Abstract
High-speed atomic force microscopy (HS-AFM) is becoming a reference tool for the study of dynamic biological processes. The spatial and time resolutions of HS-AFM are on the order of nanometers and milliseconds, respectively, and allow structural and functional characterization of biological processes at the single-molecule level. In this work we present contact-mode HS-AFM movies of purple membranes containing two-dimensional arrays of bacteriorhodopsin (bR). In high-resolution movies acquired at a 100 ms frame acquisition time, the substructure on individual bR trimers was visualized. In regions in between different bR arrays, dynamic topographies were observed and interpreted as motion of the bR trimers. Similarly, motion of bR monomers in the vicinity of lattice defects in the purple membrane was observed. Our findings indicate that the bR arrays are in a mobile association-dissociation equilibrium. HS-AFM on membranes provides novel perspectives for analyzing the membrane diffusion processes of nonlabeled molecules.
Introduction
The atomic force microscope is a powerful tool for the characterization of biological samples. It can provide high-resolution images of biological molecules, such as DNA complexes (1) or membrane proteins (2,3), and enables imaging in physiological buffers at room temperature and under atmospheric pressure (4) with an outstanding signal/noise ratio (5). Because of these features, atomic force microscopy (AFM) is believed to have great potential for observing the dynamics of individual biological molecules. Unfortunately, however, the slowness of image acquisition (one to several minutes per image) by conventional AFM has hampered that application. To observe the dynamics of biological molecules, image acquisition times of ≤1 s must be attained. Therefore, a new generation of high-speed (HS) atomic force microscopes needed to be developed. The main limiting factor of the imaging rate of AFM is the response speed of the moving components of the atomic force microscope, i.e., the probe and the scanner. The response speed of these components is a function of the inverse of the square root of their mass (). Therefore, to increase the imaging rate of AFM, the use of lighter probes and scanners was required, and consequently the control electronics and optical cantilever detection path of the atomic force microscope had to be adapted. Since about 2000, HS-AFM has been developed by three groups, known as the Hansma (6,7), Miles (8,9), and Ando (10,11) groups. Because of its fast feedback loop capacities, the Ando type of HS-AFM proved to be particularly adequate for observing single biomolecules at work (12,13).
In conventional AFM and HS-AFM systems, precise control of the force applied by the probe on the sample is crucial for successfully imaging biological samples. It has been observed (in conventional AFM) that applied forces higher than ∼300 pN applied during the imaging of soft biological samples typically distort or damage the samples (14). In a good case, when the force can be precisely controlled to minimal values (50–100 pN), a resolution high enough to observe details on individual biological molecules with dimensions of ∼1 nm can be achieved (5). In conventional AFM, two strategies have been used to image biological molecules at high resolution with controlled minimal force: 1), in contact mode, soft probes of low spring constant (0.01–0.1 N/m) are used that provide a high deflection/force ratio (16–19); and 2), in oscillating mode, operation probes of ∼0.1 N/m are used and oscillated, reducing the friction between the probe and the sample (20–22). In conventional AFM, the contact mode is more commonly used for high-resolution imaging, probably because the surface contact time is ∼9 times larger (at the same scan rate) compared to the oscillating mode, where only the lower part of the sinusoidal tip movement makes contact with the sample. So far, HS-AFM imaging of biological molecules at high resolution has only been achieved in oscillating mode, probably because the mass-reduced probes in HS-AFM have spring constants of ∼0.1 N/m, and because intuitively the lateral forces due to friction and the feedback loop delay may increase during fast scanning.
In this work we show that by carefully controlling the applied force, it is possible to obtain high-resolution images of membrane proteins in contact-mode HS-AFM. We show movies of individual bacteriorhodopsin (bR) molecules at a 100 ms frame acquisition time. These movies reveal the structural and positional stability of bR inside the bR lattices, and molecule diffusion events at the bR lattice edges. We demonstrate the power of HS-AFM for directly visualizing the membrane diffusion processes of nonlabeled molecules.
Materials and Methods
Sample preparation
Purple membranes (PMs) from Halobacterium salinarum containing wild-type bR were isolated according to Oesterhelt and Stoeckenius (23). PM patches were deposited on freshly cleaved mica, prepared as previously described (24). To check the cleavage quality, the mica was first imaged in 50 μL of an adsorption buffer containing 10 mM Tris-HCl (pH 7.6), 150 mM KCl, 25 mM MgCl2. Subsequently, 3 μL of PM solution were injected into the adsorption buffer drop on the mica surface. After 15 min the sample was rinsed with 10 volumes of a recording buffer containing 10 mM Tris-HCl (pH 7.6), 150 mM KCl.
