Abstract
It has been established that a long DNA molecule exhibits a large discrete conformational change from a coiled state to a highly folded state in aqueous solution, depending on the presence of various condensing agents such as polyamines. In this study, T4 DNA labeled with fluorescent dyes was encapsulated in a cell-sized microdroplet covered with a phospholipid membrane to investigate the conformational behavior of a DNA molecule in such a confined space. Fluorescence microscopy showed that the presence of Mg2+ induced the adsorption of DNA onto the membrane inner-surface of a droplet composed of phosphatidylethanolamine, while no adsorption was observed onto a phosphatidylcholine membrane. Under the presence of spermine (tetravalent amine), DNA had a folded conformation in the bulk solution. However, when these molecules were encapsulated in the microdroplet, DNA adsorbed onto the membrane surface accompanied by unfolding of its structure into an extended coil conformation under high concentrations of Mg2+. In addition, DNA molecules trapped in large droplets tended not to be adsorbed on the membrane, i.e., no conformational transition occurred. A thermodynamic analysis suggests that the translational entropy loss of a DNA molecule that is accompanied by adsorption is a key factor in these phenomena under micrometer-scale confinement.
Introduction
It has been revealed that long DNA molecules exhibit a large discrete change in conformation from a coiled state to a highly compact folded state, such as a toroid or rod structure, in aqueous solution, depending on the presence of various condensing agents including cellular environmental factors such as polyamines (spermine and spermidine) and polyphosphates (1–5). This characteristic is based on the fact that DNA is a negatively-charged semiflexible polymer. Single-molecule observation studies have revealed that the conformational transition of a long DNA molecule larger than several tens of kilo basepairs (kbp) is discrete at the single-molecule level, and can be described in terms of a first-order phase transition under the criterion of Landau (6,7).
In a cell, DNAs and other molecules are present in a micrometer-scale confined space. To understand the characteristics and driving forces of the molecular events that occur within a cell, studies under conditions of cell-sized confinement, in addition to those in a bulk system, are needed. Recently, it has become possible to create the giant phospholipid vesicles (diameter > 1 μm) that encapsulate DNAs, proteins, and other biochemical reaction components using several methods, and artificial cell model studies in which various reactions such as transcription and translation are reconstituted in the vesicle has been progressing (8–13). We have also studied the formation of cell-sized aqueous droplets coated with a phospholipid membrane in oil (i.e., water-in-oil microdroplet; see Fig. 1) (14).
Figure 1.
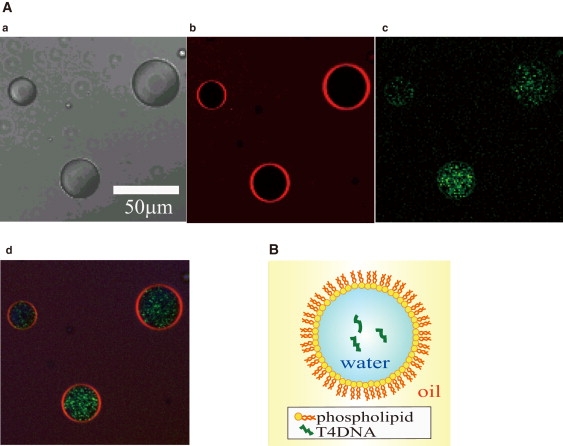
Water-in-oil microdroplet encapsulating T4 DNA. (A) Confocal laser scanning microscopy images of the prepared DOPE water-in-oil microdroplets. (a) Bright-field image, (b) fluorescence image (pseudo-color) of a fluorescent phospholipid, TRITC-DHPE, (c) fluorescence image (pseudo-color) of T4 DNA labeled with YOYO-1, and (d) merger of panels b and c. (B) A schematic representation of the prepared droplet; the hydrophilic headgroup of a DOPE molecule faces the aqueous solution phase and the hydrophobic alkyl chain faces oil, and DOPE molecules are aligned to form a phospholipid layer (membrane), which covers the aqueous solution phase inside. The droplet encapsulates T4 DNA molecules.
In this study, to elucidate the characteristics of the conformational behavior of DNA molecules in a confined space, a large DNA (bacteriophage T4 DNA of 166 kbp labeled with fluorescent dyes) was encapsulated in a water-in-oil microdroplet, and the distribution and conformation of DNA within the droplet was investigated under fluorescence microscopy. The presence of Mg2+ induced the adsorption of DNA onto the membrane inner-surface of the droplet when it was composed of DOPE phospholipid, while no adsorption was observed when the membrane was composed of eggPC phospholipid. We also found that under the presence of spermine, adsorption was accompanied by a conformational transition of the DNA molecule from the folded state to the unfolded coil state on the membrane surface. Moreover, DNA molecules trapped in large droplets tended not to be adsorbed on the membrane surface and instead be in the folded state in the aqueous phase. A thermodynamic analysis of these observations suggests that the translational entropy loss of the DNA molecule caused by adsorption (from confinement in a three-dimensional space to that on a two-dimensional surface) works as a key factor that determines the conformational behavior of DNA molecules under confinement within a micrometer-scale space.
Materials and Methods
Labeling of T4 DNA with the fluorescent dye YOYO-1
Bacteriophage T4 DNA was purchased from Nippon Gene (Tokyo, Japan). To label T4 DNA with YOYO-1 iodide (Molecular Probes, Eugene, OR), an equal volume of 1.5 μM YOYO-1 in water was added to 10 ng/μL of T4 DNA (molar ratio of basepairs to YOYO-1 was 10), and the mixture was allowed to stand overnight at 4°C. For the observation of T4 DNA molecules under a microscope, T4 DNA/YOYO-1 solution was diluted 10- to 100-fold in 10 mM Tris-HCl, pH 7.4, 100 mM KCl, and various concentrations of MgCl2 and spermine (Nakarai Tesque, Kyoto, Japan). To avoid the intermolecular aggregation of T4 DNA molecules induced by the addition of spermine, we used T4 DNA at a very low concentration (0.1–1.0 ng/μL).
