, L384 and M388 on the last pair of pseudo-HEAT repeats of PP2A’s B’ subunit. This interaction is strengthened by two salt bridges between K374 and K385, which are conserved within the B’ subfamily, and Sgo1b’s D80 and Sgo1a’s E88 respectively. Structural superposition of the AB’C-Sgo1 complex and the ABC family PP2A holoenzyme (Xu et al., 2008) indicates that Sgo1 cannot interact with both the C subunit and the PP2A B subunit, which explains why Sgo1 does not interact with the ABC family PP2A holoenzymes (Figure S6).
The effect of mutations on PP2A binding and Sgo1 dimerization
To assess the contribution of key residues to Sgo1 dimerization and its interaction with PP2A, we measured the effects of mutating them, mainly to alanine. Substitution of Sgo1’s N61 by either alanine or isoleucine, which causes precocious loss of sister centromere cohesion in Drosophila (Tang et al., 1998), abolishes binding to PP2A (Figure 3D and data not shown). Substitutions at L64 and L68, which participate in dimer formation as well as hydrophobic interactions with PP2A’s C subunit, have a similar effect. L64A reduces PP2A binding while L68A abolishes it (Figure 3D). Substitution by alanine of a conserved leucine within the Sgo1 dimer interface (L85) has a moderate effect. Mutations of individual residues specifically involved in PP2A binding, including Y57A, N60A and K62A, have little effect on PP2A binding, though Y57A causes a modest reduction. In contrast, several double substitutions, namely Y57A/N60A, Y57A/E69A, and N60A/K62A, greatly reduce PP2A binding under our experimental conditions (Figure 3D).
To test the effects on Sgo1 dimerization, we purified mutant proteins with or without a FLAG tag. To distinguish them in SDS PAGE gels, the tagged and untagged proteins contained different amounts of Sgo1 sequence, namely 47-105 and 51-105 respectively. Equal amounts of tagged and untagged proteins containing the same mutation were mixed, the tagged protein pulled down using anti-FLAG beads, and the abundance of tagged and untagged protein compared using SDS PAGE. The Y57A/N60A, Y57A/K62A and N60A/K62A double mutations, which should only affect interaction with PP2A, have no effect on dimer formation using this assay. According to the crystal structure, L68 is involved in Sgo1a-Sgo1b interaction and has little direct involvement in PP2A interaction. Its substitution by alanine (L68A) abolishes dimer formation as well as interaction with PP2A. This result, together with our observation that both Sgo1 helices interact extensively with the same PP2A complex, imply that Sgo1 homo-dimerization is a prerequisite binding to PP2A. Surprisingly, N61I forms dimers as efficiently as wild type. N61 sidechains pack tightly at the center of the coiled-coil interface and introduction of a larger sidechain (as occurs after substitution by isoleucine) might have been expected to disrupt dimer formation. Remaining interactions between the Sgo1a and Sgo1b helices must therefore be sufficient for dimerization. Why, if N61 sidechains are not required for dimer formation and make no direct contacts with PP2A, does N61I disrupt binding of PP2A and cause a dramatic phenotype in flies? Introduction of a larger sidechain (as in substitution by isoleucine) or simply loss of key hydrogen bonds (as in substitution by alanine) presumably disrupts the coiled-coil’s conformation, affecting the interaction with PP2A of neighboring residues such as Y57, N60 and K62. In summary, our mutagenesis is consistent with the crystal structure and with amino acid conservation. Crucially, we realized one of our primary goals, namely to design mutant shugoshins defective in PP2A interaction without changing other properties of the protein (that we know about).
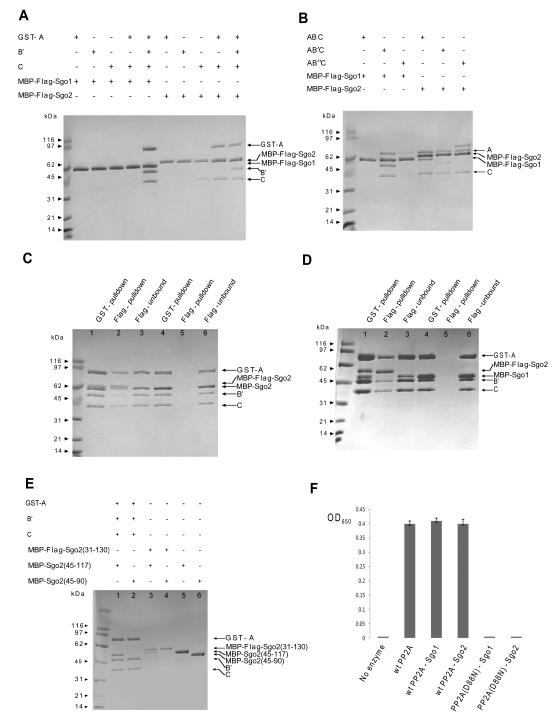
Sgo2 interacts as a dimer with all PP2A forms
Mammalian cells express two major types of shugoshin, called Sgo1 and Sgo2. The finding that mice lacking Sgo2 are viable (Llano et al., 2008) is inconsistent with the claims that this protein has an important role during mitosis (Huang et al., 2007; Kitajima et al., 2006) and it is likely that Sgo1 alone is necessary to protect centromere cohesion during mitosis (McGuinness et al., 2005; Salic et al., 2004). What about meiosis? RNA interference studies have come to conflicting conclusions asto Sgo1’s role (Lee et al., 2008; Yin et al., 2008). What is clear is that sgo2Δ/sgo2Δ mice are infertile and defective in retaining cohesin at centromeres after meiosis I (Llano et al., 2008). This suggests that mammalian Sgo2 and, possibly not Sgo1, fulfills the function performed by proteins called Sgo1 or Mei-S332 in yeast and flies respectively. Does hSgo2 also bind PP2A? Like hSgo1, it contains an N-terminal coiled-coil region (residues 31-130), which we found to be sufficient for PP2A interaction. In contrast to hSgo1, which interacts with the AB’C holoenzyme in vivo (Kitajima et al., 2006; Riedel et al., 2006; Tang et al., 2006) and in vitro (Figure 1) but not with individual PP2A A, B, C subunits or the AC core complex, mouse Sgo2 forms a stable complex with the PP2A’s C subunit (Figure 4A). It interacts with all three types of PP2A holoenzyme as well as with core PP2A AC complex. This demonstrates that mammalian Sgo1 and Sgo2 form distinct shugoshin-PP2A complexes and may therefore have distinct substrate specificities at centromeres.
