Abstract
We investigated the specificity of interaction of a new type A lantibiotic, clausin, isolated from Bacillus clausii, with lipid intermediates of bacterial envelope biosynthesis pathways. Isothermal calorimetry and steady-state fluorescence anisotropy (with dansylated derivatives) identified peptidoglycan lipids I and II, embedded in dodecylphosphocholine micelles, as potential targets. Complex formation with dissociation constants of ∼0.3 μM and stoichiometry of ∼2:1 peptides/lipid intermediate was observed. The interaction is enthalpy-driven. For the first time, to our knowledge, we evidenced the interaction between a lantibiotic and C55-PP-GlcNAc, a lipid intermediate in the biosynthesis of other bacterial cell wall polymers, including teichoic acids. The pyrophosphate moiety of these lipid intermediates was crucial for the interaction because a strong binding with undecaprenyl pyrophosphate, accounting for 80% of the free energy of binding, was observed. No binding occurred with the undecaprenyl phosphate derivative. The pentapeptide and the N-acetylated sugar moieties strengthened the interaction, but their contributions were weaker than that of the pyrophosphate group. The lantibiotic decreased the mobility of the pentapeptide. Clausin did not interact with the water-soluble UDP-MurNAc- and pyrophosphoryl-MurNAc-pentapeptides, pointing out the importance of the hydrocarbon chain of the lipid target.
Introduction
Infectious diseases are the second leading cause of death worldwide and the third leading cause of death in developed countries (1). The escalating rate of bacterial multiresistance to commonly used antibiotics as a result of the widespread use of these drugs has highlighted the urgent need for new effective antibacterial agents (2,3). This bacterial resistance to antibiotics will lead to treatment failure unless new drugs are discovered. The search for and characterization of new antibacterial drugs is becoming a priority for public health. Some bacterial strains produce substances with antibacterial activity, such as bacteriocins often acting against closely related species. Lantibiotics, a class I of bacteriocins, are peptides encoded by structural genes and synthesized on ribosomes. Then, they undergo posttranslational modifications resulting in the presence of a large proportion of unusual amino acids, such as thioether amino acids (lanthionine and β-methyllanthionine) and dehydrated amino acids (dehydroalanine and dehydrobutyrine) (4–9). These amino acids contribute to the formation of several thioether rings, as shown initially for nisin (10).
Many lantibiotics target the peptidoglycan lipid II intermediate (Scheme 1 A) (11–14). Some, such as nisin, form pores in the cytoplasmic membrane, leading to bacterial lysis (7,15). Bacterial peptidoglycan consists of a network of N-acetylglucosaminyl-β-1,4-N-acetylmuramyl (GlcNAc-MurNAc) disaccharide units cross-linked by short peptide chains (16–20). This macromolecular structure enables the cell to resist lysis due to high internal osmotic pressure. It defines the shape of the bacterium, is involved in cell growth and division, and serves as a platform for the anchoring of other cell envelope components. Peptidoglycan biosynthesis occurs in three stages. The first stage, biosynthesis of the nucleotide precursors UDP-GlcNAc and UDP-MurNAc-pentapeptide by a set of highly specific enzymes, occurs in the cytoplasm. Two membrane enzymes, MraY and MurG, then catalyze the transfer of the MurNAc-pentapeptide and GlcNAc motifs to the undecaprenyl phosphate carrier lipid, generating the undecaprenyl-pyrophosphoryl-MurNAc-pentapeptide (lipid I) and the undecaprenyl-pyrophosphoryl-MurNAc-(pentapeptide)-GlcNAc (lipid II), respectively (21,22). The final step is the polymerization of the disaccharide pentapeptide motif, catalyzed by glycosyl-transferases and transpeptidases on the outer side of the cytoplasmic membrane. The precursors for peptidoglycan biosynthesis are essential for bacterial viability and growth. Inhibition of the synthesis of these precursors or their sequestration leads to bacterial lysis and death. New antibiotics interfering with this biosynthesis pathway would therefore be of considerable interest.
Scheme 1.
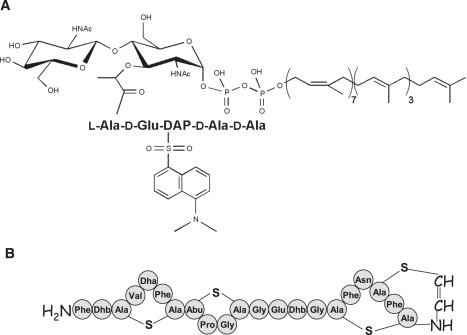
(A) Structure of peptidoglycan lipid II bearing a DNS group on the DAP residue. (B) Primary structure of the lantibiotic clausin.
A new type A lantibiotic, clausin, was recently isolated from Bacillus clausii (23). This peptide displays 75% sequence identity with mutacin-1140 and a much lower one with nisin (30%). It also retains the classical motif of two A/B lanthionine rings (Scheme 1 B) and is active against various gram-positive bacteria (24). We investigated the specificity of the interaction of this new lantibiotic with lipid intermediates essential for the biosynthesis of peptidoglycan and other bacterial cell wall polymers. This interaction was monitored in dodecylphosphocholine (DPC) surfactant micelles, used as membrane mimics. The parameters of clausin binding to these lipid intermediates were determined by isothermal titration microcalorimetry (ITC) and steady-state fluorescence anisotropy, using 1-(dimethylamino)-5-naphthalenesulfonyl derivatives of lipids I and II (DNS-lipids I/II). The use of these fluorescent derivatives made it possible to monitor environment and dynamic changes and accessibility of the pentapeptide moiety to solvent, through time-resolved fluorescence intensity and anisotropy measurements.
