Abstract
Histone acetylation plays an important role in the regulation of gene expression. A DNA aptamer generated by in vitro selection to be highly specific for histone H4 protein acetylated at lysine 16 was used as a recognition element for atomic force microscopy-based recognition imaging of synthetic nucleosomal arrays with precisely controlled acetylation. The aptamer proved to be reasonably specific at recognizing acetylated histones, with recognition efficiencies of 60% on-target and 12% off-target. Though this selectivity is much poorer than the >2000:1 equilibrium specificity of the aptamer, it is a large improvement on the performance of a ChIP-quality antibody, which is not selective at all in this application, and it should permit high-fidelity recognition with repeated imaging. The ability to image the precise location of posttranslational modifications may permit nanometer-scale investigation of their effect on chromatin structure.
Introduction
It is widely believed that posttranslational modifications of histone proteins imprint chromatin with a gene-control language called the “histone code” (1,2). Furthermore, at least one type of biologically important modification, acetylation of histone H4 at lysine 16 (H4-K16Ac), has a dramatic effect on chromatin structure in the presence of Mg2+ (3). It would therefore be valuable to develop a technique that could report on chromatin structure on the nanometer scale and at the same time map the chemical modifications of the histone proteins that compose the core of nucleosomes.
Recognition imaging is a new technique that enables the simultaneous collection of topographical images and chemical information on a nanometer scale with an atomic force microscope (AFM) operated in aqueous electrolyte solutions (4). The recognition signal is generated by means of an affinity ligand (such as an antibody) attached to the end of an AFM probe by means of a short, flexible polyethyleneglycol (PEG) tether. Binding of the ligand causes a small displacement of the upper part of the cantilever swing, and this displacement signal is recorded simultaneously with the topographical image to generate a map of the sites where the ligand is bound.
Recognition imaging is limited primarily by the recognition ligand itself, and antibodies can suffer from batch-to-batch variability and significant cross-reactions in certain cases (5). Furthermore, the specificity of the interaction between the antibody and its antigen can be greatly reduced when the interaction is sensed over the short timescale that is characteristic of AFM imaging (S. Lindsay, unpublished data). These problems can be exacerbated when the target is distinguished only by small chemical modifications. To overcome this problem, aptamers have been developed as a replacement for antibodies (see Lin et al. (12)). Aptamers are nucleic acid molecules that behave like antibodies by adopting a well-folded tertiary structure that is complementary in shape and charge to the antigen target (6,7). However, unlike antibodies, aptamers are small, easy to engineer, and can be generated quickly using standard in vitro selection techniques. In recent years, aptamers have been developed to bind a diverse set of targets ranging from small molecules to large proteins and even whole cells (8,9). Once generated, aptamers represent an inexhaustible supply of high-quality affinity reagents that do not change composition or specificity over time. Most striking, and relevant to the detection of posttranslational modifications, is the fact that aptamers have been shown to detect small chemical changes in target molecules. Theophylline and caffeine, for example, differ in their chemical structures by one methyl group, yet a theophylline-binding aptamer has 10,000-fold weaker affinity for caffeine than theophylline (10). Whether aptamers can be evolved to distinguish key epigenetic modifications is unclear, as the large positive charge associated with histone proteins might result in a dominance of nonspecific binders. However, we have found that we can select high-affinity DNA aptamers to histone H4 using capillary electrophoresis systematic evolution of ligands through exponential enrichment (11) at reasonably high (300 mM) salt concentrations (12). We previously showed that aptamers can function as replacements for antibodies in recognition imaging microscopy (13).