Figure 4. Comparison and characterization of Sgo1 and Sgo2 binding with PP2A.
A. PP2A C subunit is sufficient for Sgo2, but not for Sgo1 binding. PP2A A, B’, C individual subunit, the AC core enzyme and the AB’C holoenzyme were incubated with MBP-FLAG-Sgo1(51-105) or MBP-FLAG-Sgo2(45-117) for one hour followed by FLAG affinity pull down. Bead bound proteins were eluted in a buffer containing FLAG peptide and were resolved on a SDS-PAGE gel.
B. Interaction of Sgo1 or Sgo2 with PP2A holoenzyme containing different B-type regulatory subunits. MBP-FLAG-Sgo1(51-105) or MBP-FLAG-Sgo2(45-107) was incubated with ABC, AB’C or AB”C for one hour followed by FLAG affinity pull down. Bead bound proteins were eluted and resolved on a SDS-PAGE gel. Sgo1 interacts specifically with the AB’C PP2A only, whereas Sgo2 can interact with all PP2A isoforms.
C. At least two Sgo2 molecules bind to the same PP2A complex simultaneously. MBP-Sgo2(45-117) was incubated either with equal amount of (lanes 1, 2 and 3) or without (lanes 4, 5 and 6) MBP-FLAG-Sgo2(45-117) and PP2A GST-A, B and C subunits prior to GST affinity pull down. Bead bound proteins were eluted in a buffer containing glutathione. Eluted proteins were incubated with FLAG affinity beads before elution in a buffer containing FLAG peptide. Eluates and unbound fractions were resolved on a SDS-PAGE gel.
D. Sgo1 and Sgo2 peptides can form stable heterodimers (data not shown), but they cannot bind to PP2A as stably as the homodimers. MBP-Sgo1(51-105) was incubated either with equal amount of (lanes 1, 2 and 3) or without (lanes 4, 5 and 6) MBP-FLAG-Sgo2(45-117) and PP2A GST-A, B and C subunits prior to GST affinity pull down. Bead bound proteins were eluted in a buffer containing glutathione. Eluted proteins were incubated with FLAG affinity beads before elution in a buffer containing FLAG peptide. Eluates and unbound fractions were resolved on a SDS-PAGE gel.
E. Sgo2 requires a coiled-coil region longer than that of Sgo1 for dimer formation and PP2A binding. MBP-Sgo2(45-90) and MBP-Sgo2(45-117) were tested for binding to PP2A as described in B (lanes 1-2). Dimerization abilities of these fragments were determined by testing their binding to the longer MBP-FLAG-Sgo2(31-130). After FLAG affinity pull down, bead bound proteins were eluted in a buffer containing FLAG peptide and the eluates were resolved on the SDS-PAGE gel (lanes 3-4). Individual MBP-Sgo2(45-117) and MBP-Sgo2(45-90) samples were loaded on the lanes 5 and 6, respectively.
F. Sgo1 binding does not change PP2A enzymatic activity. Phospatase activities of wild type PP2A, PP2A-Sgo1, PP2A-Sgo2, PP2A(D88N)-Sgo1 and PP2A(D88N)-Sgo2 were analyzed by a standard Ser/Thr Phosphatase Assay Kit. The reaction without PP2A was used as a negative control.
In the presence of PP2A, FLAG-tagged Sgo2 pulls down untagged Sgo2, implying that PP2A-Sgo2 complexes contain two Sgo2 molecules and a single PP2A (Figure 4C). Sgo2’s dimerization differs however from that of Sgo1. Thus, FLAG-Sgo2(31-130) pulls down Sgo2(45-117) but not Sgo2(45-90) (Figure 4E). Indeed, a failure to form dimers explains why MBP-Sgo1(51-96) pulls down PP2A (Figure 1B) while the corresponding fragment of Sgo2 (residues 45-90) cannot do so (data not shown). Sequence alignments show that L53, L54, I81 and I82 of Sgo1, which are involved in dimer formation, are replaced by small or hydrophilic residues in Sgo2, which would explain why Sgo2 dimer formation requires a longer coiled-coil. Indeed, Sgo2’s predicted coiled-coil containing periodic L/I residues extends further in a C-terminal direction than that of Sgo1. Sgo2’s tight interaction with PP2A’s C subunit might nevertheless involve only the N-terminal part of this extended coiled-coil. Finally, we found that MBP-Sgo1 forms a stable heterodimer with MBP-Sgo2 (data not shown) that binds PP2A, albeit less efficiently than homodimers (Figure 4D).
Sgo1 does not alter PP2A’s catalytic activity
We measured phosphatase activities of wild type and catalytically-inactive (D88N) PP2A in the presence and absence of purified MBP-Sgo1 or MBP-Sgo2 proteins. While the PP2A-C(D88N) mutation abolishes activity, neither Sgo1 nor Sgo2 has an effect on the dephosphorylation of peptide substrate (Figure 4F), which fits with the finding that Sgo1 does not induce a change in the conformation of PP2A’s active site. Shugoshins might nevertheless affect substrate recruitment in vivo.
The shugoshin-PP2A interaction protects centromeric cohesin in yeast
To address the physiological significance of shugoshin’s interaction with PP2A, we turned to the budding yeast S. cerevisiae whose sole shugoshin, scSgo1, is essential for protecting centromeric cohesin from separase at meiosis I (Katis et al., 2004; Marston et al., 2004). scSgo1 possesses the conserved N-terminal coiled-coil within which scN51 corresponds to hN61 (Figure S8). Mutation to alanine of three surface residues expected to contact PP2A, namely Y47, Q50, and S52 (equivalent to Y57, N60, and K62 in hSgo1) abolishes co-localization of PP2A’s B’ subunit Rts1 with centromeric Ndc10 in chromosome spreads from pachytene cells (Figure 5A). Replacement of N51 to isoleucine (N51I) has a similar effect (data not shown). Importantly, these mutations have no effect either on Sgo1’s steady state levels (i.e. stability; data not shown) or on its recruitment to centromeres (Figures 5B and S9). These findings both validate the Sgo1:PP2A crystal structure and demonstrate that interaction between PP2A and shugoshin is essential for the PP2A’s centromere localization, at least during yeast meiosis.