Materials and Methods
Chemicals
Dodecylphosphocholine was purchased from Avanti Polar Lipids (Alabaster, AL) and Triton X-100 from Sigma-Aldrich Chimie (Lyon, France). Undecaprenyl phosphate (C55-P) and undecaprenyl pyrophosphate (C55-PP) were provided by the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences, Warsaw, Poland. UDP-MurNAc-pentapeptide and pyrophosphoryl-MurNAc-pentapeptide were prepared as described previously (25,26). UDP-MurNAc-pentapeptide bearing a DNS group on the meso-diaminopimelic acid (DAP) residue was kindly provided by Dr. D. Le Beller (27). These nucleotide precursors were then successfully converted into the corresponding lipids I and II (unlabeled and fluorescent) in the presence of C55-P and purified MraY and MurG enzymes (0.5–1 μmol scale), as described previously (28). Lipids I and II were purified on a DEAE-cellulose column (Whatman DE32, Whatman Schleicher et Schuell, Versailles, France) equilibrated with chloroform/methanol/water (2:3:1, v/v/v). The column was washed with chloroform/methanol/ammonium bicarbonate 0.3 M (2:3:1, v/v/v) (buffer A) at a flow rate of 0.5 ml × min−1. Buffer A supplemented with 10% acetic acid was used for elution. The fractions collected from the DEAE-cellulose column were analyzed by quantitative aminosugar analysis after sample hydrolysis in 6 M HCl for 16 h at 95°C. The fractions containing the lipid were pooled and evaporated to dryness. The residue was dissolved in chloroform-methanol (2:1, v/v). C55-PP-GlcNAc lipid was produced by enzymatic synthesis, using the WecA enzyme (29,30). All other materials were reagent grade and obtained from commercial sources.
Clausin production and purification
B. claussi OC strain was grown as published previously (24) and the culture supernatants were used to purify the lantibiotic (17). Briefly, supernatants were loaded onto a Sep-pak C-18 plus reverse phase cartridge (Waters SA, St. Quentin/Yvelines, France) and the antimicrobial substance was eluted with 100% methanol. In the second-purification step the antimicrobial substance was loaded in a C4 RP-HPLC semipreparative column (Synchrom, Lafayette, LA) and fractions presenting activity were pooled. Finally, reverse phase HPLC on a C1 Prontosil column (Bischoff, Germany) was used to purify the semipreparative extract. The purity of the compound obtained at the end of this stage is >90%. All purification steps were carried out with a 600E HPLC system (Waters SA). Clausin stock solutions (1 mM) were prepared in 20 mM HEPES buffer, pH 7.2, containing 2% DPC.
Steady-state fluorescence measurements
Fluorescence emission spectra were recorded with a Cary Eclipse spectrofluorimeter (Varian, Les Ulis, France) with slit widths of 10 nm and 5 nm for excitation and emission, respectively. Samples were contained in microcuvettes: aliquots of clausin stock solution were added to DNS-lipids I/II solutions in 20 mM HEPES buffer, pH 7.2, containing 1% DPC in a final volume of 140 μL. Steady-state fluorescence anisotropy measurements were carried out on a customized spectrofluopolarimeter, using the sample compartment optics of a T-format SLM 8000 fluorimeter. The excitation source was a 75 W PTI Xenon arc lamp. An excitation wavelength of 335 nm was selected with a Jobin-Yvon double grating monochromator (JY UV-DH10, Jobin Yvon, Longjumeau, France) (slit width 4 nm) and vertically polarized with a Glan-Thomson prism. The emitted light was collected through Glan-Thomson polarizers, oriented either vertically or horizontally, with Schott KV 450 cutoff filter and measured with photon-counting Hamamatsu photomultipliers (H3460-54) monitoring each optical channel. The output signals of the photon-counting heads were plugged into the A and B inputs, respectively, of an Ortec 944 dual counter/timer interfaced with a microcomputer.
Time-resolved fluorescence measurements
Fluorescence intensity and anisotropy decays were obtained from the polarized Ivv(t) and Ivh(t) components, measured by the time-correlated single-photon counting technique, with an instrument described elsewhere (31). A diode laser (LDH 370 from Picoquant, Berlin-Adlershof, Germany; maximal emission at 375 nm) operating at 5 MHz was used as an excitation source and a Hamamatsu single-photon fast photomultiplier (model R3235-01) was used for detection. The emission wavelength was selected with a Jobin-Yvon H10 monochromator (band width 8 nm) and a Schott KV450 cutoff filter (Schott, Mainz, Germany). The instrumental response function (full width at half-maximum ∼600 ps) was automatically collected by measuring the light scattering of a glycogen solution. Samples were contained in microcuvettes (140 μL).
Fluorescence intensity decay analyses were carried out with MEM, using a multiexponential model: I(t) = , as described previously (32,33). A classical anisotropy model: A(t) = , in which any rotational correlation time (θ) is coupled with each lifetime (τ), was used to resolve polarized fluorescence decays (34). Calculations were carried out with a set of 150 or 100 independent variables (equally spaced on a logarithmic scale) for intensity and anisotropy, respectively. The programs, including the MEMSYS 5 subroutines (MEDC, Cambridge, UK), were written in double-precision FORTRAN 77.
ITC measurements of clausin binding
ITC experiments were carried out with a VP-ITC isothermal titration calorimeter from MicroCal (Northampton, MA). The experiments were carried out at 20°C. The lantibiotic concentration in the microcalorimeter cell (1.4323 mL) varied from 10 to 20 μM. In total, 28 injections of 10 μL (or 50 injections of 5 μL) of lipid solution (concentration from 170 to 300 μM) in 1% DPC were carried out at 240-s intervals, with stirring at 270 rpm. Theoretical titration curve software supplied by MicroCal (ORIGIN) was used to fit titration curves to the experimental data. This software generates titration curves based on the relationship between the heat generated by each injection and ΔH (enthalpy change in kcal × mol−1), Kd (the dissociation binding constant in M), n (the number of binding sites), total peptide concentration and free and total lipid concentrations.
Determination of binding parameters by steady-state fluorescence anisotropy
Two solutions A and B were prepared and mixed at the appropriate ratio. Solution A contains clausin (5 μM) in 20 mM HEPES, pH 7.2, and DPC (1%). Solution B was prepared as solution A but supplemented by DNS-lipid II (8.5 μM). Total lipid final concentrations ranged from 0.5 μM up to 8.5 μM. The bound (αb) and free (αf) DNS-lipid II mole fractions (the subscripts b and f refer to the bound and free ligand forms, respectively) were calculated from the resulting fluorescence anisotropy values of the DNS probe for each species during the titration. The resulting steady-state anisotropy (r) of a mixture of fluorescent components is classically expressed as (35)
| (1) |
where rf and rb are the anisotropy values for each form. rb was estimated at very low concentrations of DNS-lipid II and in conditions of saturating peptide concentration. rf was estimated in DPC micelles in the absence of lantibiotic. τb and τf are the amplitude-averaged excited state lifetime values of DNS-lipid II for each form, measured in the same conditions as for the anisotropy rf and rb. Under normalization conditions (αf + αb = 1), Eq. 1 becomes
| (2) |
Thus, from the experimental anisotropy value (r) obtained for each concentration of DNS-lipid II, the free and bound mole fractions were calculated and the dissociation constant and stoichiometry of the equilibrium were estimated from classical Scatchard plots.