In the work presented here, we used a DNA aptamer as a recognition ligand in single-molecule AFM imaging of chromatin containing histone H4 acetylated at lysine 16. We used aptamers that are highly specific for histone H4 proteins modified with an acetyl group at position 16 (H4-K16Ac). This target was chosen based on the known modulation of higher-order chromatin structure and function by this specific acetylation (3). One of the sequenced high-affinity binders, clone 4.20, which adopts the most intricate secondary structure, was found to have the highest target specificity. Full details of the selection and characterization of these aptamers have been published elsewhere (see Williams et al. (17)). This aptamer was used as a recognition element for recognition imaging of synthesized arrays with specific acetylation. The aptamer proved to be reasonably specific at recognizing acetylated histones, with recognition efficiencies of 60% on-target and 12% off-target. In contrast, a standard ChIP-quality anti-H4-K16Ac antibody failed to differentiate acetylated nucleosomes from nonacetylated nucleosomes, with recognition efficiencies of 79% on-target and 72% off-target.
Materials and Methods
Histone H4-K16Ac synthesis, nucleosome reconstitution, and recognition imaging
Biochemical methods for acetylation of histones can result in both unpredictable levels of acetylation and modifications at more than one site on the histones. To test the possibility of recognition imaging of histone acetylation, it was necessary to generate synthetic samples using recombinant histones that were either fully acetylated at H4-K16 or completely unacetylated. Fully unacetylated Xenopus laevis H4 histone was generated recombinantly and purified according to standard protocols (14). A fully acetylated population was generated by chemical ligation of a synthetic peptide tail to a recombinant histone H4 fragment lacking amino acid residues 1–22, a procedure described in detail elsewhere (3). Briefly, the modified histone H4 peptide AGRGKGGKGLGKGGAK(Ac)RHRKVL, containing residues 1–22 of the N-terminal tail of histone H4 was chemically synthesized (Protein Chemistry Core Laboratory, Baylor College of Medicine, Houston, TX). After cleavage from the resin, the side-chain protected peptide (5 mM, final) was dissolved in DMSO, activated at the C-terminus with DCC (100 mM), and reacted with benzyl mercaptan (100 mM) at 25°C for 3 h. After side-chain deprotection was completed, the crude reaction mixture was purified by C18 reverse-phase high-performance liquid chromatography. Product purity and identity were confirmed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectroscopy. The thioester peptide was ligated to recombinant H4 histone fragment (minus amino acids 1–22 with R23C mutation) under denaturing conditions and isolated by cation exchange chromatography. Purified histone fractions were validated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and MALDI-TOF mass spectroscopy, and used directly in subsequent experiments.
Nucleosomes were reconstituted by salt dialysis (14). The control sample used recombinant Xenopus H2A, H2B, H3, and H4 histones (generously provided by Karolin Luger, Colorado State University) reconstituted in high ionic strength buffer and purified by gel filtration chromatography. The acetylated sample was reconstituted with substitution of H4-K16Ac for the Xenopus H4. A 1.9 kb single-copy DNA fragment containing the mouse mammary tumor virus (MMTV) promoter (a generous gift of Gordon Hager, National Institutes of Health, Bethesda, MD) was used as a template for nucleosome formation (15). The DNA and histone octamers were incubated on ice for 30 min at a ratio of 3:4 (w/w) in TE buffer (10 mM Tris, pH 7.5, 1 mM EDTA) containing 1 mM DTT and 1 M NaCl. The mixture was dialyzed stepwise with a 6–8 kDa MWCO membrane into 0.8 M, 0.6 M, and 0.15 M NaCl buffer supplemented with 1 × TE. The nucleosome-DNA complex was cross-linked with 0.1% glutaraldehyde (v:v). Nucleosomal arrays were diluted to 0.25 ng/μL for AFM experiments.