HS-AFM
The bR molecules were observed in contact mode using Ando-type HS-AFM (10). A high-sensitivity photodetector was implemented and the photodetector signal was low-pass filtered (pass ≤ 5 MHz). Probes designed for HS-AFM (Olympus, Tokyo, Japan) were used with a spring constant of 0.1–0.2 N/m, a resonance frequency of 0.8–1.2 MHz, and a quality factor of ∼2 in solution (25). In contrast to the usual operation of Ando-type HS-AFM in oscillating mode, the probes did not feature any additional electron beam deposition (EBD) tip at the end of the microfabricated pyramidal tip (26). The sensitivity of the AFM system to probe deflection was 0.1 V/nm. The scan areas were 660 × 660 and 1136 × 1136 (Å)2, and the number of pixels per frame was 200 × 200. The imaging rate was 10 frames/s.
Data analysis
Individual frames were flattened and the frame shift was corrected by cross correlation for movie assembly using the WSxM software (Nanotec, Madrid, Spain) (27). All image analysis was performed using self-written routines for the ImageJ image processing package (5,28,29). Averaging of aligned molecule images was achieved by addition and normalization of pixels in the image matrix according to , where N is the number of molecules averaged, and xij stands for the gray value of pixel i in particle image j. Concomitantly, a standard deviation (SD) map was calculated according to the following equation: .
Results
High-resolution contact-mode HS-AFM was performed on the extracellular surface of native PMs containing two-dimensional (2D) arrays of trimeric bR, a light-driven proton pump of the seven-transmembrane-helix protein family (e.g., mammalian rhodopsin and GPCRs). Image acquisition was performed at a 100 ms frame frequency on a 660 Å square field scanned at a 200 pixel edge width (Fig. 1 A). Fig. 1 shows fifty consecutive image frames (5 s of a longer movie; see Movie S1 in the Supporting Material) that reveal similar image quality, and in which the individual bR molecules are resolved. We were able to perform contact-mode HS-AFM imaging at minimal loading forces (∼50 pN); therefore, imaging was nondestructive to the membrane during long-term experiments. At 100 ms imaging frequency and a scan size of 660 Å at a sampling of 200 pixels, the AFM probes each pixel, (3.3 Å)2 in size, during only 2.5 μs. This is faster by ∼2 and 3 orders of magnitude compared to conventional contact-mode or oscillating-mode imaging, respectively. The load per area and time is consequently also diminished by such a factor, rendering the acquisition less invasive. Individual frames (Fig. 1 B, top panel) reveal imaging quality comparable to that of conventional slow contact-mode (30) and oscillating-mode (21) images of the extracellular surface of bR. As can be readily seen in raw data topographs, averaging and concomitant calculation of SD maps revealed the trimeric architecture of the extracellular surface of the bR (Fig. 1 C, top panel). Despite the fast sampling rate on each pixel, the height and SD values are similar to those acquired in low-speed high-resolution imaging (31). Similarly, the majority of the molecules found in the image have a cross-correlation value (CCV (5)) between 0.6 and 0.74, with a noise-free ensemble average used as reference (Fig. 1 D, top panel). The symmetry of the trimers is also nicely preserved, with the majority of the molecules revealing an internal symmetry (IS) (5) between 0.7 and 0.8 (Fig. 1 E, top panel). To learn more about the influence of the scan and sampling rates, we merged two, five, and 10 subsequent images in the movie, resulting in image (and pixel) sampling times of 200 ms (5 μs), 500 ms (12.5 μs), and 1 s (25 μs), respectively (Fig. 1 B, bottom three panels). The corresponding CCV (Fig. 1 C, bottom three panels) and IS (Fig. 1 D, bottom three panels) analysis of the molecules revealed that both characteristics increased with longer acquisition times toward a CCV and IS of 0.8. From this, we learn that our image frames at the 100 ms rate contain some minor noise, which we may learn to eliminate in the future. Nevertheless, the best molecules in the 100 ms acquisition time images revealed a CCV of 0.75 and IS of 0.8, indicating the feasibility of high-speed, low-noise image acquisition. Whatever the sampling rate and the quantity of noise present in the images, the averages all show the same features, and we estimate that all averages have the same lateral resolution of ∼20 Å, using our stringent resolution criterion (5). Indeed, the averages show the same features as averages presented by subnanometer resolution contact-mode and oscillating-mode imaging (21,30). Another aspect that merits close examination is the image stability. Distortions may be introduced on the bR lattice, for three major reasons: 1), piezo drift, creep, and imperfections can distort the entire image; 2), lattice disruption due to the scan mechanism can alter the distance between the molecules; and 3), real protein movement may also result in distance changes between molecules. To analyze this, we performed a pair correlation function (PCF) analysis of five images (Fig. 1 F). PCF analysis graphs the probability of finding another molecule as a function of the distance within an image; it is therefore sensitive to short- and long-range variations between molecules. In Fig. 1 F we plot the average (black line) and SD (gray lines) of the intermolecular distances found. In all analyzed images, a sharp high-probability peak appears at 63 Å, corresponding to the nearest neighbors in the bR lattice (a = b = 62.5 Å, γ = 120° (32)); also, the long-distance peaks (multiples of the lattice) are well preserved. This analysis illustrates the stability of the image acquisition, corroborates the preservation of the bR lattice under the scan conditions employed here, and affirms the high lateral stability of the bR molecules within their 2D array. Finally, we also analyzed the IS of the average (n = 46) of frame 038 (shown at the top of Fig. 1 B), which peaks at 0.86 for threefold symmetry, indicating a minor tip asymmetry (Fig. 1 G).