Results and Discussion
Selectivity of the interaction of clausin with lipid I and lipid II: steady-state fluorescence intensity and anisotropy study with DNS-lipid I/II
Lipid intermediates in the synthesis of bacterial cell wall peptidoglycan often constitute specific targets for peptide antibiotics of the lantibiotic family (11,12). The conservation of the pyrophosphate (PP) binding motif (lanthionine A/B rings) (36,37) in clausin (23) suggests that this molecule may recognize the peptidoglycan precursors lipids I and II. We investigated this interaction, using fluorescent derivatives of lipid I and lipid II bearing a DNS group on the DAP residue of their pentapeptide moiety, dispersed in DPC micelles. We made use of the high sensitivity of DNS group fluorescence emission to changes in the polarity and dynamics of its microenvironment (38–42) to detect the clausin-lipid I/II interaction in the submicromolar to micromolar concentration ranges.
The addition of clausin to DPC micelles loaded with either DNS-lipid II or DNS-lipid I induced a significant and progressive quenching of DNS fluorescence emission (Fig. 1 for DNS-lipid II), without significant shift (528–530 nm). Fluorescence intensity decreased to a plateau value of ∼70% of the integrated initial intensity (maximum quenching efficiency of ∼30%) (Fig. 1, inset). Clausin had similar effects on the fluorescence intensity and maximum emission wavelength of DNS-lipid I (data not shown). Moreover, within the same concentration range, clausin triggered a doubling of DNS steady-state fluorescence anisotropy (Fig. 2). The steady-state anisotropy parameter, which principally reflects the rotational mobility of the DNS group, was therefore more sensitive to the interaction than fluorescence intensity. Clausin induced no change in DNS fluorescence parameters for the water-soluble UDP-MurNAc-pentapeptide-DNS (maximum emission at 555 nm).
Figure 1.
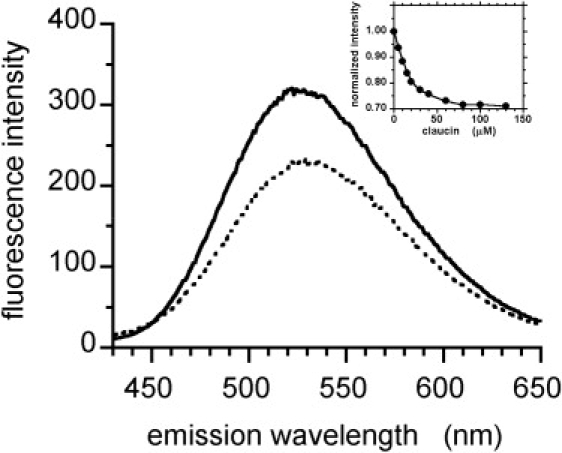
Clausin binding to lipid II. Fluorescence emission spectrum of DNS-lipid II (5 μM) embedded in 1% DPC (solid line); DNS-lipid II 5 μM + clausin 80 μM (dotted line). Inset: Normalized fluorescence intensity as a function of clausin concentration. Excitation wavelength, 350 nm.
Figure 2.
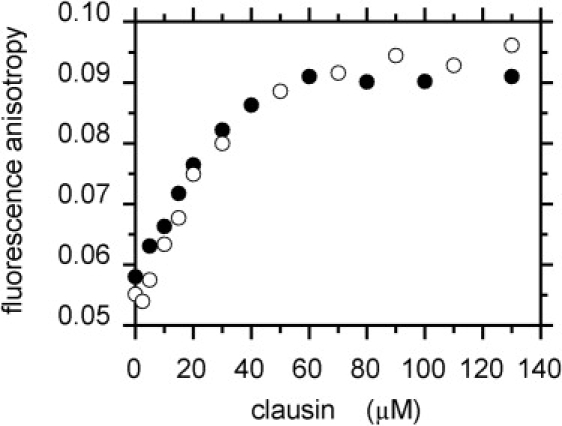
Clausin binding to lipid II. Steady-state fluorescence anisotropy of DNS-lipid I (5 μM) (○), and DNS-lipid II (5 μM) (●) in 1% DPC as a function of clausin concentration.
Steady-state anisotropy experiments were carried out to evaluate competition for interaction with clausin between DNS-lipid II and the nonfluorescent metabolic intermediates, C55-P and C55-PP. Control competition experiments between unlabeled and DNS-lipid II for binding to clausin showed a sharp decrease in DNS fluorescence anisotropy to the levels observed for free DNS-lipid II. This is consistent with the unlabeled lipid II progressively displacing the fluorescent derivative in the clausin-DNS-lipid II complex formed on the DPC micelles (Fig. 3). The mideffect occurred at ∼1 μM. Competition was detected with C55-PP-GlcNAc, a lipid intermediate involved in the biosynthesis of several bacterial cell wall polymers, including teichoic acids. Competition with C55-PP was also observed in the same experimental conditions, but it required much higher concentrations (mideffect ∼20 μM) (Fig. 3). By contrast, no competition with C55-P was observed (Fig. 3).
Figure 3.
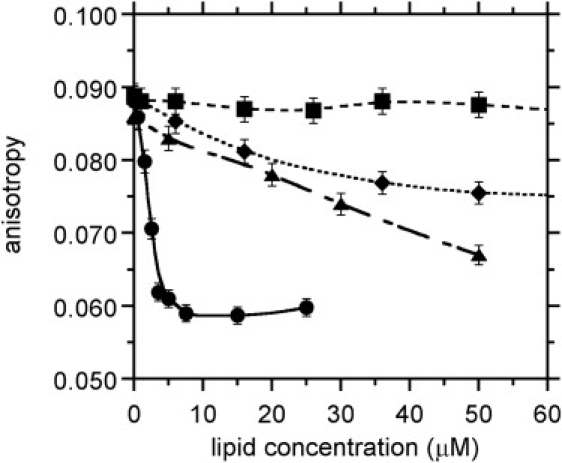
Competition experiments with DNS-lipid II. Lipid II (●), C55-PP-GlcNAc (▴), C55-PP (♦), and C55-P (■). DNS-lipid II concentration, 1 μM; clausin concentration, 20 μM.