Preparation of water-in-oil microdroplet encapsulating T4 DNA
1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and L-α-lysophosphatidylcholine from chicken egg (eggPC) were obtained from Avanti Polar Lipids (Alabaster, AL), and n-(6-tetramethylrhodamineehiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TRITC-DHPE) from Molecular Probes. The water-in-oil microdroplet (Fig. 1) was prepared as described previously (14). Briefly, a dry film of phospholipid (DOPE or eggPC) was made on the bottom of a glass tube, and mineral oil (Nakarai Tesque) was added to the phospholipid film to give a final concentration of 1.0 mM of phospholipid in mineral oil. To observe the phospholipid distribution in the prepared sample, 1 mol % of TRITC-DHPE was mixed with DOPE in chloroform solution, and the mixture was dried to a film on the bottom of a glass tube. The sample was sonicated at 50°C for 60 min to disperse the phospholipid in oil. Next, 5–10% (V/V) aqueous solution (10 mM Tris-HCl pH 7.4, 100 mM KCl with various concentrations of MgCl2 and spermine) containing T4 DNA molecules labeled with YOYO-1 was added to the lipid/oil solution and mixed well by pipetting. The resulting emulsion contained many aqueous droplets coated with phospholipid thin layers, with diameters that ranged from 5 to 200 μm (mainly 20–60 μm) (Fig. 1 A).
Confocal laser scanning microscopy
An aliquot of the emulsion was placed on a silicon-coated coverslip. Bright-field and fluorescence microscope images were obtained by a model No. LSM510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany). The samples were excited with a model No. LGK7812ML4 argon laser (488 nm; Lasertechnik, Jena, Germany) and the images were obtained through a 505–530-nm bandpass filter for YOYO-1-labeled T4 DNA molecules. A model No. LGK 7786P helium-neon laser (543 nm; Lasertechnik) with a 560-nm long-path filter was used for the imaging of TRITC-DHPE.
Single molecule observation of T4 DNA under fluorescence microscopy
Fluorescence images of each T4 DNA molecule were observed and recorded under a model No. TE2000U microscope with a ×100 objective (Plan Fluor 100/1.30 Oil Ph3 DLL; Nikon, Tokyo, Japan) and a model No. C7190-23 EB-CCD camera (Hamamatsu, Hamamatsu City, Japan). Excitation was performed with a model No. C-SHG1 Hg lamp (Nikon), and fluorescence images through a model No. DVS-20 ARGUS-10 image processor (Hamamatsu) were recorded on a model No. RD-XS38 DVD recorder (Toshiba, Tokyo, Japan). The image of each DNA molecule was recorded for >5 s. On the recorded image, the conformation of each T4 DNA molecule was classified as coiled (unfolded) or compact (folded), and the apparent long-axis length L, which was defined as the longest distance in the outline of the DNA image during the 5 s of image-acquisition, was measured. Since there is a blurring effect of ∼0.3 μm on the observed fluorescence images due to the technical characteristics of the resolution limit (5), the observed DNA images are slightly larger than the actual size of the DNAs.
Results
Confinement of DNA molecules in a water-in-oil microdroplet
As previously reported (15), a T4 DNA molecule (166 kbp) has a free coiled conformation in an aqueous solution. We encapsulated DNA molecules, labeled with YOYO-1, within water-in-oil microdroplets composed of DOPE. Fig. 1 shows confocal laser scanning microscopy images of the prepared droplets. In the fluorescence images (pseudo-color) in Fig. 1 A, red indicates a fluorescent lipid analog, TRITC-DHPE, and green indicates YOYO-1, which bound to T4 DNA molecules. The images show that our preparation made droplets with diameters of mostly 20 to 60 μm, each of which was coated with phospholipid thin layers, and these encapsulated T4 DNA molecules. In the preparation, 0.1–1.0 ng/μL of T4 DNA aqueous solution was mixed with the lipid-dispersion-in-oil solution, which corresponds to 18–180 T4 DNA molecules in a droplet 40 μm in diameter. We analyzed droplets with diameters of 20–60 μm below, unless otherwise indicated.
DNA is adsorbed onto the DOPE membrane surface in the presence of Mg2+
First, we investigated the distribution of T4 DNA molecules within a microdroplet (Fig. 2). In a droplet composed of DOPE, DNA molecules were diffusely distributed in the aqueous phase of the droplet in the absence of Mg2+ (Fig. 2 a). When Mg2+ was present at 10 mM, almost all of the DNA molecules were located on the inner-surface of the droplet (phospholipid membrane surface) (Fig. 2 b). The localization of DNA molecules on the membrane surface in the presence of Mg2+ is a phospholipid headgroup-dependent phenomenon. When the droplets were prepared with eggPC instead of DOPE, T4 DNA molecules were distributed in the aqueous phase and were not bound to the membrane surface even in the presence of 10 mM Mg2+ (B).
Figure 2.