Figure 1.
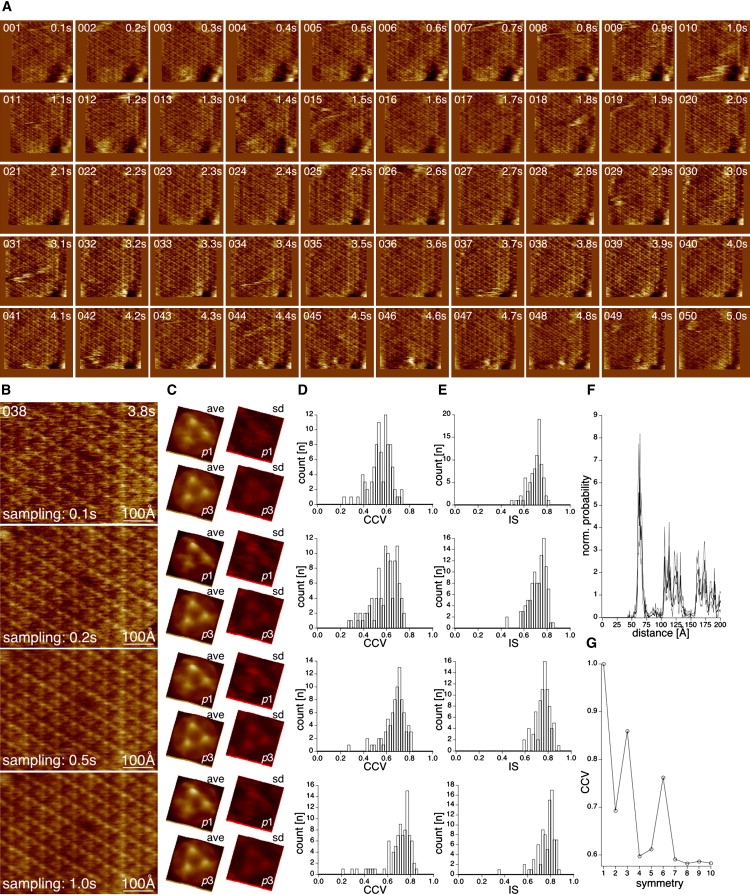
High-resolution contact-mode HS-AFM of the extracellular surface of individual bacteriorhodopsin (bR) molecules. (A) Fifty aligned images (660 Å × 660 Å; 200 pixel × 200 pixel) recorded at 100 ms time intervals, from 5 s of a longer HS-AFM movie. The image quality is consistent and depicts the trimeric assembly of individual bR molecules in the purple membrane (PM). (B) From top to bottom: one individual frame (frame sampling time: 100 ms; pixel sampling time: 2.5 μs), and merge of two, five, and 10 subsequent frames with 200 ms, 500 ms, and 1 s acquisition sampling time. (C) Averages and SD maps (n = 46) corresponding to the images shown in B; the averages have a full color range of 0.5 Å < height < 6.7 Å, and the SD maps have a full color scale of 0.5 Å < SD < 2.3 Å, 0.5 Å < SD < 2.1 Å, 0.4 Å < SD < 1.8 Å, and 0.4 Å < SD < 1.6 Å, respectively. (D) Histogram showing the number of individual molecules found in the images shown in B as function of their CCV with a noise-free ensemble average. (E) Histogram showing the number of individual molecules found in the images shown in B as function of their IS. (F) Average (black line) and SD (gray lines) of the PCF between bR trimers, peaking at 63 Å (FWHH: 5 Å) of five individual image frames, showing the high precision and stability of the scanner. (G) Symmetry analysis of the average calculated from one image frame (B and C, top) peaking for threefold symmetry at 0.85.