Parameters of clausin binding to the various bacterial cell wall lipid intermediates: steady-state fluorescence anisotropy and isothermal titration microcalorimetry
The saturation curves for fluorescence intensity and anisotropy observed with increasing concentrations of clausin suggested that this interaction could be considered to be in equilibrium. The association constant and stoichiometry of binding were measured by both steady-state fluorescence anisotropy and isothermal calorimetry. Scatchard plots of steady-state anisotropy data for DNS-lipid II were linear (Fig. 4). A Kd value of 0.66 μM was obtained, with a stoichiometry of 2.4 clausin molecules per DNS-lipid II. For nisin, the most studied member of this lantibiotic family, a 1:1 complex was found in DMSO (37), whereas an 8:4 nisin/lipid II ratio was estimated for pore formation in membrane bilayers (15). These results suggest that the complex consists of two clausin molecules and one lipid.
Figure 4.
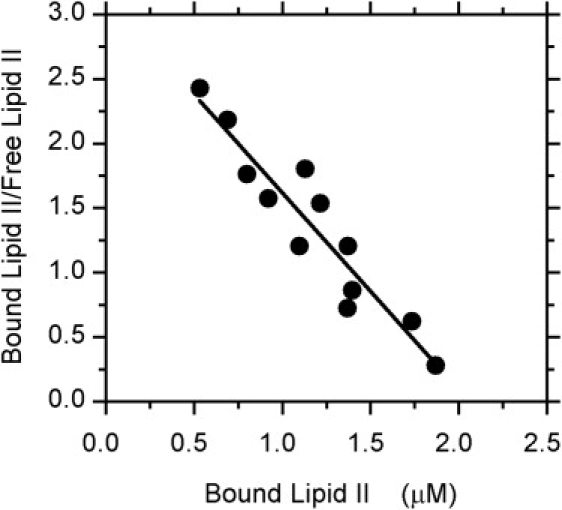
Scatchard plot of clausin binding to DNS-lipid II, as monitored by steady-state fluorescence anisotropy measurements. A Kd value of 0.66 μM was obtained, with a stoichiometry of 2.4 clausin molecules per DNS-lipid II. Clausin concentration, 5 μM.
ITC results for the binding of clausin to lipid II are shown in Fig. 5. The calculated values of the binding parameters (Table 1) are similar to those obtained by fluorescence anisotropy measurements. A 2:1 clausin/lipid I/II stoichiometry was also found. The 2.5-fold higher Kd value obtained with fluorescence anisotropy measurements corresponds to a 6% decrease in binding free energy, indicating that the dansyl moiety interferes with complex formation only slightly. The binding parameters for lipids I and II were very similar, both in the subnanomolar concentration range (Table 1), whereas the Kd for C55-PP was 20 times higher than those for lipids I and II (Table 1). An intermediate situation was observed for C55-PP-GlcNAc (Table 1). No interaction was detected with C55-P, UDP-MurNAc-pentapeptide or pyrophosphoryl-MurNAc-pentapeptide.
Figure 5.
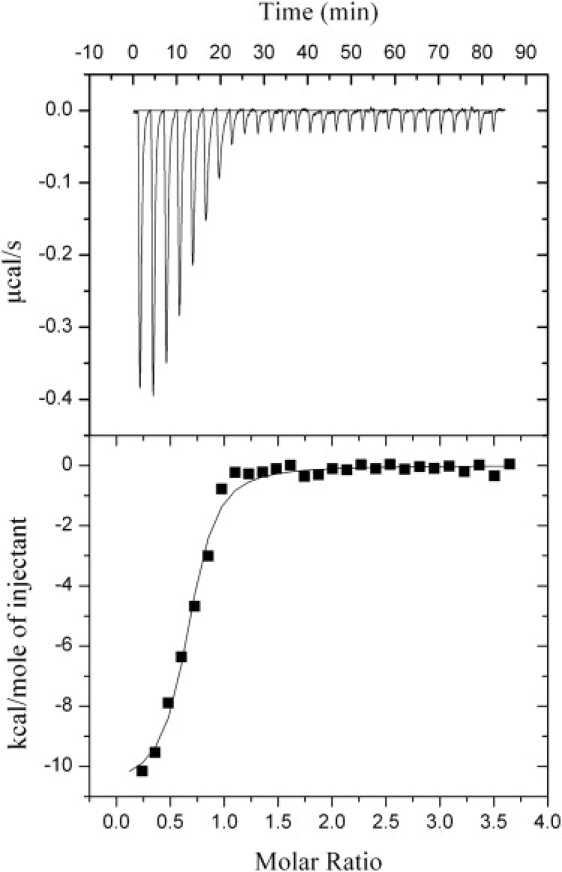
Binding of clausin to lipid II, as monitored by ITC. The titration cell was loaded with a mixture containing clausin (10 μM). Twenty-eight injections of 10 μL of a mixture containing lipid II (170 μM) were carried out at 240 s intervals. Temperature, 20°C.
Table 1.
Clausin binding parameters, the dissociation constant (Kd), and binding capacity (n), as determined by ITC
| Kd (μM) | n (clausin/lipid) | |
|---|---|---|
| Lipid I | 0.30 ± 0.05 | 1.9 ± 0.6 |
| Lipid II | 0.26 ± 0.05 | 1.9 ± 0.6 |
| C55-PP-GlcNAc | 2.6 ± 0.3 | 2.0 ± 0.2 |
| C55-PP | 5.7 ± 0.4 | 1.6 ± 0.2 |
The thermodynamic parameters of the interaction are presented in Table 2. For all compounds, the enthalpy term made a major contribution to the free energy of binding (ΔG). This is consistent with the absence of an interaction between C55-P and clausin and indicates that the interaction observed with the other lipids involves principally electrostatic or hydrogen bonding via the PP group. However, the value of the enthalpy/entropy ratio clearly depends on the size of the lipid intermediate, the enthalpic term being minimal for the C55-PP compound, for which the entropic term makes a favorable contribution. This suggests that for C55-PP, hydrophobic interactions also contribute to the binding whereas extra electrostatic or hydrogen bond interactions, in addition to those existing with the PP moiety, are involved in the binding to lipids I and II.
Table 2.