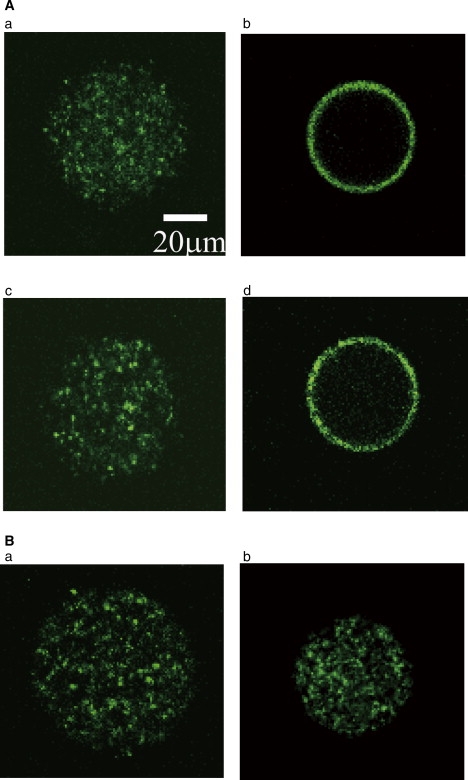
Distribution of T4 DNA molecules in a microdroplet. Confocal fluorescence microscope images of T4 DNA labeled with YOYO-1. (A) DOPE droplet. (a) Mg2+ 0 mM, spermine 0 mM, (b) Mg2+ 10 mM, spermine 0 mM, (c) Mg2+ 0 mM, spermine 1.5 mM, and (d) Mg2+ 10 mM, spermine 1.5 mM. The preparations were repeated three times, and >60 droplets with diameter of 20-60 μm were observed for each condition. In 0 mM Mg2+ (a and c), DNA was located on the inner-surface in none of the droplets. In 10 mM Mg2+ (b and d), in ∼95 % of the droplets, almost all of the DNA molecules were located on the inner-surface of the droplet, as shown. (B) eggPC droplet. (a) Mg2+ 0 mM, spermine 0 mM, (b) Mg2+ 10 mM, spermine 0 mM.
Next, the conformation of individual T4 DNA molecules in the droplet was examined under a fluorescence microscope (Fig. 3; movies are available in the Supporting Material). We first observed and recorded DNA molecules in the bulk aqueous solution, and the sample was then encapsulated into droplets. On recorded images, the apparent long-axis length of each DNA molecule was estimated. The distributions of the long-axis length under various conditions are shown in Fig. 4 along with those in the bulk aqueous solution for comparison. For each condition, >50 DNA molecules were analyzed.
Figure 3.
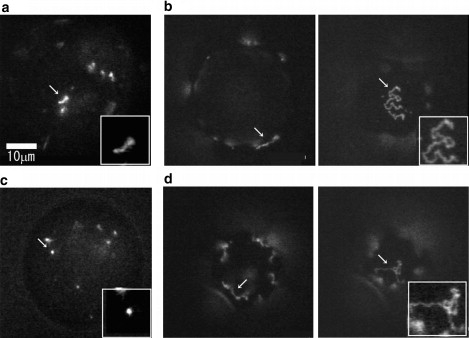
Single-molecule images of T4 DNA in a DOPE droplet. In each image, one droplet is shown. The arrow indicates one T4 DNA molecule, which is also shown separately in the inset. (a) Mg2+ 0 mM, spermine 0 mM. (b) Mg2+ 10 mM, spermine 0 mM. (Left image) The focus is on the center of the droplet in the z axis. T4 DNA molecules are seen along the periphery of the droplet. (Right image) The focus is on the bottom surface of the droplet and a T4 DNA molecule on the inner-surface of the membrane is seen. (c) Mg2+ 0 mM, spermine 1.5 mM. (d) Mg2+ 10 mM, spermine 1.5 mM. (Left image) The focus is on the center of the droplet in the z axis; (right image) the focus is on the bottom surface of the droplet.
Figure 4.
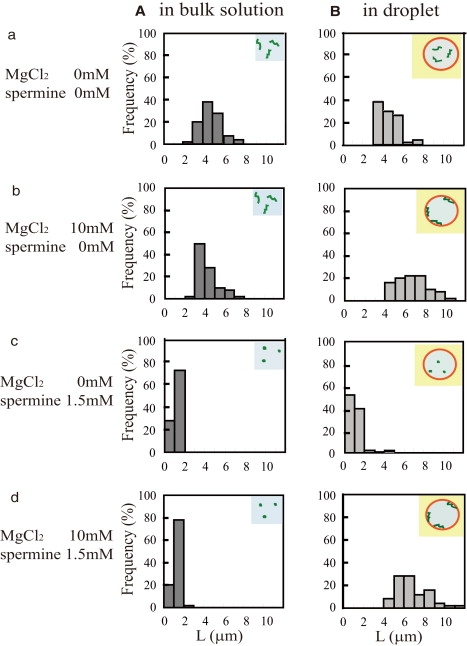
Histogram of the long-axis length (L) of T4 DNA molecules under various conditions. For each condition, >50 T4 DNA molecules were analyzed.
In the aqueous phase of the droplet in the absence of Mg2+, T4 DNA had a free coiled conformation with intramolecular chain fluctuation, and underwent relatively slow translational diffusion in the droplet (Fig. 3 a and Movie S1), almost the same as in the bulk solution (data not shown). The mean long-axis length of DNA molecules in the droplet was 4.53 ± 0.95 μm and that in the bulk solution was 4.87 ± 1.01 μm (Fig. 4 a).
In the bulk solution with 10 mM Mg2+, T4 DNA had almost the same conformation and intramolecular motion as in the absence of Mg2+ (the mean long-axis length was 4.26 ± 1.03 μm). When this sample was encapsulated into the droplet, almost all of the DNA molecules were adsorbed onto the inner surface of the droplet. The full length of the molecule was on the membrane, and the molecule exhibited a quasi-two-dimensional coiled state with intramolecular chain motion on the membrane surface (Fig. 3 b and Movie S2). The average long-axis length was 6.66 ± 1.48 μm (Fig. 4 B (b)). As we discuss later, this implies that roughly 10% of the segments of the DNA molecule were attached to the membrane surface.