One aspect that merits particular attention in the high-resolution analysis movie is the detection of a lattice vacancy consisting of one missing bR monomer (Fig. 2, cell D4). HS imaging of the area of the vacancy showed an induced instability of the surrounding bR monomers, where sporadic motion of the adjacent monomers into the vacant area is observed (Fig. 2 C, cell D4). Nonetheless, the vacancy remains unoccupied most of the time, which may suggest a structural malformation of the area of the vacancy, occupation of the vacant area with lipid molecules, or restricted rotational freedom of the molecules and consequent weak association of invading molecules within the vacancy. The sporadic motion of the adjacent monomers to the vacancy was recorded over a time span of 700 ms (Fig. 2, A–D; cells D4–D5); this measurement illustrates the power of the technique for membrane diffusion analysis.
Figure 2.
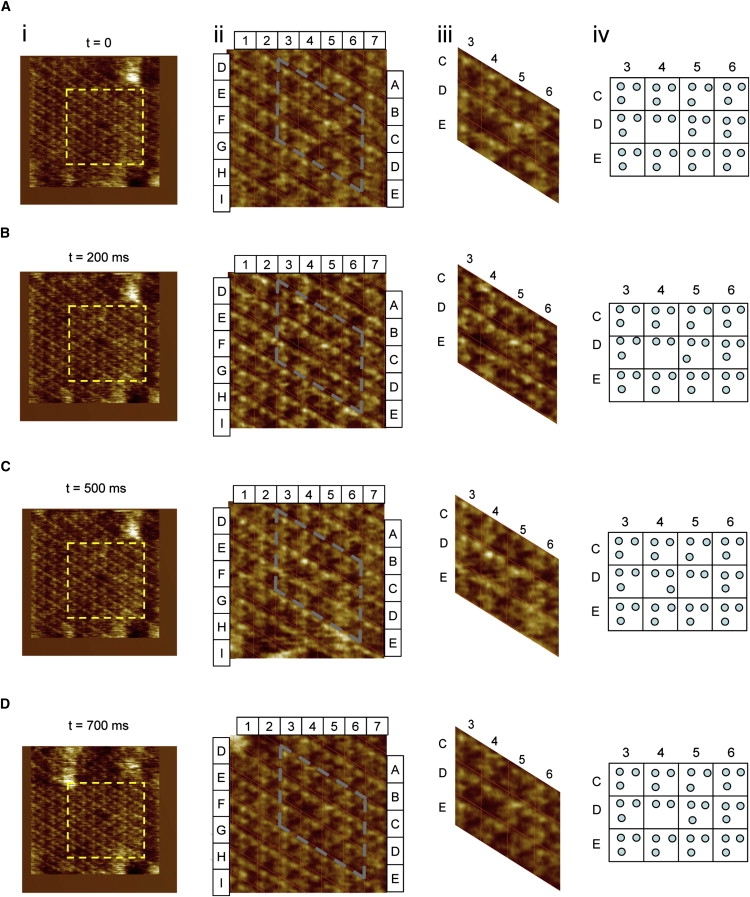
Sporadic motion of a bR monomer adjacent to a lattice vacancy. Images acquired at different times are shown: (A) t = 0, (B) t = 200 ms, (C) t = 500 ms, and (D) t = 700 ms. The four columns represent i), fully acquired images (660 Å × 660 Å; 200 pixel × 200 pixel); ii), digital zoom images in the region of the vacancy (409 Å × 409 Å; 124 pixel × 124 pixel), where a mesh is included for identifying individual bR lattice cells; iii), selection of relevant cells for the observation of the motion of bR monomers; and iv), schematic of the positions of bR monomers. From this sequence, it can be concluded that back-and-forth motion of a single bR monomer takes place between the adjacent bR trimer (cell D5) and the vacancy (cell D4).