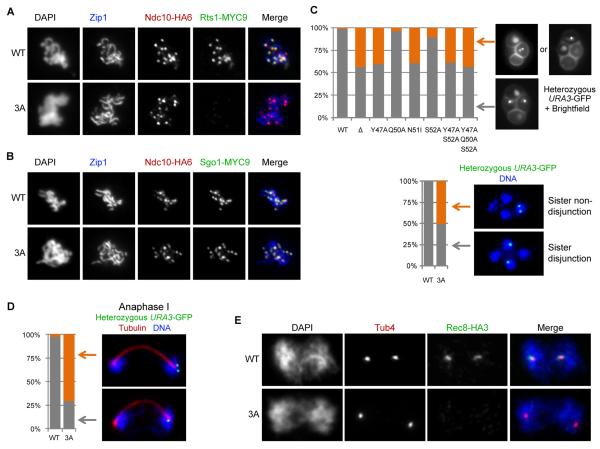
Figure 5. Sgo1-PP2A interaction is necessary for protecting centromeric cohesin.
A. Co-localization of 9xMYC epitope tagged PP2A-B’ subunit Rts1 with 6xHA-epitope tagged kinetochore protein Ndc10 on pachytene staged nuclear spreads of wild-type (WT; K16140) or sgo1-3A (3A; K16139) cells. Pachytene stage was identified by the Zip1 staining which lines the axis of synapsed chromosomes. At least 30 pachytene nuclei were scored for both strains.
B. Co-localization of 9xMYC-epitope tagged wild-type (WT; K12113) or Y47A/Q50A/S52A mutant form (3A; K16235) of scSgo1 with 6xHA-epitope tagged kinetochore protein Ndc10 on pachytene staged nuclear spreads. At least 30 pachytene nuclei were scored for both strains.
C. Diploid wild-type (WT; 16213) or mutant strains carrying the scSgo1 mutations sgo1Δ (Δ; K16214), Y47A (K16226), Q50A (K16259), N51I (K16216), S52A (K16227), Y47A/S52A (K16228), Y47A/Q50A/S52A (K16215) and heterozygous GFP-marked chromosome V at URA3 locus (URA3-GFP) were sporulated on plates and 100 tetrads produced from each strain were scored for the segregation of URA3-GFP to asci using fluorescence microscopy with brightfield illumination (upper panel). Wild-type (WT; K16213) and sgo1-3A (3A; K16215) cells were also scored after sporulation in liquid culture, formaldehyde fixation, spheroplasting and DAPI staining (lower panel).
D. Diploid wild-type (WT; K16213) and sgo1-3A (3A; K16215) cells containing heterozygous URA3-GFP as well as Pds1-myc18 and Rec8-HA3 epitope tagged proteins were sporulated in synchronous liquid cultures and samples were taken for in situ staining. For each strain 100 anaphase I staged cells, identified by two DNA masses connected with elongated spindles and low levels of Pds1 (not shown) were scored for the reductional and equational (never observed) segregation of URA3-GFP dots. Reductional segregation was further scored for precocious sister splitting.
E. Immuno-staining of anaphase I stage nuclear spreads from wild-type (WT; K16213) and sgo1-3A (3A; K16215) cells expressing Rec8-HA3 epitope tagged protein. Tub4 staining marks spindle pole bodies where split Tub4 signal with bi-lobed DNA mass indicates for anaphase I. At least 20 anaphase I spreads were scored for both strains.
Like sgo1Δ (Katis et al., 2004), sgo1 N51I and sgo1Y47A/Q50A/S52A (sgo1-3A) diploids produce inviable haploid spores while Y47A/S52A, Y47A, S52A and Q50A diploids produce spores whose viability is 8.3%, 15.8%, 90.8% and 91.3% respectively (Figure S10). To address whether the mutations cause non-disjunction of sister centromeres at meiosis II, we scored segregation within tetrads of a single chromosome V marked by Tet repressor fused to GFP bound to a tandem array of operators at the URA3 locus 35kb away from the centromere (Michaelis et al., 1997). Y47A, N51I, Y47A/S52A, and Y47A/Q50A/S52A mutations cause high rates of sister centromere (URA3) non-disjunction (Figure 5C), which presumably arises from precocious loss of sister centromere cohesion because Y47A/Q50A/S52A causes 70% of sister URA3 sequences to disjoin by the end of anaphase I (Figure 5D), unlike wild type where they only disjoin at anaphase II. None of the mutations alter the kinetics of meiosis I (data not shown) or co-segregation of sister URA3 sequences to the same pole at anaphase I (Figure 5D), implying that interaction between PP2A and scSgo1 is not required for meiosis I mono-orientation of sister kinetochores (Toth et al., 2000). To address whether the interaction is required to protect centromeric cohesin, we compared the distribution of cohesin’s Rec8 protein in chromosome spreads from wild type and triple mutant (sgo1-3A) cells. Rec8’s association with pachytene chromosomes (Figure S11) was unaffected by the mutation but its persistence at centromeres between meiotic divisions was abolished by the sgo1-3A triple mutation (Figure 5E). We conclude that protection of centromeric cohesin from separase at meiosis I depends on Sgo1’s ability to bind PP2A, a function that is possibly conserved in animal cells given the precocious loss of sister chromatid cohesion in Drosophila caused by N61I (Tang et al., 1998). Our findings extend in an important way the previous demonstration that PP2A’s Rts1 B’ subunit is required for centromeric cohesin protection (Riedel et al., 2006). PP2A has many functions during mitosis and meiosis and the effect of Rts1 inactivation on cohesin protection may have been indirect. The current data demonstrate that it is only the small pool of PP2A molecules associated with Sgo1 that is essential for cohesin protection.