Conformational transition of DNA molecule from the folded state to the unfolded state occurs in a microdroplet through interplay with the phospholipid membrane
As we previously reported (16), polyamines such as spermidine and spermine induce a conformational transition of long DNA molecules of more than several tens of kbp from a coil state to a highly compact folded state in aqueous solution. In the buffered solution examined in this study (10 mM Tris-HCl, pH 7.4, 100 mM KCl), all of the T4 DNA molecules showed a highly compact conformation in 1.5 mM spermine (data not shown).
DNA with a compact (folded) conformation was encapsulated into a DOPE microdroplet, and the distribution and conformation of DNA molecules in the droplet were investigated. In the absence of Mg2+, T4 DNAs were mainly located in the aqueous phase while some were on the membrane surface (Fig. 2 c). Single-molecule observation showed that the DNA molecules in the aqueous phase had a compact conformation (Fig. 3 c and Movie S3). The estimated long-axis length was 1.15 ± 0.74 μm, which is almost the same as that in the bulk solution (1.06 ± 0.19 μm) (Fig. 4 c). In the presence of 10 mM Mg2+ with 1.5 mM spermine, in the bulk solution, the DNA molecules had a highly compact and folded conformation and the mean long-axis length of DNA molecules was 1.15 ± 0.71 μm (Fig. 4 A (d)), which shows that the presence of Mg2+ had no apparent effect. When these DNA molecules were encapsulated within a DOPE droplet, most of them were located on the membrane surface (Fig. 2 d). On the membrane, they exhibited an unfolded coil conformation (Fig. 3 d) and underwent intramolecular chain motion above the membrane surface (Movie S4), as with the presence of Mg2+ without spermine. The mean long-axis length was 6.80 ± 1.59 μm (Fig. 4 B (d)). These results show that a DNA molecule with a compact conformation in the bulk solution unfolds its structure through interplay with the phospholipid membrane surface when encapsulated within a microdroplet.
Effects of Mg2+ and spermine: phase diagram
We investigated the distribution and conformation of DNA molecules in a DOPE droplet under various concentrations of Mg2+ (0, 3, 5, 7, 8, and 10 mM) and spermine (0, 0.5, 1.0, 1.5, and 1.8 mM). For each condition, >20 droplets were examined. Based on the results, a phase diagram was constructed with the spermine concentration as the x axis and the Mg2+ concentration as the y axis (Fig. 5). Since there is a relatively broad intermediate region for both the spermine and Mg2+ concentrations, in which DNA molecules with various states (coil and compact, on the membrane and in the aqueous phase) coexist within the same droplet, the diagram can be described only in a rough manner. Also, in the intermediate region, DNA molecules with intramolecular phase segregation (17,18), in which a coil part and a folded part coexist on a single DNA molecule, were frequently observed both in the aqueous phase and on the membrane surface (data not shown). This state emerges in the intermediate region for the coil-globule transition when the correlation length in the transition becomes shorter than the contour length of the DNA molecule under specific conditions such as a high salt concentration. This intramolecular segregation state can be considered to be equivalent to the state of the coexistence of the coil conformation and the folded conformation of a whole DNA molecule when the correlation length is longer than the contour length (17,18). The experimental condition used in this study (10 mM Tris-HCl and 100 mM KCl) is close to that (10 mM Tris-HCl and 100 mM NaCl) under which intramolecular segregation of T4 DNA molecules was reported (18).
Figure 5.
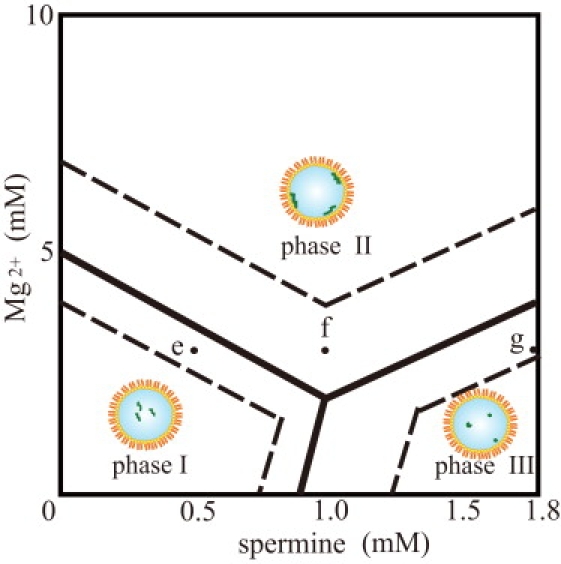
Phase diagram for the Mg2+ and spermine concentrations. Phase I: DNA has a coil conformation in the aqueous phase. Phase II: DNA is adsorbed onto the membrane surface. Phase III: DNA has a highly compact conformation in the aqueous phase. The field surrounded with broken lines is the intermediate region, in which different states of DNA molecules (coil and compact, in the aqueous phase and on the membrane surface) coexist within a single droplet.
As shown in the phase diagram, when the Mg2+ concentration was sufficiently high, DNA molecules showed an adsorbed and unfolded state on the membrane surface under all spermine concentrations ranging from 0 mM to 1.8 mM (phase II). Mg2+ works to induce and likely stabilizes the adsorption of DNA molecules onto the DOPE membrane surface, but does not induce the folding transition of DNA. When spermine was present at low concentrations under which it does not induce DNA molecules to have a folded state (phase I), the adsorption of DNA molecules onto the membrane surface occurred at a lower Mg2+ concentration as the spermine concentration increased. These results show that spermine also works for the adsorption of DNA molecules, although it prefers to work for induction of a conformational transition into a folded state. Indeed, in 0–3 mM Mg2+ with 0–0.5 mM spermine, most of the DNA molecules were present in the aqueous phase in the coiled state (e). When the spermine concentration increased to 1.0 mM, many DNA molecules were located on the membrane surface (some were in the aqueous phase) (f). When the spermine concentration further increased to 1.8 mM, most of DNAs were present in the aqueous phase in the folded state (g).