Thermodynamic parameters of the interaction (determined by ITC)
| ΔHapp (kcal/mol) | ΔG (kcal/mol) | TΔS (kcal/mol) | |
|---|---|---|---|
| Lipid I | −9.7 ± 0.4 | −8.8 ± 0.1 | −0.9 |
| Lipid II | −10.7 ± 0.3 | −8.8 ± 0.1 | −1.9 |
| C55-PP-GlcNAc | −7.9 ± 1.1 | −7.5 ± 0.1 | −0.4 |
| C55-PP | −6.2 ± 0.2 | −7.0 ± 0.1 | 0.8 |
Changes in the microenvironment and local conformation of DNS-lipid I and DNS-lipid II triggered by complex formation with clausin
The excited state lifetime of the DNS moiety is highly sensitive to the polarity and protic characteristics of the solvent, varying from ∼3 ns in water to ∼20 ns in DMF or DMSO (38,43), because DNS and its derivatives have different emission states in polar and apolar solvents (38,44). The high probability of nonradiative transition in highly protic solvents, such as water, results mostly from solute-solvent vibrational relaxation. Time-resolved fluorescence intensity measurements of DNS are therefore valuable for evaluating changes in the microenvironment of DNS.
The fluorescence intensity decays of the DNS moiety in UDP-MurNAc-pentapeptide-DNS in buffer and in DNS-lipid II in DPC micelles (in the presence or absence of clausin) were compared (Fig. 6 A). For UDP-MurNAc-pentapeptide-DNS, the dominant fluorescence intensity decay had a lifetime of 3.6 ns (Fig. 6 B and Table 3), characteristic of DNS in water (38,45), whereas for DNS-lipid II and DNS-lipid I in DPC micelles, the dominant decay had a longer lifetime, characteristic of an environment less polar than water (∼14 ns, similar to 1,5-DNS amide in methanol (38)) (Fig. 6 B and Table 3).
Figure 6.
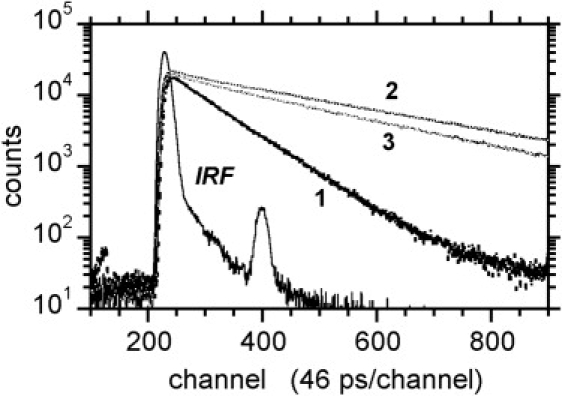
DNS fluorescence intensity decays. UDP-MurNAc-pentapeptide-DNS (curve 1), DNS-lipid II (curve 2), and DNS-lipid II in the presence of clausin (curve 3). IRF, instrument response function; excitation wavelength, 375 nm; emission wavelength, 530 nm.
Table 3.
Excited state lifetime distribution of UDP-MurNAc-pentapeptide-DNS (40 μM), DNS-lipid I, and DNS-lipid II (5 μM each)
| Sample | α1 | α2 | α3 | τ1 (ns) | τ2 (ns) | τ3 (ns) | 〈τ〉∗ (ns) |
|---|---|---|---|---|---|---|---|
| UDP-MurNAc-pentapeptide-DNS | 0.22 ± 0.02 | 0.03 ± 0.02 | 0.75 ± 0.02 | 0.20 ± 0.06 | 1.38 ± 0.53 | 3.58 ± 0.12 | 2.77 ± 0.06 |
| DNS-lipid I | 0.19 ± 0.01 | 0.06 ± 0.01 | 0.75 ± 0.02 | 0.92 ± 0.14 | 4.23 ± 0.31 | 14.28± 0.09 | 11.16 ± 0.15 |
| DNS-lipid I/clausin | 0.21 ± 0.02 | 0.14 ± 0.01 | 0.65 ± 0.02 | 0.65 ± 0.10 | 2.45 ± 0.47 | 11.88 ± 0.24 | 8.20 ± 0.05 |
| DNS-lipid II | 0.20 ± 0.01 | 0.08 ± 0.01 | 0.72 ± 0.01 | 0.66 ± 0.15 | 3.33 ± 0.38 | 14.32 ± 0.03 | 10.69 ± 0.28 |
| DNS-lipid II/clausin | 0.23 ± 0.01 | 0.12 ± 0.03 | 0.65 ± 0.03 | 0.81 ± 0.11 | 3.19 ± 1.05 | 11.98 ± 0.25 | 8.36 ± 0.25 |
Microcuvettes of 140 μL were used. Clausin concentration, 130 μM; DPC, 1%; excitation, 375 nm; emission, 525 nm + Schott KV418 cutoff filter.
Amplitude-averaged lifetime was defined as .
The presence of a subnanosecond lifetime for all compounds probably resulted from the photophysics of DNS, with subnanosecond internal excited state relaxations (change in the configuration of the dimethylamino group and/or solvent relaxation) (38,46–49). For UDP-MurNAc-pentapeptide-DNS in buffer, the short lifetime of 0.2 ns probably corresponded to the excited state internal relaxation of the dimethylamino group, water dipolar relaxation being too fast to be measured with our instruments. For DNS-lipids I and II in DPC micelles, the shortest lifetime was longer than for UDP-MurNAc-pentapeptide-DNS, probably due to both internal and solvent relaxations, as water solvent relaxation at micelle/water interfaces is much slower than that of bulk water (50,51). The excited lifetime of 3.3–4.2 ns probably corresponds to a minor conformer fully exposed to the aqueous solvent.
The effects of clausin binding were a decrease of ∼20% in the longest lifetime value for DNS, indicating stronger interaction with the aqueous solvent, and a decrease in its amplitude by ∼15%, favoring the 3–4 ns lifetime typical of solvent-exposed DNS. Thus, clausin decreased mean amplitude lifetime 〈τ〉 by 22–27% (Table 3), consistent with steady-state fluorescence quenching results. These lifetime data are consistent with a change in the conformation of the pentapeptide moiety, resulting in a higher degree of exposure to solvent.
The rotational dynamics of DNS-lipid I/II
Clausin binding decreased the mobility of the DNS group over the nanosecond timescale, as shown by fluorescence anisotropy decay measurements (Fig. 7). Two measurable rotational correlation times were obtained in the nanosecond time ranges (Table 4). The short rotational correlation time θ1 suggests probable segmental motions of the pentapeptide moiety, whereas the long correlation time θ2 was shorter than expected for pure DPC micelles (∼15 ns) (52,53). This may result from rotational/translational motions of lipid I/II within the micelles (54,55) or from micelles being nonspherical due to insertion of the long undecaprenyl hydrocarbon chain of lipid I/II. Subnanosecond rotation, describing the movement of the DNS group about its linker, is also likely to occur, as the initial anisotropy At=0 values obtained were significantly smaller than the intrinsic anisotropy A0 values obtained for immobilized DNS in pure glycerol at low temperature (A0 = 0.370 ± 0.013) (39,42).