Recruitment of PP2A to chromatin by Sgo1 blocks cohesin cleavage in oocytes
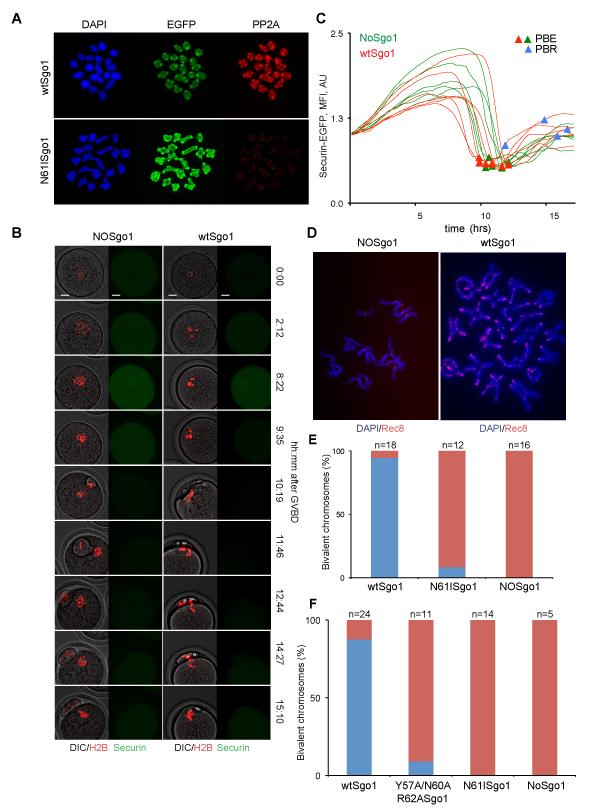
If recruitment of PP2A to centromeres were the mechanism by which shugoshins protect cohesin, then deposition of PP2A onto the arms of meiosis I bivalents should prevent their conversion to dyads. To address this, we investigated the effects of over-expressing Sgo1 in mouse oocytes. mSgo1-GFP fusion protein concentrates at centromeres after injection of modest amounts of mRNA into oocytes at the GV stage (data not shown) but decorates the arms of bivalent chromosomes after injection of larger amounts (Figure 6A). This is accompanied by a similar re-distribution of PP2A (Figure 6A), which is normally found only at centromeres (data not shown). PP2A’s artificial recruitment to the arms of bivalents is largely abolished by N61I (Figure 6A) and reduced by Y57A/N60A/R62A mutations (data not shown). Remarkably, the re-distribution of PP2A caused by mSgo1 over-expression is accompanied by a block to meiosis I chromosome segregation. The failure to segregate chromosomes is not caused by a failure to activate separase because destruction of securin-GFP protein (produced by an mRNA injected along with that of untagged mSgo1) (Figures 6B and 6C) takes place on schedule. Oocytes over-expressing mSgo1 also extrude polar bodies with normal kinetics but these are subsequently retracted (Figures 6B and C).
Figure 6. PP2A recruitment to bivalent arms block chiasmata resolution in mouse ooctes.
A. GV — stage CD1 oocytes were injected with wtSgo1-EGFP and N61ISgo1-EGFP mRNAs and matured for approximately 6 hours. Chromosome spreads were prepared from individual oocytes and stained with DAPI (blue), PP2A was detected with PP19 antibody (red) and mSgo1 was detected with EGFP (green).
B. Images from movies with CD1 strain oocytes injected with H2B-mCherry, securin-EGFP and wtSgo1 mRNAs as indicated. The merged DIC/H2B-mCherry (left) and Securin-EGFP (right) channels are shown. Bar = 10 μm.
C. Securin-EGFP levels in oocytes injected with H2B-mCherry and securin-EGFP (green) or with H2B-mCherry and securin-EGFP together with wtSgo1 (red) were measured at each timepoint. Values were normalized relative to that at GVBD (0 h), and plotted, in arbitrary units (AU), against time. The timing of polar body extrusion (PBE — green or red triangles) or retraction (PBR — blue triangles) is indicated.
D. Oocytes carrying Rec8-myc transgene were injected with H2B-mCherry mRNA only or with H2B-mCherry together with wtSgo1 mRNA. Live imaging was used to monitor a meiotic progression for approximately 18 hours and only oocytes, which extruded the polar body, were process for DNA spreads. Chromosome spreads were stained with DAPI (blue) to visualize DNA and with anti-myc antibody (red) to detect Rec8.
E. GV — stage CD1 oocytes were injected with H2B-mCherry and with wtSgo1 and N61ISgo1 mRNAs and monitored by live imaging for 18 hours. Chromosome spreads were prepared only from oocytes, which extruded polar body. Spreads were stained with DAPI and CREST and scored for the presence of bivalent chromosomes. Blue columns represent a percentage of cells containing at least 1 bivalent chromosome; red columns represent a percentage of cells containing only univalent chromosomes. Data were obtained from 4 independent experiments; numbers of cells in each group is indicated.
F. CD1 oocytes were injected with H2B-mCherry mRNA and also with wtSgo1-EGFP, N61ISgo1-EGFP and Y57A/N60A/R62ASgo1-EGFP mRNAs. Meiotic maturation and chromosome segregation was monitored by live imaging and after 18 hours oocytes, which extruded polar bodies, were assayed by Z stacks covering the entire cell volume. After 3D reconstruction cells were scored for presence of bivalent chromosomes. Blue columns represent a percentage of cells containing at least 1 bivalent chromosome; red columns represent a percentage of cells containing only univalent chromosomes. Data were collected from 6 experiments and the numbers of cells are indicated.
Chromosome spreads from MII oocytes reveal that Sgo1 over-expression blocks resolution of chiasmata and removal of cohesin’s Rec8 subunit from chromosome arms (Figures 6D and E). Crucially, N61I and Y57A/N60A/R62A mutations largely abolish mSgo1’ ability to block the chiasmata resolution (Figures 6E and 6F). These data suggest that recruitment of PP2A to chromosome arms may be sufficient to block Rec8 cleavage. It is likely that centromeric Rec8 in mouse oocytes is normally protected from separase not by mSgo1 but by mSgo2 (Llano et al., 2008). We suggest that mSgo2 performs this function, like scSgo1, by recruiting PP2A. If this proves to be case, then our experiments imply that PP2A recruitment would be sufficient to protect centromeric cohesin. No other function might be required.