Thermodynamic considerations regarding our observations
It has been established that a long DNA molecule exhibits a conformational transition between a coiled state and a folded/compact state in aqueous solution (5–7). In this study, we found that when a DNA molecule is enclosed within a microdroplet coated with a phospholipid membrane, the molecule exhibits an unfolded coil state on the membrane surface under the same experimental conditions under which it exhibits a folded state in the bulk aqueous solution. To understand the characteristics of this conformational transition of DNA molecules within a finite volume, a thermodynamic analysis was performed on the interplay between DNA adsorption and folding within such a finite volume.
A DNA molecule is assumed to have one of the following states in a droplet:
-
1.
A free coil conformation in the aqueous solution phase.
-
2.
A quasi-two-dimensional coil conformation that is adsorbed onto the phospholipid membrane surface.
-
3.
A compactly folded conformation in the aqueous solution phase.
Let us consider a single DNA molecule as a semiflexible polymer of contour length L, segment (persistence) length λ (= 100 nm, 300 basepairs), and segment number N = L/λ, which is enclosed in a volume V ≃ R3 (R is the radius of the droplet). In the absence of spermine, DNA takes a random coil conformation. By setting this state as a reference, we can write its free energy as the translational entropy of the molecule within the volume,
where k and T are the Boltzmann constant and temperature, respectively, and φ1 ≃ r13/V (r1 is the size of DNA coil in three-dimensional space). This signifies that the F1 state free energy shows logarithmic dependence on the system size (V) through the volume fraction φ1.
The adsorbed state can be characterized as follows. Movie images of DNA molecules adsorbed onto the membrane surface (Movie S2 and Movie S4) show that the adsorption is rather weak, which means that only a fraction of the segment is attached to the membrane and other segments between the attached points are free from the surface, so that they exhibit conformational fluctuations. This indicates that an adsorbed DNA molecule can be described as a two-dimensional self-avoiding walk of blobs of size ξ ≃ λ g1/2, where g is the number of segments inside a blob (19). The overall two-dimensional size of DNA is r2 ≃ ξ(N/g)3/4 ≃ λN3/4g−1/4, whereas the size of the free DNA in three-dimensional space (r1) is given by ≃λN1/2, since the excluded volume effect becomes apparent only for very long DNA due to its local stiffness. If we compare these expressions with the experimental results (long-axis length (6.66 μm) for a DNA molecule in the adsorbed state on the membrane surface and that (4.53 μm) in the free coil state in the aqueous phase (Fig. 4)), the g value can be estimated to be ∼10. The free energy of the adsorbed state may be written as
where −ɛa is the change in free energy due to the DNA-membrane surface interaction per segment, and φ2 ≃ r22/R2 is the area fraction of blobs on the surface.
The addition of spermine induces the folding transition of the DNA molecule. If we denote the change in free energy due to the folding per segment as −ɛf, a similar consideration leads to the free energy of the folded state,
where φ3 ≃ r33/V (r3 is the size of folded DNA).
Although the detailed modeling and calculations for determining the quantitative phase behaviors are beyond the scope of this article, we can determine some basic features at a qualitative level with the present phenomenological modeling. The results of such an analysis are summarized in Fig. 6 in the form of a state diagram with ɛf/kT as the x axis and ɛa/kT as the y axis. When F1 < F2 and F1 < F3, where both the Mg2+ and spermine concentrations are sufficiently low, DNA has a coil conformation in the aqueous phase. The experimental results show that Mg2+ induces the adsorption of DNA onto the membrane surface. With regard to folding, it was observed that Mg2+ exhibited some inhibitory effect in the coexistence region for relatively low concentrations of Mg2+ and spermine. This would be caused by their competitive binding to the phosphate groups of DNA. Therefore, we can assume that ɛa increases and ɛf slightly decreases as the Mg2+ concentration increases. When Mg2+ is absent or present at only a low concentration, ɛa has a negative or a small positive value, and thus F1 < F2, resulting in no adsorption. As the Mg2+ concentration increases, the ɛa value increases, and at some point (F1 = F2, line 2) adsorption takes place.
Figure 6.
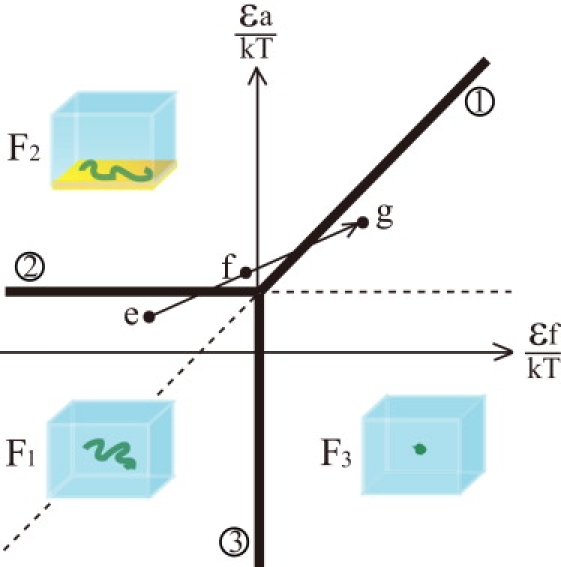
State diagram of DNA molecules in a microdroplet. Three states are assumed: F1, a free coil conformation in the aqueous solution phase; F2, a quasi-two-dimensional coil conformation adsorbed onto the surface of the phospholipid membrane; and F3, a compactly folded conformation in the aqueous solution phase. On line 1, where ɛa/kT ≃ ɛf/kT + 1/N ln(R/λ), the free energy of the F2 state is equal to that of the F3 state. On line 2, where ɛa/kT ≃ 1/N ln(R/λ), the free energy of the F1 state is equal to that of the F2 state. On line 3, where ɛf/kT ≃ 0, the free energy of the F1 state is equal to that of the F3 state.