AFM probes were modified with either clone 4.20 or anti-acetyl-histone H4-K16 antibody (catalog No. 07-329; Upstate Biotechnology, Billarica, MA) as previously described (4,12,13). Tips were washed with buffer A (150 mM NaCl, 5 mM Na2HPO4, 1 mM EDTA, pH 7.5) and PBS buffer (150 mM NaCl, 5 mM Na2HPO4, pH 7.5) three times. The control or acetylated chromatin arrays were deposited onto glutaraldehyde-aminopropyltriethoxysilane-modified mica (16), and 70 μL of a 0.25 ng/μL chromatin in 1 mM EDTA were left on the treated mica surface for 30 min. The mica was scanned in selection buffer (10 mM Na2HPO4, 300 mM NaCl, and 5 mM MgCl2, pH 7.5) for aptamer assays and in PBS for antibody assays. AFM MAC mode was used with PicoTREC (PicoPlus with picoTREC; Agilent, Chandler, AZ) for recognition imaging. Images were recorded at a speed of ∼1.6 μm/s with an oscillation amplitude of ∼6 nm (approximately equal to the PEG tether length). The oscillation frequency was 8 kHz with the amplitude set-point set at 70% of full amplitude. Recognition spots were identified by using histograms of the pixel intensity distribution as described previously (13), and the data shown here were accumulated by analyzing six to eight different scans in each case.
Results and Discussion
The aptamer, clone 4.20, with sequences of 5′-dAGACGTAAGTTAATTGGACTTGGTCGTGTGCGGCACAGCGATTGAAAT-3′, binds the H4-K16Ac peptide with a dissociation constant (Kd) of 47 nM and discriminates against the unacetylated target by ∼1200-fold, as determined by affinity capillary electrophoresis (17). We addressed the performance of this aptamer in comparison with a standard ChIP-quality antibody for recognition imaging. To compare the two affinity reagents, we used nucleosomal arrays made by reconstituting recombinant Xenopus histone octamers (wild-type arrays) or H4-K16 acetylated histones synthesized by chemical ligation together with Xenopus H3, H2A, and H2B (acetylated arrays) on a 1.9 kb DNA fragment from the MMTV.
Fig. 1 illustrates imaging of the control and acetylated nucleosomal arrays using the two affinity reagents. Topographical images (left) show individual nucleosomes appearing as white spots on a dark background, and the corresponding recognition image (right) identifies individual recognition events as dark spots on a lighter background. The morphology of the two types of chromatin, as imaged here, is not dramatically different, because deposition (but not imaging) was carried out in the absence of Mg2+ to avoid condensation so as to enhance visualization of the individual nucleosomes. We overlaid the recognition and topographical images (not shown here) to ascertain whether the recognition spots coincided with the nucleosome locations. The aptamer produces essentially no recognition of the unacetylated control (Fig. 1 a, right panel), whereas the antibody fails to distinguish between the control (Fig. 1 c, right panel) and acetylated (Fig. 1 d, right panel) samples; both samples produce recognition signals. High salt (300 mM) was required for recognition because the selection was carried out at high salt to minimize nonspecific interactions (17). Recognition images taken at low salt (the PBS buffer) showed no features, whereas strong recognition signals were obtained over a range of salt concentrations between 0.3 and 1 M. Binding assays (17) show that the nonspecific binding at low salt is weak. Addition of free target peptide to the imaging cell abolished the recognition images, but in the case of images taken with the aptamer, it also severely affected the topographical images.
Figure 1.
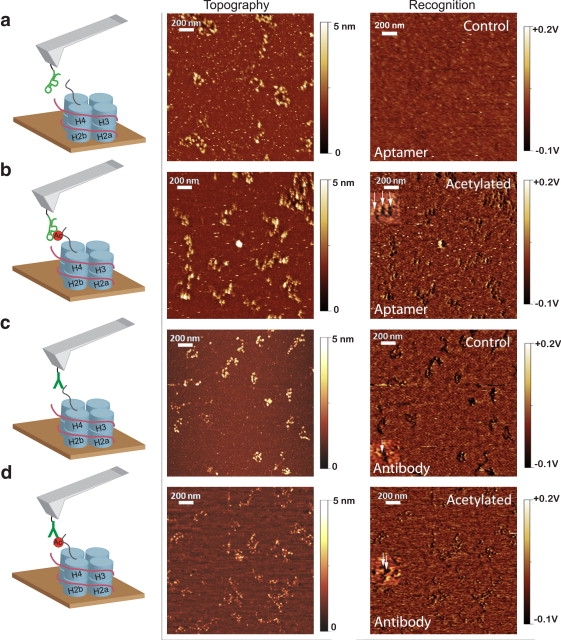
Recognition imaging of artificial nucleosomal arrays by histone H4-K16Ac specific recognition elements. Histone H4 and H4-K16Ac nucleosomal arrays were deposited onto fresh mica surfaces and imaged by AFM. Simultaneous topography (middle) and recognition (right) images were collected using a cantilever tip conjugated to (a and b) the aptamer 4.20 or (c and d) an antihistone H4-K16Ac antibody. Images a and c were taken with control samples, and images b and d were taken with acetylated samples. White features in the topography image are nucleosomes on the mica surface, and dark spots on the recognition image denote the location of nucleosomes recognized by the affinity reagent (white arrows point to 2× magnified examples of recognition spots). Recognition events coincide with nucleosomes in the topographic image.