The spindle assembly checkpoint in yeast requires binding of PP2A to shugoshin
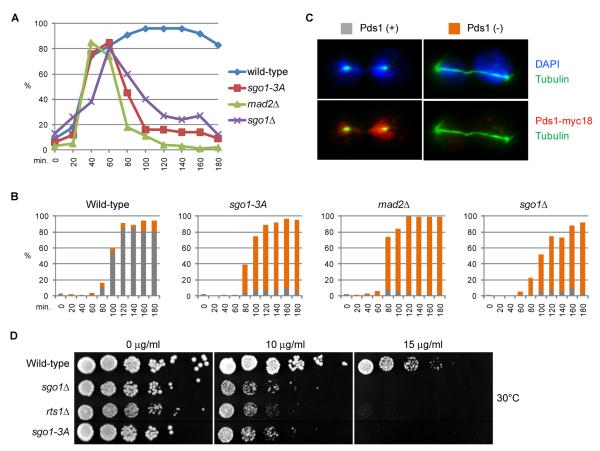
In budding yeast, which expresses only a single shugoshin, scSgo1 also has important functions in mitotic cells, one of which is to delay activation of the anaphase-promoting complex/cyclosome (APC/C) when there is a lack of tension at the inter-connection between microtubules and kinetochores (Indjeian et al., 2005). When cells with a temperature sensitive cdc15-2 mutation are released from G1 arrest at the restrictive temperature, they undergo DNA replication, securin (Pds1) destruction, and chromosome segregation but fail to exit mitosis due to a defect in the mitotic exit network (MEN) (Bardin et al., 2003). Securin destruction and stable spindle elongation does not take place in scc1-73 cdc15-2 double mutant cells, which cannot form sister chromatid cohesion (Figure 7A-C, (Severin et al., 2001)). Both phenotypes are presumably due to a spindle assembly checkpoint (SAC) induced APC/C activation block as they are suppressed by deleting MAD2 (Fig. 7A-C). Importantly, sgo1-3A as well as sgo1Δ (Fig. 7A-C) also suppresses the spindle instability and the lack of securin destruction in scc1-73 cdc15-2 cells. Binding of PP2A to Sgo1 is therefore required for inhibition of the APC/C by the SAC in response to a lack of cohesion. Consistent with this finding, sgo1-3A and RTS1 deletion cells are hypersensitive to the microtubule-destabilizing drug benomyl (Figure 7D).
Figure 7. Sgo1-PP2A interaction is required for spindle assembly check point activation in yeast.
(A-C) Haploid MATa strains carrying mutations cdc15-2, scc1-73 (wild-type; K16260), cdc15-2, scc1-73, sgo1-3A (sgo1-3A; K16261), cdc15-2, scc1-73, mad2Δ (mad2Δ; K16262), cdc15-2, scc1-73, sgo1Δ (sgo1Δ; K16315), were synchronized at G1 stage by α-factor arrest at 24°C and released into non-permissive temperature at 35.5°C. Samples of the cultures were taken at indicated time points and analyzed by FACS for cellular DNA content (Fig. S12), and by in situ immunostaining of α-tubulin and 18xMYC-epitope tagged Pds1. (A) Fraction of Pds1-myc18 positive cells. (B) Fraction of bi-nucleates that are Pds1 positive (green bar) or Pds1 negative (red bar). (C) Representative images for (B). (D) Wild-type (K15785), sgo1Δ (K15784), rts1Δ (K14165) and sgo1-3A (K15789) cells were tested for growth at indicated concentrations of benomyl at 30°C.
DISCUSSION
The myriad functions performed by PP2A, or any other phosphatase for that matter, poses a major problem: how to regulate its activity against particular substrates? Protein phosphorylation is determined by a delicate balance between kinases and phosphatases and changes in substrate phosphorylation cannot be regulated merely by raising or lowering activity of kinases and phosphatases throughout the cell. A capacity to regulate differentially specific phosphatase subpopulations that target specific substrates in specific locations would therefore be a desirable property. It was hitherto thought that this sort of specificity was conferred largely by PP2A’s regulatory B subunits, of which there are several types (B, B’, B”, B”’). However, the finding that a fraction of AB’C PP2A holoenzymes associate with Shugoshin/Mei-S332 type proteins that are concentrated mainly at centromeres (and centrosomes) raises the prospect of far greater specificity. Our structural and biochemical data show that shugoshin is an adaptor protein, which does not affect PP2A enzymatic activity but instead determines the specific PP2A form(s) to be recruited, and thus determines the PP2A substrate specificity at centromeres. In contrast to the conventional view that the targeting/regulatory B subunit is responsible for PP2A localization, the catalytic C subunit of PP2A is the primary docking site for shugoshin. Our work therefore provides a paradigm according to which PP2A targets substrates via an adaptor protein that concentrates the enzyme at a particular location in the cell.
Structural basis of the PP2A-Shugoshin interaction: the importance of Sgo dimerization
A key feature of the Sgo1-PP2A interaction is that the both strands of Sgo1’s coiled-coil interact with both catalytic C and regulatory B subunits of the same PP2A holoenzyme. This means that formation of Sgo1’s coiled-coil homodimer is essential for its interaction with PP2A, a conclusion confirmed by the effects of the L68A mutation, which disrupts formation of the coiled-coil dimer and abolishes the PP2A-Sgo1 interaction (Figures 3A, 3D and 3E). Indeed, the dramatic difference between the PP2A binding affinities of hSgo1(51-96) and hSgo1(51-103) (Figure S1) can be explained not by differences in the number of their contacts with PP2A but rather by the fact that the latter is predicted to form two extra helical turns, creating thereby a more stable Sgo1 homodimer. Likewise, mSgo2(45-107) forms a stable homodimer that interacts with PP2A while mSgo2(45-90), which shares the identical N-terminal half that is most likely responsible for interacting with PP2A’s C subunit, cannot form a homodimer and does not therefore interact with PP2A.