Figure 7.
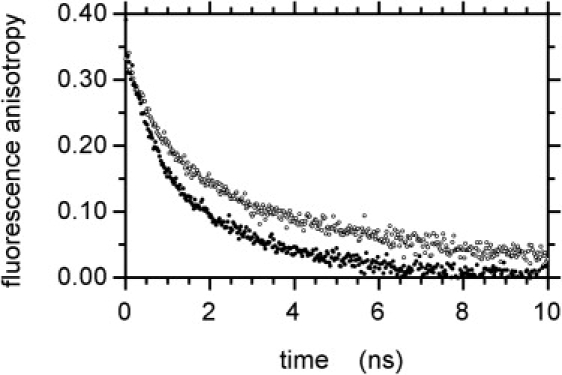
Experimental fluorescence anisotropy decay. DNS-lipid II (●) and DNS-lipid II in the presence of clausin (○). Experimental conditions were as in Fig. 6.
Table 4.
Fluorescence anisotropy decay parameters of UDP-MurNAc-pentapeptide-DNS (40 μM), DNS-lipid I, and DNS-lipid II (5 μM each)
| Sample | β1 | β2 | θ1 (ns) | θ2 (ns) | At=0 | ωmax (°) |
|---|---|---|---|---|---|---|
| UDP-MurNAc-pentapeptide-DNS | - | 0.343 ± 0.031 | - | 0.28 ± 0.02 | 0.343 ± 0.031 | - |
| DNS-lipid I | 0.092 ± 0.021 | 0.198 ± 0.004 | 0.7 ± 0.1 | 3.6 ± 0.1 | 0.289 ± 0.017 | 23 ± 3 |
| DNS-lipid I/clausin | 0.084 ± 0.008 | 0.188 ± 0.009 | 1.8 ± 0.5 | 9.4 ± 0.3 | 0.272 ± 0.009 | 26 ± 2 |
| DNS-lipid II | 0.105 ± 0.020 | 0.182 ± 0.008 | 0.8 ± 0.2 | 4.5 ± 0.2 | 0.287 ± 0.048 | 23 ± 3 |
| DNS-lipid II/clausin | 0.079 ± 0.005 | 0.174 ± 0.002 | 1.7 ± 0.1 | 8.9 ± 0.1 | 0.253 ± 0.003 | 28 ± 2 |
Clausin binding did not significantly affect the amplitude of the subnanosecond rotation, as shown by the values of the semi-angle of the wobbling-in-cone motion ωmax (Table 4). It increased θ2 to a value closer to that expected for DPC micelle rotation (52,53), indicating that the rotational/translational motions of lipid I/II within micelles were significantly hindered in the complex with clausin.
Conclusions
This study shows that clausin, a new class A lantibiotic isolated from B. clausii (23), can target lipid intermediates of bacterial peptidoglycan biosynthesis. It also provides what we believe is the first demonstration of interaction between a lantibiotic and C55-PP-GlcNAc. This lantibiotic may therefore interfere with the biosynthesis of other cell wall components, such as teichoic acid, in gram-positive bacteria. In this respect, it has been proposed that nisin could target the lipid intermediate of pseudomurein biosynthesis in Methanobacterium species (56). A combination of calorimetric and spectroscopic approaches was used for detailed characterization of the interaction between clausin and several bacterial cell wall lipid intermediates. A stoichiometry of two clausin molecules per lipid target was observed for this interaction, suggesting that the active form of clausin may be a dimer. The PP moiety was absolutely required for complex formation, as no interaction was observed with C55-P. Interaction with the PP moiety accounted for 80% of the free energy of binding. The pentapeptide moiety made a weaker contribution to binding. The fluorescence emission spectra of DNS-lipid I or DNS-lipid II in DPC micelles was blue-shifted by ∼30 nm with respect to that of UDP-MurNAc-pentapeptide-DNS in buffer, indicating an interfacial location of the fluorescent tag and, therefore, of the pentapeptide moiety (57). DNS-lipid I or DNS-lipid II was also highly mobile over the nanosecond timescale. Formation of the complex with clausin reduced its mobility and increased its exposure to solvent, as deduced from time-resolved measurements, but no direct interaction of clausin with the central part of the peptide was observed. The complex with clausin therefore probably involves only the first few amino acids of the pentapeptide. The terminal sugar moiety of lipid II is not involved in the interaction, because lipid I, which lacks the GlcNAc moiety and lipid II had similar affinities for clausin. Clausin did not bind to the water-soluble UDP-MurNAc-pentapeptide or the pyrophosphoryl-MurNAc-pentapeptide. This study suggests that the clausin/lipid I/II complex occurs at the micelle/water interface. The orientation and/or mobility of the binding moieties and their hydration are probably important parameters. Whether the clausin/lipid target complex is organized in pores or in larger clusters, as demonstrated for other lantibiotics (58,59), remains to be investigated.
As illustrated in this study, members of the lantibiotic A family may target not only peptidoglycan lipid II intermediates, but also sugar-lipids involved in the biosynthesis of other bacterial cell wall polymers.
Acknowledgments
We acknowledge the European Community for the financial support of the EUR-INTAFAR integrated project within the 6th PCRDT framework (contract LSHM-CT-2004-512138) and for a postdoctoral fellowship to Bayan Al-Dabbagh.
This work was supported by the Centre National de la Recherche Scientifique (UMR 8619, UMR 5248).
Contributor Information
Ahmed Bouhss, Email: ahmed.bouhss@u-psud.fr.
Jacques Gallay, Email: jacques.gallay@u-psud.fr.