At medium resolution, we imaged two differently oriented bR lattices and a nonordered membrane region in between. Image acquisition was performed at a 100 ms frame frequency on a 1136 Å square field scanned at a 200 pixel edge width (Fig. 3 A). The fifty consecutive image frames (pixel sampling (5.7 Å)2) represent 5 s of a longer movie (Movie S2). The high imaging rate did not disturb the features, and all images in Fig. 2 A and throughout the movie reveal a similar topography. Most of the individual image frames depict the different lattice orientations of the bR lattice in the top left and the one on the bottom of the frame (Fig. 3 B). Lateral alignment of all image frames with respect to each other allowed the calculation of an average image (Fig. 3 C). In this figure, although the features are somewhat blurred by the averaging process, the two independent bR lattices in the membrane, the lattice edges, and the area in between are well visible. At a 100 ms frame acquisition time, movements of proteins can be observed by HS-AFM (13,33). Knowing that except for some defects, the bR molecules inside the 2D lattice are essentially immobile (34), we focused on the membrane region in between the two bR arrays (Fig. 3, B and C; yellow dashed outline). Indeed, in this region fine topographical changes as a function of time are visible in the images acquired at the 100 ms frame rate (Fig. 3 D), as documented in the corresponding movie (Movie S3). For example, in the frames shown in Fig. 3 D, a topographical feature appears in frames 5–8. Although our resolution is currently insufficient to assign this topography unambiguously to bR, its dimension of ∼60 Å corresponds well to the size of a bR trimer. Furthermore, the presence of the topographical feature clearly localized during four subsequent frames (400 ms) is a strong argument that we are not looking at imaging noise, but at a real topographic feature in the PM. We hypothesize that individual bR trimers diffuse between the edges of the two bR lattices, and this can be observed by high-resolution contact-mode HS-AFM. This finding indicates that the native bR 2D arrays in the membranes are in association-dissociation equilibrium with neighboring bR arrays, as was recently shown in a study using oscillating-mode HS-AFM (35).
Figure 3.
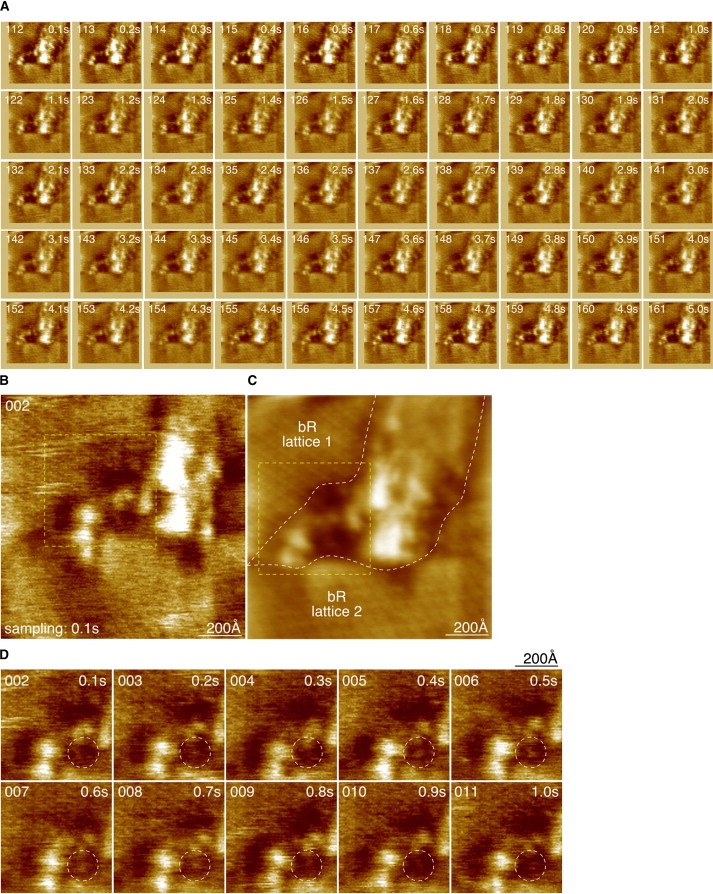
Medium-resolution contact-mode HS-AFM of the extracellular surface of bR lattices and their interface. (A) Fifty aligned images (1136 Å × 1136 Å; 200 pixel × 200 pixel) recorded at 100 ms time intervals, from 5 s of a longer HS-AFM movie. The image quality is consistent and reveals two differently oriented bR lattices interspaces by protrusions and depressions. (B) One individual frame (frame sampling time: 100 ms; pixel sampling time: 2.5 μs) of the movie. The different orientations of the two lattices (top left and bottom of the image) are visible. (C) Average of the aligned images in the movie depicts with stronger contrast the two bR lattices (white dashed outlines) with different orientation and the interspaces comprising a protruding and a lipid region. (D) Ten aligned images of the region (yellow dashed outline in B and C) in between the two bR lattices. Protein movement is visible in frames 5–8 (white dashed circles).