mSgo2 forms complexes with multiple types of PP2A holoenzyme
mSgo2 also interacts with PP2A using its coiled-coil domain. That it does so in a manner similar to hSgo1 is suggested by sequence conservation and the finding that hSgo1 and mSgo2 form heterodimers capable of interacting with PP2A, at least in vitro. In contrast to hSgo1, which only interacts with the AB’C family trimeric holoenzymes, hSgo2 can form stable complexes with PP2A’s catalytic C subunit in the absence of A and B subunits. As a consequence, hSgo2 interacts with AC core complexes and with trimeric holoenzymes containing a variety of B subunits. Indeed, hSgo2’s coiled-coil appears to bind more tightly with ABC and AB”C holoenzymes than with the AB’C holoenzymes. Because regulatory B subunits have an important role in substrate specificity (Cho and Xu, 2007; Janssens and Goris, 2001; Lechward et al., 2001; Sontag, 2001; Xu et al., 2006; Yu, 2007), it is therefore conceivable that PP2A recruited to centromeres by hSgo2 might regulate phosphorylation of a different set of substrates to those regulated by hSgo1. This could explain how different types of shugoshin have different functions despite having similar cellular locations. In mitotic mammalian cells, for example, Sgo1 and Sgo2 are both concentrated at centromeres but only the former is necessary to protect cohesin from the prophase dissociation pathway (Llano et al., 2008; McGuinness et al., 2005). Likewise, in fission yeast, both spSgo1 and spSgo2 are associated with centromeres during meiosis I but only the former is required to protect cohesin from separase at the onset of anaphase I (Rabitsch et al., 2004; Riedel et al., 2006; Vanoosthuyse et al., 2007; Vaur et al., 2005). Thus, the finding that PP2A is still present at centromeres after depletion of hSgo1 in Hela cells (Lee et al., 2008) does not necessarily imply that hSgo1 does not protect centromeric cohesin by recruiting PP2A because the PP2A remaining at centromeres in the depleted cells may have different properties to PP2A putatively associated with hSgo1. Whether or not binding of PP2A to hSgo1 is necessary to protect centromeric cohesin from the prophase pathway remains therefore an open question. Importantly, our crystal structure enables this to be addressed in a rigorous manner by testing the phenotype of mutations that abolish specifically PP2A binding. The sequences flanking the coiled-coil regions of hSgo1 and hSgo2 are predicted to be unstructured (Figure S7). This potentially flexible region is considerably larger in hSgo2, which might therefore bind to a different set of centromeric proteins, further differentiating its activity from that of hSgo1.
Recruitment of PP2A to centromeres by shugoshin protects cohesin from separase
A key finding made possible by the crystal structure of the hSgo1-PP2A complex is that mutation (within yeast Sgo1) of key residues involved in PP2A binding abolishes recruitment of the Rts1 AB’C holoenzyme to centromeres, causes the precocious loss of cohesin from this location at the onset of anaphase I and massive nondisjunction at meiosis II. Mutation of the highly conserved asparagine (equivalent to N61 in humans) has a similar phenotype in yeast (this work) and in flies (Tang et al., 1998). In this case, however, the mutation may affect more than just PP2A binding as it probably alters the coiled-coil’s conformation. Our observations extend the previous finding that inactivation of Rts1 causes precocious loss of sister centromere cohesion (Riedel et al., 2006). Crucially, we now know that it is the small population of PP2A recruited to centromeres by scSgo1 that is responsible for protecting centromeric cohesin and not merely global AB’C holoenzyme activity.
How might PP2A associated with scSgo1 protect centromeric cohesin? More specifically, what is the process whose deregulation causes precocious centromeric cohesin loss when Sgo1 cannot bind PP2A? There are two possibilities: cleavage of Rec8 by separase or cleavage- independent dissociation. The finding that non-cleavable Rec8 blocks both meiotic divisions (Buonomo et al., 2000; Kitajima et al., 2003) suggests that cleavage-independent mechanisms analogous to the mitotic prophase pathway are not capable of destroying cohesion. PP2A must therefore regulate Rec8 cleavage. It could do this either by regulating separase or its target Rec8. The finding that cleavage of Rec8’s mitotic counterpart Scc1 cannot be protected by Sgo1 when expressed in meiotic cells (Toth et al., 2000), the recent identification of Rec8 phosphorylation sites essential for its cleavage along chromosome arms at meiosis I, and the finding that phospho-mimicking Rec8 mutants cause precocious loss of sister centromere cohesion (Katis and Nasmyth, unpublished observations) suggests that PP2A targets Rec8 and not separase. We suggest that PP2A’s preferential accumulation at centromeres leads to de-phosphorylation of centromeric Rec8, which cannot therefore be cleaved by separase. Our finding that over-expression of hSgo1 induces the recruitment of PP2A to the arms of bivalent chromosomes in oocytes and that this is accompanied by a block to chiasmata resolution indicates that the above model applies to meiosis in mammals as well as yeast.
Role of PP2A in the spindle assembly checkpoint
Our finding that sgo1-3A mutants are defective in blocking APC/C activation in cells lacking sister chromatid cohesion suggests that binding of PP2A to Sgo1 has a crucial role in the “tension-dependent” spindle assembly checkpoint (SAC). Previous work has implicated PP2A’s B regulatory subunit (Cdc55) in the SAC (Wang and Burke, 1997; Tang and Wang, 2006). However, cdc55 mutants in yeast have rather pleiotropic effects on the cell cycle and it has never been clear how direct a role PP2A has in the SAC. Our finding that both sgo1-3A and rts1 (PP2A B’) mutants are similarly sensitive to benomyl together with the observation that Sgo1 has hitherto only been co-purified with its B’ (Rts1) regulatory subunit (albeit so far only in meiotic yeast cells) indicates that Sgo1’s SAC function is mediated by AB’C PP2A and not the ABC form. Nevertheless, it is conceivable that Sgo1 might also bind ABC PP2A in vivo in mitotic cells, a possibility strengthened by our finding that human Sgo2 binds the ABC as well as the AB’C form. PP2A’s mechanistic role in the SAC remains enigmatic; that is, what are its substrates? Interestingly, orderly homolog disjunction during meiosis I, which is dependent on Mad2 (Shonn et al., 2003), is not affected by the sgo1-3A mutation. This implies that the SAC’s mechanism differs between mitosis, where Sgo1’s association with PP2A may be important, and meiosis I, where it is probably not.