References
- 1.Fauci A.S. Infectious diseases: considerations for the 21st century. Clin. Infect. Dis. 2001;32:675–685. doi: 10.1086/319235. [DOI] [PubMed] [Google Scholar]
- 2.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 3.Courvalin P., Davies J. Antimicrobials: time to act! Curr. Opin. Microbiol. 2003;6:425–426. [Google Scholar]
- 4.Ingram L.C. Synthesis of the antibiotic nisin: formation of lanthionine and beta-methyl-lanthionine. Biochim. Biophys. Acta. 1969;184:216–219. doi: 10.1016/0304-4165(69)90121-4. [DOI] [PubMed] [Google Scholar]
- 5.Jung G. In: Lantibiotics: a survey. Jung G., Sahl H.-G., editors. Escom; Leiden, The Netherlands: 1991. [Google Scholar]
- 6.Jack R., Bierbaum G., Heidrich C., Sahl H.G. The genetics of lantibiotic biosynthesis. Bioessays. 1995;17:793–802. doi: 10.1002/bies.950170909. [DOI] [PubMed] [Google Scholar]
- 7.Brotz H., Josten M., Wiedemann I., Schneider U., Gotz F. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 8.van Kraaij C., de Vos W.M., Siezen R.J., Kuipers O.P. Lantibiotics: biosynthesis, mode of action and applications. Nat. Prod. Rep. 1999;16:575–587. doi: 10.1039/a804531c. [DOI] [PubMed] [Google Scholar]
- 9.Guder A., Wiedemann I., Sahl H.G. Posttranslationally modified bacteriocins-the lantibiotics. Biopolymers. 2000;55:62–73. doi: 10.1002/1097-0282(2000)55:1<62::AID-BIP60>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 10.Gross E., Morell J.L. The structure of nisin. J. Am. Chem. Soc. 1971;93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 11.de Kruijff B., van Dam V., Breukink E. Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot. Essent. Fatty Acids. 2008;79:117–121. doi: 10.1016/j.plefa.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Breukink E., de Kruijff B. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 13.Bonelli R.R., Schneider T., Sahl H.G., Wiedemann I. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 2006;50:1449–1457. doi: 10.1128/AAC.50.4.1449-1457.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiedemann I., Bottiger T., Bonelli R.R., Wiese A., Hagge S.O. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 2006;61:285–296. doi: 10.1111/j.1365-2958.2006.05223.x. [DOI] [PubMed] [Google Scholar]
- 15.Hasper H.E., de Kruijff B., Breukink E. Assembly and stability of nisin-Lipid II pores. Biochemistry. 2004;43:11567–11575. doi: 10.1021/bi049476b. [DOI] [PubMed] [Google Scholar]
- 16.Rogers H.J. Biogenesis of the wall in bacterial morphogenesis. Adv. Microb. Physiol. 1979;19:1–62. doi: 10.1016/s0065-2911(08)60197-6. [DOI] [PubMed] [Google Scholar]
- 17.van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 18.Vollmer W., Blanot D., de Pedro M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 19.Barreteau H., Kovac A., Boniface A., Sova M., Gobec S. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008;32:168–207. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 20.Bouhss A., Trunkfield A.E., Bugg T.D., Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 21.Al-Dabbagh B., Henry X., El Ghachi M., Auger G., Blanot D. Active site mapping of MraY, a member of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily, catalyzing the first membrane step of peptidoglycan biosynthesis. Biochemistry. 2008;47:8919–8928. doi: 10.1021/bi8006274. [DOI] [PubMed] [Google Scholar]
- 22.Crouvoisier M., Auger G., Blanot D., Mengin-Lecreulx D. Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis. Biochimie. 2007;89:1498–1508. doi: 10.1016/j.biochi.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Bressolier, P., M.A. Brugo, P. Robineau, M. Schmitter, M. Sofeir, et al. 2007. Peptide compound with biological activity, its preparation and application. WIPO PCT/IB2007/0022003. France.
- 24.Urdaci M.C., Bressollier P., Pinchuk I. Bacillus clausii probiotic strains: antimicrobial and immunomodulatory activities. J. Clin. Gastroenterol. 2004;38:S86–S90. doi: 10.1097/01.mcg.0000128925.06662.69. [DOI] [PubMed] [Google Scholar]
- 25.Bouhss A., Crouvoisier M., Blanot D., Mengin-Lecreulx D. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J. Biol. Chem. 2004;279:29974–29980. doi: 10.1074/jbc.M314165200. [DOI] [PubMed] [Google Scholar]
- 26.Maillard A.P., Biarrotte-Sorin S., Villet R., Mesnage S., Bouhss A. Structure-based site-directed mutagenesis of the UDP-MurNAc-pentapeptide-binding cavity of the FemX alanyl transferase from Weissella viridescens. J. Bacteriol. 2005;187:3833–3838. doi: 10.1128/JB.187.11.3833-3838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stachyra T., Dini C., Ferrari P., Bouhss A., van Heijenoort J. Fluorescence detection-based functional assay for high-throughput screening for MraY. Antimicrob. Agents Chemother. 2004;48:897–902. doi: 10.1128/AAC.48.3.897-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Ghachi M., Bouhss A., Barreteau H., Touzé T., Auger G. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 2006;281:22761–22772. doi: 10.1074/jbc.M602834200. [DOI] [PubMed] [Google Scholar]
- 29.Al-Dabbagh B., Blanot D., Mengin-Lecreulx D., Bouhss A. Preparative enzymatic synthesis of polyprenyl-pyrophosphoryl-N-acetylglucosamine, an essential lipid intermediate for the biosynthesis of various bacterial cell envelope polymers. Anal. Biochem. 2009;391:163–165. doi: 10.1016/j.ab.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Al-Dabbagh B., Mengin-Lecreulx D., Bouhss A. Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA. J. Bacteriol. 2008;190:7141–7146. doi: 10.1128/JB.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoegy F., Celia H., Mislin G.L., Vincent M., Gallay J. Binding of iron-free siderophore, a common feature of siderophore outer membrane transporters of Escherichia coli and Pseudomonas aeruginosa. J. Biol. Chem. 2005;280:20222–20230. doi: 10.1074/jbc.M500776200. [DOI] [PubMed] [Google Scholar]
- 32.Livesey A.K., Brochon J.C. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys. J. 1987;52:693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent M., Brochon J.C., Merola F., Jordi W., Gallay J. Nanosecond dynamics of horse heart apocytochrome c in aqueous solution as studied by time-resolved fluorescence of the single tryptophan residue (Trp-59) Biochemistry. 1988;27:8752–8761. doi: 10.1021/bi00424a010. [DOI] [PubMed] [Google Scholar]