Discussion
Because of the speed of the analog feedback control and the high sensitivity of the photodiode (10,26), it is possible to precisely control (precision: ∼10 pN) the force applied to the tip during contact-mode HS-AFM imaging with cantilevers with a spring constant of ∼0.1 N/m. Using high-resolution contact-mode HS-AFM, not only are we able to observe individual bR molecules in the PM, we can also detect the motion of the bR molecules. Investigators have attempted to use conventional AFM to analyze the movement of membrane proteins (36,37), but at a frame acquisition time of ∼1 min only snapshots of stable states can be acquired. Now, with the use of HS-AFM, protein motion can be visualized, and the myosin motor movement (11), the chaperonin cycle of GroEL (38), and the association-dissociation process of streptavidin in 2D arrays (33) have been observed. Here we present evidence for the observation of membrane protein diffusion using contact-mode HS-AFM. The contact-mode HS-AFM methodology introduced here was successfully tested on PMs. This methodology is appropriate for samples with low corrugation; in these samples, blunt tips provide the best results because they distribute the applied force over a broader area due to electrostatic balancing (14). This situation contrasts with oscillating-mode HS-AFM studies of biological samples, in which probes featuring high-aspect-ratio EBD tips provided the best results (26). However, these probes are inconvenient in that growing the EBD tips is a time-consuming process that demands sophisticated electron microscope technology and expertise. Finally, in our study, the aspect ratio of the EBD tips proved to be unsuitable for contact-mode HS-AFM. These probes, when used in contact mode, often lead to destruction of the membranes during imaging.
The imaging conditions implemented (100 × 100 nm2, 100 ms per frame) open the door to diffusion tracking of multiple membrane proteins. Considering 〈r2〉 = 4Dt (39), where 〈r2〉 is the mean-square displacement, D is the diffusion coefficient, and t is the time, we find that the fastest diffusion coefficient observable at an image rate of 100 ms would be 2.5 × 104 nm2/s (0.025 μm2/s). Proteins with diffusion coefficients below this limit include the cystic fibrosis transmembrane conductance regulator (D = 0.005 μm2/s (40)), a glycosylphosphatidylinositol-anchored protein (D = 0.0035–0.0800 μm2/s (41)), the transferrin receptor (D = 0.005μm2/s (42)), and the glycoproteins in macrophages (D = 0.003μm2/s (43)). Therefore, we are confident that HS-AFM will soon provide the first structural movies of nonlabeled membrane protein diffusion.
Supporting Material
Three movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01152-7.
Supporting Material
Acknowledgments
This study was supported by Agence National de la Recherche grants ANR-08-NANO-010 and ANR-PCV08_343399, and by the Ville de Paris (Appel à projets de la Ville de Paris en Recherche Médicale et en Santé).
References
- 1.Lecam E., Frechon D., Barray M., Fourcade A., Delain E. Observation of binding and polymerization of Fur repressor onto operator-containing DNA with electron and atomic force microscopes. Proc. Natl. Acad. Sci. 1994;91:11816–11820. doi: 10.1073/pnas.91.25.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schabert F.A., Henn C., Engel A. Native Escherichia coli OmpF porin surfaces probed by atomic force microscopy. Science. 1995;268:92–94. doi: 10.1126/science.7701347. [DOI] [PubMed] [Google Scholar]
- 3.Scheuring S., Sturgis J.N. Chromatic adaptation of photosynthetic membranes. Science. 2005;309:484–487. doi: 10.1126/science.1110879. [DOI] [PubMed] [Google Scholar]
- 4.Drake B., Prater C.B., Weisenhorn A.L., Gould S.A.C., Albrecht T.R. Imaging crystals, polymers, and processes in water with the atomic force microscope. Science. 1989;243:1586–1588. doi: 10.1126/science.2928794. [DOI] [PubMed] [Google Scholar]
- 5.Fechner P., Boudier T., Mangenot S., Jaroslawski S., Sturgis J.N. Resolution, noise and structural information in high-resolution AFM topographs. Biophys. J. 2009;96:3822–3831. doi: 10.1016/j.bpj.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viani M.B., Pietrasanta L.I., Thompson J.B., Chand A., Gebeshuber I.C. Probing protein-protein interactions in real time. Nat. Struct. Biol. 2000;7:644–647. doi: 10.1038/77936. [DOI] [PubMed] [Google Scholar]
- 7.Hansma P.K., Schitter G., Fantner G.E., Prater C. High-speed atomic force microscopy. Science. 2006;314:601–602. doi: 10.1126/science.1133497. [DOI] [PubMed] [Google Scholar]