Implications of the structure for Shugoshin and PP2A localization
Shugoshins recruit PP2A to chromosomes, both in yeast and in mouse oocytes. Might PP2A in turn facilitate the chromosomal association of shugoshins? We find no evidence that this is the case during yeast meiosis, where mutations that abolish PP2A binding have little or no effect on scSgo1’s accumulation at centromeres. However, the finding that N61I abolishes hSgo1’s accumulation at centromeres in Hela cells raises the possibility that PP2A bound to Sgo1 might facilitate Sgo1’s association with centromeres in mitotic cells (Tang et al., 2006). Whether this is indeed the case will require analysis of mutations that affect residues specifically involved in PP2A interaction. It is certainly not inconceivable because at least two different mitotic kinases have been implicated in reducing hSgo1’s association with chromatin. Phosphorylation of Mei-S332 by Polo Kinase has been implicated in dissociation from centromeres during anaphase (Clarke et al., 2005) while inhibition of Aurora B leads to increased association of hSgo1 with chromosome arms (Kueng et al., 2006; Lipp et al., 2007). In Drosophila, the chromosome passenger complex and Aurora B phosphorylation are needed for proper localization of MEI-S332 to the centromere as opposed to the chromosome arms (Resnick et al., 2006). If phosphorylation of shugoshins does indeed reduce its affinity for chromatin, then hSgo1’s interaction with PP2A would plausibly counteract this process and if the dephosphorylation of hSgo1 were largely mediated by PP2A associated with a separate hSgo1 molecule, then the interaction between hSgo1 and PP2A would tend to “sharpen” differences between arms and centromeres in the concentration of hSgo1. Our discovery that shugoshins only bind to PP2A as dimers might contribute to such a sharpening process because as hSgo1 levels drop along chromosome arms, dimer formation might decrease, leading to less PP2A binding and thereby to yet less hSgo1. In addition, if the two tails of shugoshin dimers bind to different but neighboring chromatin domains, then it is conceivable that stretching, either within or between chromatids, could lead to disruption of the coiled-coil and thereby to dissociation of PP2A. Whether or not dynamical and mechanical processes of this nature contribute to the formation of shugoshin and PP2A concentration gradients along chromosomes can now be tested using insights into the mechanism by which shugoshins bind PP2A.
Our work demonstrates that shugoshin’s most conserved domain forms a coiled-coil that is both necessary and sufficient for PP2A binding. Because disruption of this coiled-coil structure abolishes PP2A binding, it is conceivable that it might be used to convert mechanical tension on chromosomes (Indjeian et al., 2005) into a change in the local concentration of PP2A, thereby changing the susceptibility of cohesin to cleavage by separase, also turning off the SAC. Whether this is indeed the case and if so how awaits future experiments.
Experimental procedures
Data collection and structure determination
Details of protein preparation, crystallization and the phosphatase assay, are described in Supplementary data. There was one complex per asymmetric unit with a solvent content of approximately 65 %. A 2.7 Å data set was collected at ALS beamline 5.0.2. These data sets were integrated and scaled using HKL2000 (Otwinowski and Minor, 1997). Statistics for the collected data are summarized in Table 1. The structure was solved by molecular replacement using a molecule of PP2A holoenzyme (PDB ID code 2IAE) (Cho and Xu, 2007) as a search model. After rigid-body refinement, model building and refinement was performed using COOT (Emsley and Cowtan, 2004) and Refmac restrained refinement in CCP4 package (Murshudov et al., 1997), respectively. The electron density maps from (2Fo —Fc) and (Fo —Fc) calculations were used for model building. The final model contained residues 7-589 of the PP2A A subunit, 26-426 of the PP2A B’ subunit, 2-309 of the PP2A C subunit and 51-96 of Sgo1, with a working R factor of 22.8% and a free R factor of 27.7%. In the Ramachandran plot, 91.5%, 8.5% and 0% of all residues in PP2A-Sgo1 complex fall within most favored, allowed and disallowed regions, respectively.
PP2A-Shugoshin in vitro binding assay
In all in vitro binding experiments highly purified PP2A and shugoshin proteins were used. To test the binding of PP2A to various Sgo1 mutants, 2 μg of GST tagged PP2A A was incubated with extra amount of PP2A B’, C subunits and MBP-Sgo1 fusion protein for one hour on ice. Following the incubation, GST affinity beads were added to the mixture for GST-A pull down. Washed beads were boiled, subjected to SDS-PAGE analysis using Coomassie Blue staining. To investigate whether more than one Sgo1 molecule bind to the same PP2A complex , GST tagged PP2A-A was incubated with excess of PP2A B’ and C subunits, and Sgo1 orSgo2 peptides or MBP fusions with or without a FLAG-tag. The protein mixture was incubated with GST affinity beads for one hour at 4 °C. After washing 3 times with binding buffer (150mM NaCl 20mM Tris, pH=7.5), proteins were eluted with the binding buffer containing 20mM glutathione. Eluates were then incubated with FLAG affinity beads for one hour at 4 °C. Afterwards, bead bound proteins were eluted using FLAG peptide (Sigma) and subjected to SDS-PAGE analysis using Coomassie Blue staining. Other FLAG or GST tagged pull down experiments were done using the same strategy.
Yeast strains, growth and sporulation conditions
The yeast strains used in this study are listed in Table S1. Sgo1 mutant strains were generated by integrating PCR-based mutagenized SGO1 open reading frames cloned in pAG32 plasmids (Goldstein and McCusker, 1999) into endogenous SGO1 locus in sgo1Δ strains that have been published previously (Katis et al., 2004). Sporulation was performed at 30°C as described before (Buonomo et al., 2000).
Immunofluorescence staining of yeast cells and nuclear spreads
In situ immunofluorescence staining of whole cells and surface spread nuclei were performed as described in (Katis et al., 2004). DNA was visualized using 4′,6-diamidino-2-phenylindole (DAPI). Images were taken using the Zeiss Axio fluorescent microscope with appropriate filter sets that is attached to a Coolsnap HQ2 CCD camera and analyzed using Metamorph Software (Molecular Devices).
Mouse Strains
GV oocytes used in this study were isolated from CD1 strain (Harlan, UK). A transgenic line (C57BL/6J) that expresses Rec8 from a bacterial artificial chromosome (BAC), with nine tandem copies of the human c-myc epitope at its C terminus was described previously (Kudo et al., 2006). Mice were housed in animal facilities at the University of Oxford and all procedures were approved by local Ethical Review Committees and licensed by the Home Office under the Animal (Scientific Procedures) Act 1986.