We determined the specificity of aptamer 4.20 for acetylated nucleosomes by measuring the fraction of acetylated nucleosomes detected relative to the total number of nucleosomes present on the surface. Aptamer 4.20 recognized 126 of the 1050 unmodified nucleosomes (Fig. 1 a) and 649 of the 1082 acetylated nucleosomes (Fig. 1 b). These values correspond to overall recognition efficiencies of 12% and 60%, respectively. By comparison, the commercial antibody recognized 425 of the 591 nonacetylated (Fig. 1 c; 72% efficiency) and 442 of the 559 acetylated (Fig. 1 d; 79% efficiency) nucleosomes. Thus, although the antibody recognized the acetylated histone target with higher overall efficiency, the high specificity of clone 4.20 makes the aptamer a superior affinity reagent for recognizing posttranslational modification.
To quantify the recognition process further, we studied a number of samples, including arrays reconstituted with a mixture of modified and unmodified histones. The results are summarized in Table 1. Based on ∼800 molecules for each nucleosome array target, the recognition efficiency of clone 4.20 against the acetylated nucleosome array is 60.3%, with 12% false positives recorded on the control arrays (wild-type nucleosome arrays). On arrays reconstituted with an equimolar mix of H4 and H4-K16 Ac, the recognition is exactly the expected 36% (0.5 × 60 + 0.5 × 12).
Table 1.
Recognition imaging efficiency with aptamer against MMTV arrays
| Recognition efficiency | H4-K16 Ac | Mixed | Controls |
|---|---|---|---|
| Cln 4.20 | 60.28 ± 2.3% | 36.30 ± 8.2% | 12 ± 7.8% |
H4-K16 Ac: Synthesized MMTV arrays with acetylated K16 in the histone H4 tails.
Mixed: Arrays reconstituted with an equimolar mixture of H4 and H4-K16 Ac in the tails.
Controls: Wild-type MMTV arrays.
One of the aptamers selected for the unacetylated H4 target (12), clone 4.13, was used as a control in this recognition imaging study. The recognition efficiency of the nonacetylated H4 aptamer clone 4.13 against the mixed array (acetylated and nonacetylated nucleosomes) is 33.0%, the recognition efficiency against the wild-type nucleosome array is 57.0%, and the recognition efficiency against the acetylated nucleosome array is 10.5%. Thus, although it is not subject to negative selection against the acetylated peptide, this aptamer is clearly also sensitive to acetylation, in the sense that recognition is reduced considerably when the histone is acetylated.
To ensure that the recognition imaging results obtained with the commercial antibody were due to poor specificity and not poor quality, we measured the binding affinity of the antibody to the H4 and H4-K16Ac peptide sequences by surface plasmon resonance (SPR), which revealed that the antibody binds both acetylated and nonacetylated targets with dissociation constants of 6 and 72 nM, respectively. This shows that the antibody has a low intrinsic specificity. In contrast, aptamer clone 4.20 binds the H4-K16Ac peptide with an affinity (Kd) of 47 nM and discriminates against the unacetylated target by ∼1200-fold as determined by affinity capillary electrophoresis measurements verified using a Biacore (Picastaway, NJ) T-100 SPR instrument. Full details are given elsewhere (17).