- 8.Humphris A.D.L., Miles M.J., Hobbs J.K. A mechanical microscope: high-speed atomic force microscopy. Appl. Phys. Lett. 2005;86:034106. [Google Scholar]
- 9.Picco L.M., Bozec L., Ulcinas A., Engledew D.J., Antognozzi M. Breaking the speed limit with atomic force microscopy. Nanotechnology. 2007;18:044030. [Google Scholar]
- 10.Ando T., Kodera N., Takai E., Maruyama D., Saito K. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kodera N., Kinoshita T., Ito T., Ando T. High-resolution imaging of myosin motor in action by a high-speed atomic force microscope. Adv. Exp. Med. Biol. 2003;538:119–127. doi: 10.1007/978-1-4419-9029-7_11. [DOI] [PubMed] [Google Scholar]
- 12.Ando T., Uchihashi T., Kodera N., Yamamoto D., Taniguchi M. High-speed atomic force microscopy for observing dynamic biomolecular processes. J. Mol. Recognit. 2007;20:448–458. doi: 10.1002/jmr.843. [DOI] [PubMed] [Google Scholar]
- 13.Ando T., Uchihashi T., Kodera N., Yamamoto D., Taniguch M. Invited review: high-speed AFM and nano-visualization of biomolecular processes. Eur. J. Phys. 2008;456:211–225. doi: 10.1007/s00424-007-0406-0. [DOI] [PubMed] [Google Scholar]
- 14.Müller D.J., Fotiadis D., Scheuring S., Müller S.A., Engel A. Electrostatically balanced subnanometer imaging of biological specimens by atomic force microscopy. Biophys. J. 1999;76:1101–1111. doi: 10.1016/S0006-3495(99)77275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reference deleted in proof.
- 16.Milhiet P., Gubellini F., Berquand A., Dosset P., Rigaud J. High-resolution AFM of membrane proteins directly incorporated at high density in planar lipid bilayer. Biophys. J. 2006;91:3268–3275. doi: 10.1529/biophysj.106.087791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reviakine I., Bergsma-Schutter W., Brisson A. Growth of protein 2-D crystals on supported planar lipid bilayers imaged in situ by AFM. J. Struct. Biol. 1998;121:356–361. doi: 10.1006/jsbi.1998.4003. [DOI] [PubMed] [Google Scholar]
- 18.Scheuring S., Seguin J., Marco S., Levy D., Robert B. Nanodissection and high-resolution imaging of the Rhodopseudomonas viridis photosynthetic core-complex in native membranes by AFM. Proc. Natl. Acad. Sci. USA. 2003;100:1690–1693. doi: 10.1073/pnas.0437992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller D.J., Heymann J.B., Oesterhelt F., Moller C., Gaub H. Atomic force microscopy of native purple membrane. Biochim. Biophys. Acta. 2000;1460:27–38. doi: 10.1016/s0005-2728(00)00127-4. [DOI] [PubMed] [Google Scholar]
- 20.Kienberger F., Stroh C., Kada G., Moser R., Baumgartner W. Dynamic force microscopy imaging of native membranes. Ultramicroscopy. 2003;97:229–237. doi: 10.1016/S0304-3991(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 21.Möller C., Allen M., Elings V., Engel A., Müller D.J. Tapping mode atomic force microscopy produces faithful high-resolution images of protein surfaces. Biophys. J. 1999;77:1050–1058. doi: 10.1016/S0006-3495(99)76966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark M., Möller C., Müller D.J., Guckenberger R. From images to interactions: high-resolution phase imaging in tapping-mode atomic force microscopy. Biophys. J. 2001;80:3009–3018. doi: 10.1016/S0006-3495(01)76266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oesterhelt D., Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fraction into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 24.Schabert F.A., Engel A. Reproducible acquisition of Escherichia coli porin surface topographs by atomic force microscopy. Biophys. J. 1994;67:2394–2403. doi: 10.1016/S0006-3495(94)80726-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitazawa M., Shiotani K., Toda A. Batch fabrication of sharpened silicon nitride tips. Jpn. J. Appl. Phys. 2003;42:4844–4847. [Google Scholar]
- 26.Ando T., Uchihashi T., Kodera N., Miyagi A., Nakakita R., Yamashita H., Sakashita M. High-speed atomic force microscopy for studying the dynamic behavior of protein molecules at work. Jpn. J. Appl. Phys. 2006;45:1897–1903. [Google Scholar]
- 27.Horcas I., Fernández R., Gómez-Rodríguez J., Colchero J., Gómez-Herrero J. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007;78:13705–13708. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 28.Scheuring S., Boudier T., Sturgis J.N. From high-resolution AFM topographs to atomic models of supramolecular assemblies. J. Struct. Biol. 2007;159:268–276. doi: 10.1016/j.jsb.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 29.Rasband, W.S. 1997–2005. ImageJ.U.S. National Institutes of Health, Bethesda, Maryland. http://rsb.info.nih.gov/ij/.