Oocyte culture, microinjection and live imaging
Techniques used for oocyte culture, micromanipulation and microinjection were described previously (Kudo et al., 2006; McGuinness et al., 2009). Zeiss LSM510 META confocal microscope equipped with incubator, maintaining stable temperature and CO2 and with Plan-Neofluor 20×/0.5, C-Apochromat 40×/1.2 NA W Corr and C-Apochromat 63×/1.2 NA W Corr lenses, was used for live imaging. For detection of Cascade-Blue® dextran, EGFP and mCheery signal we used 405 nm, 488 nm and 561 nm excitation and LP 420, BP 505-550 and LP 575 filters. Chromosomes were tracked using H2B-mCherry signal and EMBL-developed tracking software (Rabut and Ellenberg, 2004) adapted to our microscope by Annelie Wuensche (EMBL Heidelberg). Image stacks were captured every 5-15 min for 16-20 h. Quantification was performed using ImageJ software (http://rsb.info.nih.gov/ij/). The area for measuring securin—EGFP signal was defined manually in each frame, and the mean fluorescence intensity of the signal was normalized to the value at the time of GVBD.
Supplementary Material
Acknowledgements
We thank Dr. Zhiyi Wei for help with crystallographic computation, Dr. Julia Zhu for providing purified active PP2A samples, Dr. Vittorio L. Katis for his advice on yeast meiotic cultures and Dr. Andrew Murray for his advice about using cdc15 mutants. We also appreciate the support from the staff at ALS beamlines 5.0.2 and 8.2.2. This work was supported by NIH grant CA 129509 to W.X.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers Coordinates and structure factors have been deposited in the Protein Data Bank (PDB; http://www.rcsb.org/pdb) under ID code 3FGA.
References
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Boselli MG, Amon A. Mitotic exit regulation through distinct domains within the protein kinase Cdc15. Mol Cell Biol. 2003;23:5018–5030. doi: 10.1128/MCB.23.14.5018-5030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- Clarke AS, Tang TT, Ooi DL, Orr-Weaver TL. POLO kinase regulates the Drosophila centromere cohesion protein MEI-S332. Dev Cell. 2005;8:53–64. doi: 10.1016/j.devcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Goldstein LS. Mechanisms of chromosome orientation revealed by two meiotic mutants in Drosophila melanogaster. Chromosoma. 1980;78:79–111. doi: 10.1007/BF00291909. [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GK, Yen TJ. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Miyazaki Y, Yamamoto M, Watanabe Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. Embo J. 2003;22:5643–5653. doi: 10.1093/emboj/cdg527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Kudo NR, Wassmann K, Anger M, Schuh M, Wirth KG, Xu H, Helmhart W, Kudo H, McKay M, Maro B, et al. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 2006;126:135–146. doi: 10.1016/j.cell.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Lechward K, Awotunde OS, Swiatek W, Muszynska G. Protein phosphatase 2A: variety of forms and diversity of functions. Acta Biochim Pol. 2001;48:921–933. [PubMed] [Google Scholar]
- Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- Lipp JJ, Hirota T, Poser I, Peters JM. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J Cell Sci. 2007;120:1245–1255. doi: 10.1242/jcs.03425. [DOI] [PubMed] [Google Scholar]
- Llano E, Gomez R, Gutierrez-Caballero C, Herran Y, Sanchez-Martin M, Vazquez-Quinones L, Hernandez T, de Alava E, Cuadrado A, Barbero JL, et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 2008;22:2400–2413. doi: 10.1101/gad.475308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Anger M, Kouznetsova A, Gil-Bernabe AM, Helmhart W, Kudo NR, Wuensche A, Taylor S, Hoog C, Novak B, Nasmyth K. Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr Biol. 2009;19:369–380. doi: 10.1016/j.cub.2009.01.064. [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Vol 276. Academic Press; New York: 1997. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Rabut G, Ellenberg J. Automatic real-time three-dimensional cell tracking by fluorescence microscopy. J Microsc. 2004;216:131–137. doi: 10.1111/j.0022-2720.2004.01404.x. [DOI] [PubMed] [Google Scholar]
- Resnick TD, Satinover DL, MacIsaac F, Stukenberg PT, Earnshaw WC, Orr-Weaver TL, Carmena M. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev Cell. 2006;11:57–68. doi: 10.1016/j.devcel.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Severin F, Hyman AA, Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J Cell Biol. 2001;155:711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn MA, Murray AL, Murray AW. Spindle checkpoint component Mad2 contributes to biorientation of homologous chromosomes. Curr Biol. 2003;13:1979–1984. doi: 10.1016/j.cub.2003.10.057. [DOI] [PubMed] [Google Scholar]
- Sontag E. Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell Signal. 2001;13:7–16. doi: 10.1016/s0898-6568(00)00123-6. [DOI] [PubMed] [Google Scholar]
- Tang TT, Bickel SE, Young LM, Orr-Weaver TL. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Wang Y. Pds1/Esp1-dependent and -independent sister chromatid separation in mutants defective for protein phosphatase 2A. Proc Natl Acad Sci U S A. 2006;103:16290–16295. doi: 10.1073/pnas.0607856103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, Prykhozhij S, Hardwick KG. Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis. Mol Biol Cell. 2007;18:1657–1669. doi: 10.1091/mbc.E06-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaur S, Cubizolles F, Plane G, Genier S, Rabitsch PK, Gregan J, Nasmyth K, Vanoosthuyse V, Hardwick KG, Javerzat JP. Control of Shugoshin function during fission-yeast meiosis. Curr Biol. 2005;15:2263–2270. doi: 10.1016/j.cub.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Vogt E, Kirsch-Volders M, Parry J, Eichenlaub-Ritter U. Spindle formation, chromosome segregation and the spindle checkpoint in mammalian oocytes and susceptibility to meiotic error. Mutat Res. 2008;651:14–29. doi: 10.1016/j.mrgentox.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Wang Y, Burke DJ. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:620–626. doi: 10.1128/mcb.17.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Chen Y, Zhang P, Jeffrey PD, Shi Y. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol Cell. 2008;31:873–885. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E, Yu JW, Strack S, Jeffrey PD, Shi Y. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- Yin S, Ai JS, Shi LH, Wei L, Yuan J, Ouyang YC, Hou Y, Chen DY, Schatten H, Sun QY. Shugoshin1 may play important roles in separation of homologous chromosomes and sister chromatids during mouse oocyte meiosis. PLoS ONE. 2008;3:e3516. doi: 10.1371/journal.pone.0003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. PP2A as a mercenary for warring kinases in the egg. Proc Natl Acad Sci U S A. 2007;104:17245–17246. doi: 10.1073/pnas.0708582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.