Spermine strongly induces the folding of DNA, which occurs by the binding of spermine to the phosphate groups of the DNA molecule (20,21). It was observed that spermine also has some inducible effect on adsorption of DNA onto the membrane surface. As the spermine concentration increases, ɛf would increase and ɛa would also somewhat increase, although in most cases the effect on the induction of folding surpasses that on the induction of adsorption. When the spermine concentration is low, ɛf is negative because of electrostatic repulsion between the phosphate groups of DNA. As the spermine concentration increases, the ɛf value increases due to more neutralization of the phosphate groups' negative charges, and the folding transition takes place at ɛf ≃ 0 (F1 = F3, line 3).
Under the coexistence of relatively high concentrations of Mg2+ and spermine, one can see the interplay between ɛa and ɛf. A high Mg2+ concentration gives a sufficiently high ɛa value so that F2 < F3 (above line 1), i.e., the state of adsorption is more stable than the folding state, and thus a DNA molecule is adsorbed onto the membrane surface accompanied by unfolding of its structure. In the intermediate concentration region for both Mg2+ and spermine, one can see the case that a DNA molecule in the F1 state transitions to the F2 state and finally goes to the F3 state as the spermine concentration increases. In (e) in the diagram, the Mg2+ concentration is not high enough for adsorption. When the spermine concentration increases, ɛf increases, but this increase is not sufficient for the folding transition to take place. Spermine can also increase ɛa to become larger than the value on line 2, and DNA moves to the F2 state (f). When the spermine concentration increases further, ɛf becomes sufficiently high so that the folding state is more stable than the adsorption state at that Mg2+ concentration (g). In the experiments, this case was actually observed in 3–5 mM Mg2+, as described in the previous section (Fig. 5).
In summary, Mg2+ and spermine act cooperatively for adsorption and competitively for folding. Thus, our theoretical diagram is in good agreement with the experimental phase diagram.
Droplet-size dependence
The state diagram in Fig. 6 shows that the adsorption line (line 1 and line 2) depends on the droplet radius R through 1/N ln(R/λ). The expression ln(R/λ) represents the translational entropy loss of a DNA molecule caused by adsorption (kT ln(φ2/φ1), kT ln(φ2/φ3)), i.e., from a confined state in three-dimensional space to that on a two-dimensional surface. This means that the adsorption of a DNA molecule confined within a larger volume is accompanied by larger translational entropy loss of the molecule, which gives a higher adsorption line along the y axis for DNA molecules trapped in a larger droplet.
To examine the above consideration experimentally, we investigated the distribution of DNAs (on membrane surface or in aqueous phase) in 200 droplets with diameters ranging from 20 μm to 200 μm in 10 mM Mg2+ and 1.5 mM spermine. Fig. 7 shows the dependence of the DNA distribution on droplet-size when 1.0 ng/μL of DNA solution was encapsulated (180 and 22,500 DNA molecules in 40 μm and 200 μm droplets, respectively). Almost the same results were obtained when 0.1 ng/μL of DNA solution was used. When the droplet size was <60 μm in diameter, adsorption of almost all of the DNA molecules onto the membrane surface was observed in >90% of droplets. When the diameter became larger than 80 μm, there were droplets in which some DNA molecules were adsorbed and others were in the aqueous phase with a folded conformation. In these droplets, it is considered that the translational entropy loss of a DNA molecule by adsorption (kT ln(R/λ)) is comparable to the difference in the free energy gain between DNA-membrane surface interaction and segment interaction within a folded DNA molecule (Nɛa-Nɛf). In most of the droplets with a diameter larger than 100 μm, either all of the DNA molecules were in the aqueous phase or the two states of DNA coexisted. To summarize, the results show that DNA molecules trapped within a larger-size droplet tend to be in the folded state in the aqueous phase, and this corresponds well to the prediction derived from the thermodynamic analysis, which indicates that the confinement volume is an effective parameter for the conformational transition from a folding state to an unfolded state adsorbed on a membrane surface.
Figure 7.
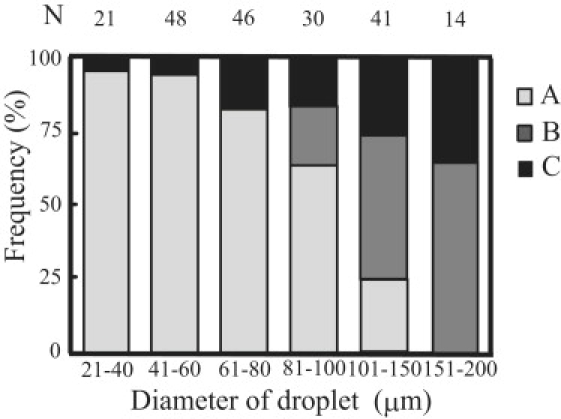
Dependence of the DNA distribution on droplet-size in DOPE microdroplets. DOPE microdroplets encapsulating T4 DNA with 10 mM Mg2+ and 1.5 mM spermine were observed under confocal microscopy. Droplets with diameters ranging from 20 μm to 200 μm were classified into three types, based on the DNA distribution in the droplet. (A) Almost all of the DNAs are located on the membrane surface. (B) Some DNAs are on the membrane surface and others are in the aqueous phase. (C) Almost all of the DNAs are in the aqueous phase. The sample preparations were repeated three times, and 200 droplets were observed. N for each bar indicates the number of droplets observed.