- 30.Müller D.J., Schabert F.A., Büldt G., Engel A. Imaging purple membranes in aqueous solutions at subnanometer resolution by atomic force microscopy. Biophys. J. 1995;68:1681–1686. doi: 10.1016/S0006-3495(95)80345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller D.J., Fotiadis D., Engel A. Mapping flexible protein domains at subnanometer resolution with the AFM. FEBS Lett. 1998;430:105–111. doi: 10.1016/s0014-5793(98)00623-1. [DOI] [PubMed] [Google Scholar]
- 32.Henderson R., Baldwin J.M., Downing K.H., Lepault J., Zemlin F. Structure of purple membrane from halobacterium halobium: Recording measurement and evaluation of electron micrographs at 3.5Å resolution. Ultramicroscopy. 1986;19:147–178. [Google Scholar]
- 33.Yamamoto D., Uchihashi T., Kodera N., Ando T. Anisotropic diffusion of point defects in two-dimensional crystal of streptavidin observed by high-speed atomic force microscopy. Nanotechnology. 2008;19:509901. doi: 10.1088/0957-4484/19/38/384009. [DOI] [PubMed] [Google Scholar]
- 34.Gibson N.J., Cassim J.Y. Nature of forces stabilizing the transmembrane protein bacteriorhodopsin in purple membrane. Biophys. J. 1989;56:769–780. doi: 10.1016/S0006-3495(89)82724-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita H., Voïtchovsky K., Uchihashi T., Antoranz Contera S., Ryan J.F. Dynamics of bacteriorhodopsin 2D crystal observed by high-speed atomic force microscopy. J. Struct. Biol. 2009 doi: 10.1016/j.jsb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Müller D.J., Engel A., Matthey U., Meier T., Dimroth P. Observing membrane protein diffusion at subnanometer resolution. J. Mol. Biol. 2003;327:925–930. doi: 10.1016/s0022-2836(03)00206-7. [DOI] [PubMed] [Google Scholar]
- 37.Scheuring S., Buzhynskyy N., Jaroslawski S., Gonçalves R.P., Hite R.K. Structural models of the supramolecular organization of AQP0 and connexons in junctional microdomains. J. Struct. Biol. 2007;160:385–394. doi: 10.1016/j.jsb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Yokokawa M., Wada C., Ando T., Sakai N., Yagi A. Fast-scanning atomic force microscopy reveals the ATP/ADP-dependent conformational changes of GroEL. EMBO J. 2006;25:4567–4576. doi: 10.1038/sj.emboj.7601326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxton, M.J. 1993. Lateral diffusion in an archipelago. Single-particle diffusion. 64:1766–1780. [DOI] [PMC free article] [PubMed]
- 40.Haggie P., Kim J., Lukacs G., Verkman A. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Mol. Biol. Cell. 2006;17:4937–4945. doi: 10.1091/mbc.E06-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheets E., Lee G., Simson R., Jacobson K. Transient confinement of a glycosylphosphatidylinositol-anchored protein in the plasma membrane. Biochemistry. 1997;36:12449–12458. doi: 10.1021/bi9710939. [DOI] [PubMed] [Google Scholar]
- 42.Sako Y., Kusumi A. Barriers for lateral diffusion of transferrin receptor in the plasma membrane as characterized by receptor dragging by laser tweezers: fence versus tether. J. Cell. Biol. 1995;129:1559–1574. doi: 10.1083/jcb.129.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheetz M., Turney S., Qian H., Elson E. Nanometre-level analysis demonstrates that lipid flow does not drive membrane glycoprotein movements. Nature. 1989;340:284–288. doi: 10.1038/340284a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