- 34.Vincent M., Gallay J. The interactions of horse heart apocytochrome c with phospholipid vesicles and surfactant micelles: time-resolved fluorescence study of the single tryptophan residue (Trp-59) Eur. Biophys. J. 1991;20:183–191. doi: 10.1007/BF01561141. [DOI] [PubMed] [Google Scholar]
- 35.Lakowicz J.R. Principles of Fluorescence Spectroscopy. 3rd ed. Kluwer Academic, Plenum Publishers; New-York: 2006. Fluorescence Anisotropy; pp. 291–318. [Google Scholar]
- 36.Bonev B.B., Breukink E., Swiezewska E., De Kruijff B., Watts A. Targeting extracellular pyrophosphates underpins the high selectivity of nisin. FASEB J. 2004;18:1862–1869. doi: 10.1096/fj.04-2358com. [DOI] [PubMed] [Google Scholar]
- 37.Hsu S.T.D., Breukink E., Tischenko E., Lutters M.A.G., de Kruijff B. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 2004;11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 38.Li Y.H., Chan L.M., Tyer L., Moody R.T., Himel C.M. Study of solvent effects on fluorescence of 1-(dimethylamino)-5-naphthalenesulfonic acid and related compounds. J. Am. Chem. Soc. 1975;97:3118–3126. [Google Scholar]
- 39.Weber G. Polarization of the fluorescence of macromolecules. II. Fluorescent conjugates of ovalbumin and bovine serum albumin. Biochem. J. 1952;51:155–167. doi: 10.1042/bj0510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuldiner S., Spencer R.D., Weber G., Weil R., Kaback H.R. Lifetime and rotational relaxation time of dansylgalactoside bound to the Lac carrier protein. J. Biol. Chem. 1975;250:8893–8896. [PubMed] [Google Scholar]
- 41.Brochon J.C., Wahl P. Measurements of fluorescent anisotropy decline of gamma-globulin and its fragments Fab, Fc, F(ab) 2 labeled with 1-sulfonyl-5-dimethyl-aminonaphthalene. Eur. J. Biochem. 1972;25:20–32. doi: 10.1111/j.1432-1033.1972.tb01662.x. [DOI] [PubMed] [Google Scholar]
- 42.Weber G., Borris D.P., De Robertis E., Barrantes F.J., La Torre J.L. The use of a cholinergic fluorescent probe for the study of the receptor proteolipid. Mol. Pharmacol. 1971;7:530–537. [PubMed] [Google Scholar]
- 43.Bailey M.F., Thompson E.H., Millar D.P. Probing DNA polymerase fidelity mechanisms using time-resolved fluorescence anisotropy. Methods. 2001;25:62–77. doi: 10.1006/meth.2001.1216. [DOI] [PubMed] [Google Scholar]
- 44.Zhong D.P., Pal S.K., Zewail A.H. Femtosecond studies of protein-DNA binding and dynamics: histone I. Chem. Phys. Chem. 2001;2:219–227. doi: 10.1002/1439-7641(20010417)2:4<219::AID-CPHC219>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 45.Martinez K.L., Corringer P.J., Edelstein S.J., Changeux J.P., Merola F. Structural differences in the two agonist binding sites of the Torpedo nicotinic acetylcholine receptor revealed by time-resolved fluorescence spectroscopy. Biochemistry. 2000;39:6979–6990. doi: 10.1021/bi992811p. [DOI] [PubMed] [Google Scholar]
- 46.Ghiggino K.P., Lee A.G., Meech S.R., O'Connor D.V., Phillips D. Time-resolved emission spectroscopy of the dansyl fluorescence probe. Biochemistry. 1981;20:5381–5389. doi: 10.1021/bi00522a005. [DOI] [PubMed] [Google Scholar]
- 47.Kimura Y., Ikegami A. Local dielectric properties around polar region of lipid bilayer membranes. J. Membr. Biol. 1985;85:225–231. doi: 10.1007/BF01871517. [DOI] [PubMed] [Google Scholar]
- 48.Sarkar R., Ghosh M., Pal S.K. Ultrafast relaxation dynamics of a biologically relevant probe dansyl at the micellar surface. J. Photochem. Photobiol. B. 2005;78:93–98. doi: 10.1016/j.jphotobiol.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Stubbs C.D., Meech S.R., Lee A.G., Phillips D. Solvent relaxation in lipid bilayers with dansyl probes. Biochim. Biophys. Acta. 1985;815:351–360. doi: 10.1016/0005-2736(85)90361-x. [DOI] [PubMed] [Google Scholar]
- 50.Bhattacharyya K., Bagchi B. Slow dynamics of constrained water in complex geometries. J. Phys. Chem. A. 2000;104:10603–10613. [Google Scholar]
- 51.Vincent M., de Foresta B., Gallay J. Nanosecond dynamics of a mimicked membrane-water interface observed by time-resolved stokes shift of LAURDAN. Biophys. J. 2005;88:4337–4350. doi: 10.1529/biophysj.104.057497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coïc Y.M., Vincent M., Gallay J., Baleux F., Mousson F. Single-spanning membrane protein insertion in membrane mimetic systems: role and localization of aromatic residues. Eur. Biophys. J. 2005;35:27–39. doi: 10.1007/s00249-005-0002-1. [DOI] [PubMed] [Google Scholar]
- 53.Vincent M., Gallay J., Jamin N., Garrigos M., de Foresta B. The predicted transmembrane fragment 17 of the human multidrug resistance protein 1 (MRP1) behaves as an interfacial helix in membrane mimics. Biochim. Biophys. Acta. 2007;1768:538–552. doi: 10.1016/j.bbamem.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 54.Sen S., Sukul D., Dutta P., Bhattacharyya K. Fluorescence anisotropy decay in polymer-surfactant aggregates. J. Phys. Chem. A. 2001;105:7495–7500. [Google Scholar]
- 55.Frezzato D., Polimeno A., Ferrarini A., Moro G.J. Stochastic modelling of roto-translational motion of dyes in micellar environment. Theor. Chem. Acc. 2007;117:1017–1027. [Google Scholar]
- 56.Breukink E., de Kruijff B. The lantibiotic nisin, a special case or not? Biochim. Biophys. Acta. 1999;1462:223–234. doi: 10.1016/s0005-2736(99)00208-4. [DOI] [PubMed] [Google Scholar]
- 57.Bernik D.L., Negri R.M. Local polarity at the polar head level of lipid vesicles using dansyl fluorescent probes. J. Colloid. Interface. Sci. 1998;203:97–105. [Google Scholar]
- 58.Hasper H.E., Kramer N.E., Smith J.L., Hillman J.D., Zachariah C. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science. 2006;313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- 59.Smith L., Hasper H., Breukink E., Novak J., Cerkasov J. Elucidation of the antimicrobial mechanism of mutacin 1140. Biochemistry. 2008;47:3308–3314. doi: 10.1021/bi701262z. [DOI] [PubMed] [Google Scholar]
- 60.Kinosita K.J., Kawato S., Ikegami A. A theory of fluorescence polarization decay in membranes. Biophys. J. 1977;20:289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