Discussion
In this study, we observed that a long DNA is adsorbed and has an unfolded conformation on a PE membrane surface in the presence of Mg2+ in a microdroplet (Figs. 2–4). The interaction of DNA with cationic lipids has been extensively investigated, and the binding of DNA to the membrane or an interface that includes these cationic lipids has been reported (22–24). For a zwitterionic phospholipid, such as PE or PC, it has been shown that these phospholipids do not interact with DNA directly, but the interaction can be mediated by divalent cations such as Ca2+ and Mg2+ (25–30). This is considered to be due to the fact that divalent cations bridge the negative phosphate moieties of the lipid headgroups and the lipid layer becomes positively charged (25,26). In the case observed in this study, the interaction between the DNA molecule and the PE membrane surface is considered to be rather weak. Indeed, a DNA molecule was attached to the membrane at several to 10 points within the molecule, and other parts of the molecule between the attachment points were free from the membrane surface (Movie S2 and Movie S4). In addition, DNA molecules were not adsorbed onto an eggPC membrane even in the presence of 10 mM Mg2+ (Fig. 2). Considering the previous reports (27–29), the electrostatic property of the PC membrane surface of the present microdroplet would not be much different from that of the PE membrane in the presence of Mg2+, and the surfaces of both the PE membrane and the PC membrane would be slightly positively charged. Therefore, we can assume that the ɛa value for the PE membrane in the Mg2+ concentration is not so much larger than that for the PC membrane. If this assumption is valid, although there is only a small difference in the free energy gain due to the interaction of DNA with the PE membrane surface and that with the PC membrane surface, DNAs are adsorbed onto the PE membrane but not onto the PC membrane in the microdroplet.
Translational entropy loss by adsorption, i.e., from a state confined in three-dimensional space to that on two-dimensional surface, becomes much smaller when a molecule is confined within a micrometer-scale space. Under the cases that the translational entropy loss by adsorption is comparable to the free energy gain due to the interaction between DNA and membrane, the small difference in the electrostatic property of the membrane surface and/or confinement volume should determine whether or not the adsorption takes place. The experimental finding that DNA molecules trapped within larger droplets tend to be in the aqueous phase (Fig. 7) suggests that this delicate balance is at work in this system. Under the specific conditions of the presence of a condensing agent (spermine) for DNA, the confinement in tens of micrometer-scale space even causes the conformational transition in DNA molecules from a folded state to an unfolded state with the help of interaction with the phospholipid membrane (Figs. 3 and 4).
A cell is a micrometer-scale confined space that includes many kinds of macro- and small molecules, within which various reactions and events occur. Most of these molecular events proceed accompanied by the formation of macromolecule complexes and the binding of small molecules such as Ca2+, Mg2+, and NTPs to the macromolecules. To reveal the characteristics and driving forces of these molecular events and their regulations occurring within a cell, they should be experimentally investigated under confinement within a cell-sized space as well as in a bulk system. Studies in a single living cell, which have been extensively developed over the past decade (31–33), is one approach to determine the cell-sized characteristics of these molecular events. Another approach is to experimentally reconstitute a simple cell model and to study the reconstituted molecular events in the cellular environment, which has also emerged in recent years (6–14). In this study, we observed a confinement effect; a conformational transition of DNA that depends on the confinement volume, in a cell-sized droplet covered with a phospholipid membrane. These microdroplets can also be used to encapsulate various proteins and biochemical reaction components (14). The approach we have taken in this study, to prepare cell-sized droplets and to study molecular events in the cellular environment, should lead to new insights and a deeper understanding of the characteristics of living cellular systems.
Acknowledgments
The authors thank Prof. Yasunori Morimoto, Josai University, for his helpful discussion and support during the work.
This work was partly supported by Japan Society for the Promotion of Science (JSPS) under Giant-in-Aid for Creative Scientific Research (Project No.18GS0421).
Footnotes
Takahiro Sakaue's present address is Department of Physics, Graduate School of Science, Kyushu University, Fukuoka, Japan.
Supporting Material
References
- 1.Bloomfield V.A. Condensation of DNA by multivalent cations: consideration on mechanism. Biopolymers. 1991;31:1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- 2.Bloomfield V.A. DNA condensation. Curr. Opin. Struct. Biol. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 3.Marquet R., Houssier C. Thermodynamics of cation-induced DNA condensation. J. Biomol. Struct. Dyn. 1991;9:159–167. doi: 10.1080/07391102.1991.10507900. [DOI] [PubMed] [Google Scholar]
- 4.Wisdom J., Baldwin R.L. Monomolecular condensation of DNA induced by cobalt hexamine. Biopolymers. 1983;22:1595–1620. doi: 10.1002/bip.360220612. [DOI] [PubMed] [Google Scholar]
- 5.Yoshikawa K. Controlling the higher-order structure of giant DNA molecules. Adv. Drug Deliv. Rev. 2001;52:235–244. doi: 10.1016/s0169-409x(01)00210-1. [DOI] [PubMed] [Google Scholar]