This leaves open the question of why the specificity is only some 5× in recognition imaging, versus the 1200× observed in equilibrium binding measurements. One possibility is that the force exerted by the PEG tether increases the off-rate substantially. Another possibility is that the equilibrium bound state is not achieved over the timescale of recognition imaging, and thus the specificity in recognition imaging reflects some intermediate, nonequilibrium complex. We have studied this issue for another ligand receptor pair, and found, in that case, that this was the dominant effect. In addition, with the target bound to a surface, the recognition sites may not be fully exposed, and this may contribute to the <100% positive recognition. It is also possible that some aptamer refolding occurs under imaging conditions. Nonetheless, the aptamer offers enough discrimination to enable identification of histone acetylation on targets of unknown origin, provided that an adequate statistical sample is analyzed. Clearly, an antibody could not be used in recognition imaging in this application.
Conclusions
In summary, we found that aptamers represent an effective alternative to antibodies as affinity reagents in recognition imaging microscopy. The efficient manner in which these molecules can be generated, coupled with their high target affinity and specificity, makes aptamers excellent candidates for detecting specific histone modifications. Further development of this class of recognition elements should help facilitate a better understanding of the histone code and the functional role of histone modifications in gene activation and silencing.
Acknowledgments
We thank Dr. Neal Woodbury for helpful discussion.
This study was supported by grants from the Agilent Foundation to S.M.L., the National Cancer Institutes (R21 CA125510) to J.C.C. and S.M.L., and the National Institutes of Health (GM79663) to M.S.K.
References
- 1.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Shogren-Knaak M., Ishii H., Sun J.-M., Pazin M.J., Davie J.R. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 4.Stroh C., Wang H., Bash R., Ashcroft B., Nelson J. Single-molecule recognition imaging microscopy. Proc. Natl. Acad. Sci. USA. 2004;101:12503–12507. doi: 10.1073/pnas.0403538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bash R., Wang H., Anderson C., Yodh J., Hager G. AFM imaging of protein movements: histone H2A–H2B release during nucleosome remodeling. FEBS Lett. 2006;580:4757–4761. doi: 10.1016/j.febslet.2006.06.101. [DOI] [PubMed] [Google Scholar]
- 6.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Wilson D.S., Szostak J. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 9.Bunka D.H.J., Stockley P.G. Aptamers come of age—at last. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 10.Jenison R.D., Gill S.C., Pardi A., Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 11.Mendonsa S.D., Bowser M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 2004;126:20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 12.Lin L., Hom D., Lindsay S., Chaput J. In vitro selection of histone H4 aptamers for recognition imaging microscopy. J. Am. Chem. Soc. 2007;129:14568–14569. doi: 10.1021/ja076488m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin L., Wang H., Liu Y., Yan H., Lindsay S.M. Recognition imaging with a DNA aptamer. Biophys. J. 2006;90:4236–4238. doi: 10.1529/biophysj.105.079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luger K., Rechsteiner T.J., Richmond T.J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 15.Bash R., Wang H., Yodh J., Hager G., Lindsay S.M. Nucleosomal arrays can be salt reconstituted onto a single copy MMTV promoter DNA template: their properties differ in several ways from those of comparable 5s concatemeric arrays. Biochemistry. 2003;42:4681–4690. doi: 10.1021/bi026887o. [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Bash R., Yodh J.G., Hager G.H., Lohr D. Glutaraldehyde modified mica: a new surface for atomic force microscopy of chromatin. Biophys. J. 2002;83:3619–3625. doi: 10.1016/S0006-3495(02)75362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams B.A., Lin L., Lindsay S.M., Chaput J.C. Evolution of a histone H4-K16 acetyl-specific DNA aptamer. J. Am. Chem. Soc. 2009;131:6330–6331. doi: 10.1021/ja900916p. [DOI] [PMC free article] [PubMed] [Google Scholar]