- 6.Mel'nikov S.M., Sergeyev V.G., Yoshikawa K. Discrete coil-globule transition of large DNA induced by cationic surfactant. J. Am. Chem. Soc. 1995;117:2401–2408. [Google Scholar]
- 7.Yoshikawa K., Takahashi M., Vasilevskaya V.V., Khokhlov A.R. Large discrete transition in a single DNA molecule appears continuous in the ensemble. Phys. Rev. Lett. 1996;76:3029–3031. doi: 10.1103/PhysRevLett.76.3029. [DOI] [PubMed] [Google Scholar]
- 8.Luisi P.L., Ferri F., Stano P. Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften. 2006;93:1–13. doi: 10.1007/s00114-005-0056-z. [DOI] [PubMed] [Google Scholar]
- 9.Murtas G., Kuruma Y., Bianchini P., Diaspro A., Luisi P.L. Protein synthesis in liposomes with a minimal set of enzymes. Biochem. Biophys. Res. Commun. 2007;363:12–17. doi: 10.1016/j.bbrc.2007.07.201. [DOI] [PubMed] [Google Scholar]
- 10.Chen I.A., Salehi-Ashtani K., Szostak J.W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005;127:13213–13219. doi: 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanczyc M.M., Fujikawa S.M., Szostak J.W. Experimental models of primitive cellular compartments: encapsulation, growth, and division. Science. 2008;302:618–622. doi: 10.1126/science.1089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kita H., Hosoda K., Sunami T., Ichihashi N., Matsuura T. Replication of genetic information with self-encoded replicase in liposomes. ChemBioChem. 2008;9:2403–2410. doi: 10.1002/cbic.200800360. [DOI] [PubMed] [Google Scholar]
- 13.Pautot S., Frisken B.J., Weitz D.A. Production of unilamellar vesicles using an inverted emulsion. Langmuir. 2003;19:2870–2879. [Google Scholar]
- 14.Hase M., Yamada A., Hamada T., Baigl D., Yoshikawa K. Manipulation of cell-sized phospholipid-coated microdroplets and their use as biochemical microreactors. Langmuir. 2007;23:348–352. doi: 10.1021/la0618521. [DOI] [PubMed] [Google Scholar]
- 15.Zinchenko A.A., Yoshikawa K. Na+ shows a markedly higher potential than K+ in DNA compaction in a crowded environment. Biophys. J. 2005;88:4118–4123. doi: 10.1529/biophysj.104.057323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi M., Yoshikawa K., Vasilevskaya V.V., Khokhlov A.R. Discrete coil-globule transition of single duplex DNAs induced by polyamine. J. Phys. Chem. B. 1997;101:9396–9401. [Google Scholar]
- 17.Yamasaki Y., Teramoto Y., Yoshikawa K. Disappearance of the negative charges in giant DNA with a folding transition. Biophys. J. 2001;80:2823–2832. doi: 10.1016/S0006-3495(01)76249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takagi S., Tsumoto K., Yoshikawa K. Intra-molecular phase segregation in a single polyelectrolyte chain. J. Chem. Phys. 2001;114:6942–6949. [Google Scholar]
- 19.de Gennes P.-G. Cornell University Press; Ithaca, NY: 1979. Scaling Concepts in Polymer Physics. [Google Scholar]
- 20.Gosule L.C., Schellman J.A. DNA condensation with polyamines. I. Spectroscopic studies. J. Mol. Biol. 1978;121:311–326. doi: 10.1016/0022-2836(78)90366-2. [DOI] [PubMed] [Google Scholar]
- 21.Wilson R.W., Bloomfield V. Counterion-induced condensation of deoxyribonucleic acid. A light-scattering study. Biochemistry. 1979;18:2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- 22.Koltover I., Wagner K., Safinya C.R. DNA condensation in two dimensions. Proc. Natl. Acad. Sci. USA. 2000;97:14046–14051. doi: 10.1073/pnas.97.26.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matulis D., Rouzina I., Bloomfield V. Thermodynamics of cationic lipid binding to DNA and DNA condensation: role of electrostatics and hydrophobicity. J. Am. Chem. Soc. 2002;124:7331–7342. doi: 10.1021/ja0124055. [DOI] [PubMed] [Google Scholar]
- 24.Tsukahara S., Suehara M., Fujiwara T. In situ measurements of the dynamics of single giant DNA molecules at the toluene-triocytlamine/water interfaces by a total internal reflection fluorescence microscopy. Langmuir. 2008;24:1673–1677. doi: 10.1021/la7027719. [DOI] [PubMed] [Google Scholar]
- 25.Lis L.J., Lis W.T., Parsegian V.A., Rand R.P. Adsorption of divalent cations to a variety of phosphatidylcholine bilayers. Biochemistry. 1981;20:1771–1777. doi: 10.1021/bi00510a010. [DOI] [PubMed] [Google Scholar]
- 26.Herbette L., Napolitano C., McDaniel R. Direct determination of the calcium profile structure for dipalmitoyllecithin multilayers using neutron diffraction. Biophys. J. 1984;46:677–685. doi: 10.1016/S0006-3495(84)84066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McManus J.J., Radler J.O., Dawson K.A. Phase behavior of DPPC in a DNA-calcium-zwitterionic lipid complex studied by small-angle x-ray scattering. Langmuir. 2003;19:9630–9637. [Google Scholar]
- 28.McLoughlin D., Dias R., Lindman B., Cardenas M., Nylander T. Surface complexation of DNA with insoluble monolayers. Influence of divalent counterions. Langmuir. 2005;21:1900–1907. doi: 10.1021/la047700s. [DOI] [PubMed] [Google Scholar]
- 29.Gromelski S., Brezesinski G. DNA condensation and interaction with zwitterionic phospholipids mediated by divalent cations. Langmuir. 2006;22:6293–6301. doi: 10.1021/la0531796. [DOI] [PubMed] [Google Scholar]
- 30.Tresset G., Cheong W.C.D., Lam Y.M. Role of multivalent cations in the self-assembly of phospholipid-DNA complexes. J. Phys. Chem. B. 2007;111:14233–14238. doi: 10.1021/jp0762830. [DOI] [PubMed] [Google Scholar]
- 31.Ying L. Single molecule biology: coming of age. Mol. Biosyst. 2007;3:377–380. doi: 10.1039/b702845h. [DOI] [PubMed] [Google Scholar]
- 32.Joo C., Balci H., Ishitsuka Y., Buranachai C., Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 33.Kim S.A., Heinze K.G., Schwille P. Fluorescence correlation spectroscopy in living cells. Nat. Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


